Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is a genetic kidney disease, caused by mutations of the PKD1 and PKD2 genes, characterized by the development of renal cysts and extrarenal complications, such as cardiac hypertrophy. Recently, a revolutionary approach, adeno-associated virus (AAV) delivered CRISPR-Cas9 gene editing, has been developed to treat inherited diseases. However, the use of this technology in kidney diseases in vivo is challenged. In this study, we adapt one of the gene editing systems, adenine base editor (ABE9), to develop a broadly expressed and a kidney-specific promoter mediated base editors, and test the effects of these two systems delivered by AAV9 on preventing disease in humanized Pkd1RC/RC mice carrying an arginine (R) to cystine (C) mutation that mimics a mutation in ADPKD patients. We show that one dose of the broadly expressed dual ABE9-AAV9 treatment corrects the pathogenic variant in kidneys, hearts and livers, and result in delaying cyst growth, decrease heart hypertrophy and improve liver function. To confirm the specificity of the base editor system in kidneys, we show that one dose of the kidney specific promoter mediated dual-ABE9-AAV9 treatment corrects the Pkd1 gene mutation in the kidney, and not in the heart, resulting in delaying cyst growth in Pkd1RC/RC kidneys, supporting a promising strategy of using base editor to target specific organs. Treatment with ABE9 base editors mediated by either the broadly expressed or kidney specific promoter increased the survival rate of Pkd1RC/null mice. These preclinical studies support a potential that single-dose genetic therapies may be through the correction of pathogenic variants to prevent ADPKD development in the clinic.
Similar content being viewed by others
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is a genetic kidney disease, caused by mutations of the PKD1 and PKD2 genes, encoding polycystin 1 (PC1) and polycystin 2 (PC2), respectively1,2. PKD gene mutations drive disease progression in both renal (kidney) and extrarenal (outside the kidney) organs such as heart3,4. Point mutations represent the largest class of known human ADPKD variants5. ADPKD is a slowly progressive disease, beginning with the formation of renal cysts in utero. Cyst formation and enlargement continue over decades with diverse mechanisms6,7. ultimately leading to kidney failure. As such, it would be advantageous to prevent the initiation of cyst formation by correcting disease-causing point mutations to stop cyst formation altogether in ADPKD kidneys.
The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-based gene editing is a powerful therapeutic tool to correct gene mutations and thus to ameliorate genetic diseases8. Adeno-associated virus-mediated (AAV-mediated) CRISPR editing, in particular the AAV delivered base editors (BEs), is a revolutionary approach for treating inherited diseases. There are two kinds of BEs including adenine base editors (ABEs) and cytidine base editors (CBEs), and both of them are comprised of a single guide RNA (sgRNA) and a nuclease-inactivated Cas9 (or nickase form) fused to a deaminase enzyme, which enables precise installation of A_T to G_C (in the case of ABEs) or C_G to T_A (in the case of CBEs) substitutions9,10. The architecture of ABEs ensures that only the targeted adenine within a specific window (usually around 4-7 nucleotides) near the protospacer adjacent motif (PAM) sequence is deaminated. Although CRISPR-Cas9 base editing has been applied in animal models of various gene mutation diseases11,12, its application in ADPKD remains untested.
In humans, the hypomorphic p.Arg3277Cys (RC) variant in exon 29 of the PKD1 gene has been linked with typical ADPKD when found in homozygosity13. Pkd1RC/RC mice have been generated to mimic human ADPKD, and include phenotypes in the kidneys and heart. In this study, we modify the ABE9 system to generate a broadly expressed and a kidney specific promoter mediated ABE9 system to genetically correct the RC disease-causing mutation in Pkd1RC/RC mice. In particularly, the kidney specific promoter mediated ABE9 base editor has better editing efficiency in the kidneys than the broadly expressed ABE9 base editor, pointing a promising strategy for using this technology to target specific organs in vivo.
Results
Develop a dual-AAV-encoded ABE9 base editor and protospacer
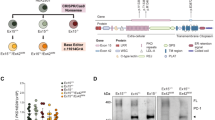
For the R3277C transition, the “AGA” codon for arginine was changed to a “TGC” cystine codon by changing two nucleotides, A > T and A > C (Fig. 1a). Both AGA and CGC encode Arg (R). Therefore, if we can convert the “T” in TGC to “C”, it should generate the “CGC” codon, also resulting in conversion of the amino acid Cys (C) to Arg (R) in Pkd1RC/RC mice (Fig. 1a). To achieve this goal, we use an adenine base editor (ABE), which can target A•T base pairs to produce G•C8. We generated a chicken-beta actin hybrid (Cbh)-AAV-ABE9 N-terminal vector by the replacement of the TadA with a TadA deaminase domain (ABE9) into the AAV-ABE7max vector (Fig. 1b), in which the ABE9 has N108Q and L145T variants to reduce both adenine and cytosine bystander editing, eliminate cytosine editing, and decrease off-target effects as described14,15. We then designed and cloned a single guide RNA (sgRNA) (5’-ACGCAAGTAAAGCGGCTACG-3’) into Cbh-AAV-ABE9 N-terminal and Cbh-AAV-ABE9 C-terminal vectors (Fig. 1a, b). This sgRNA uses a nearby ‘NGG’ PAM, which can be recognized efficiently by a Streptococcus pyogenes Cas9 variant SpCas916,17.
a Schematic of genomic sequence surrounding R3277C pathogenic variant of in exon 29 of mouse Pkd1 gene. The sgRNA (black line) and PAM (red line) are underlined. The designed protospacer placed the R3277C variant at position A5, counting from the protospacer adjacent motifs (PAM)-distal end of the protospacer. b Schematics of dual AAV vectors (not to scale) encoding an amino-terminal fragment (N-term) and carboxyl-terminal fragment (C-term) under control of the broadly expressed Cbh promoter that expresses half of the base editor and can be reconstituted to the full-length base editor within cells by trans-splicing. ITR: inverted terminal repeats. Cbh: the chicken-beta actin hybrid promoter. U6: the human Pol III promoter that efficiently transcribes guide RNAs. sgRNA: single guide RNA. c Representative histological images of kidneys from Pkd1RC/RC mice injected with 1 × 1011 or 1 × 1012 vg of AAV9-ABE9 at PN14 and PN28. Scale bar, 1 mm. d Percent cystic area relative to total kidney area of kidneys from Pkd1RC/RC mice treated with 1 × 1011 or 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14 and PN28 (n = 10 independent replicates). e–g Treatment with 1 × 1011 or 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14 and PN28 decreased KW/BW ratios (e), BUN (f), and serum creatinine levels (g). (n = 10, equal numbers of male and female mice, the colors represent female (pink) and male mice (blue)). h Western blot analysis of PC1 protein in three-month-old wild-type and age-matched Pkd1RC/RC kidneys treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 or vehicle at PN14 (n = 3 independent replicates). i The editing efficiency (%) of targeted pathogenic variant R3277C in kidneys, hearts and livers from three-month-old Pkd1RC/RC mice injected with a single dose of Cbh-ABE9-dual-AAV9 (1 × 1012 vg) at PN14 or PN28 (n = 3 independent replicates). In all panels, bars indicate mean values ± s.d., ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 or as indicated in the figure, using two-tailed t-test. Source data are provided as a Source Data file.
To evaluate the delivery efficiency of AAV9-Cbh vectors, we generated AAV9-Cbh-EGFP particles, and evaluated their distribution in organs 2 weeks after intravenous (IV) injection by an in vivo imaging system (IVIS). AAV9-Cbh-EGFP particles entered (Supplementary Fig. 1a) and GFP was expressed (Supplementary Fig. 1b) in heart, kidney, and liver in AAV9-Cbh-EGFP injected mice, in which the liver was the organ with the strongest GFP expression as examined with IVIS. By performing immunofluorescence (IF) staining in kidneys with GFP antibody and co-stained with different markers, including dolichos biflorus agglutinin (DBA, a collecting ducts marker), lotus Tetragonolobus Lectin (LTL, a proximal tube marker), Na + –K + –Cl− cotransporter 2 (NKCC2, a loop of Henle marker), and Na + -Cl− cotransporter (NCC, a distal convoluted tubule marker), we found that the GFP positive cells could be detected in collecting duct cells (about 26% in total DBA positive cells), proximal tubule cells (about 24% in total LTL positive cells), and distal convoluted tubules cells (about 10% in total NCC positive cells), but not in loop of Henle (Supplementary Fig. 1c–f). The average percentage of GFP-positive cells was about 19% among those cells in AAV9-Cbh-EGFP treated kidneys.
Treatment with Cbh-ABE9-dual-AAV9 delays cyst growth in Pkd1 RC/RC mice
We evaluate the effect of the broadly expressed Cbh promoter-directed Cbh-ABE9-dual-AAV9 in Pkd1RC/RC mice, in which renal cysts could be detected at about one month of age in the C57BL/6 J background (Supplementary Fig. 1g)13, by intravenous (IV) injection at either postnatal day 14 (PN14) or PN28. The total dose of the Cbh-ABE9-dual-AAV9 base editor was 1 × 1011 (low dosage) or 1 × 1012 vg (high dosage) per mouse, and the mice were euthanized at three month. We found that one dose of Cbh-ABE9-dual-AAV9 particles at either dose delayed cyst growth, as seen by the decrease of cyst index and kidney weight/body weight (KW/BW) ratios as well as blood urea nitrogen (BUN) and serum creatinine levels in Pkd1RC/RC mice (n = 10) compared to vehicle treated mice (n = 10) (Fig. 1c–g, Supplementary Fig. 2a). Importantly, injection at PN14 was more effective at delaying cyst growth than injection at PN28 (Supplementary Fig. 2b–d). It is consistent with the finding that PN14 is a critical time point for renal cyst development in Pkd1 mutant mouse models18. Treatment with high dose of Cbh-ABE9-dual-AAV9 through IV injection at PN14 did not induce renal cyst (Supplementary Fig. 2e) and had no effect on KW/BW ratio, BUN and serum creatinine levels in 3-month-old wild type mice (n = 10) (Fig. 1d–g) and Pkd1RC/+ mice (n = 10) (Supplementary Fig. 2f, g) compared to vehicle treated age matched wild type and Pkd1RC/+ mice (n = 10). Treatment with high dose of Cbh-ABE9-dual-AAV9 also did not induce renal cysts and heart hypertrophy in Pkd1RC/+ mice (Supplementary Fig. 2e). In addition, the editing efficiency was around 9.8% in kidneys in Pkd1RC/+ mice treated with Cbh-ABE9-dual-AAV9 (Supplementary Fig. 2h).
It has been reported that a PC1 triplet can be detected in wild type kidneys by Western blot analysis, in which the top PC1 band of the triplet represents the full length PC1, and the middle band, the N-terminal (NT) endoglycosidase H (EndoH) resistant product (NTR), which is the mature and likely surface localized PC1, and the lower band, EndoH sensitive product (NTS, 440 kDa), which may be ER localized PC1. In Pkd1RC/RC kidneys, the PC1-NTS product can be detected as that in Pkd1 wildtype kidneys, whereas the PC1-NTR product is decreased in Pkd1RC/RC kidneys, resulting in more robust cyst development19. We found that treatment with Cbh-ABE9-dual-AAV9 particles has no effect on the level of PC1 in wild type kidneys (Fig. 1h, Supplementary Fig. 2i), but could partially normalize the expression of PC1, representing by an increase of PC1-NTR product in Pkd1RC/+ and Pkd1RC/RC kidneys compared to that seen in vehicle treated age matched control kidneys (Fig. 1h, Supplementary Fig. 2j, and Supplementary Fig. 3a, b). The Pkd1 mRNA was also increased in Pkd1RC/RC kidneys treated with Cbh-ABE9-dual-AAV9 compared to vehicle-treated controls as examined by qRT-PCR analysis (Supplementary Fig. 3c).
To determine the editing efficiency of the Cbh-ABE9-dual-AAV9 base editor in vivo, we found that DNA base-editing was 7.7 ± 1.6% and 11.6 ± 1.5% in kidneys, 15.2 ± 2.2% and 21.3 ± 3.2% in hearts, and 31.9 ± 4.2% to 40.1 ± 8.7% in livers in low dosage vs. high dosage PN14-treated mice harvested at 3-month Pkd1RC/RC mice (Supplementary Fig. 3d), suggesting that high dosage injected at PN14 have the higher editing efficiency in kidneys from 3-month Pkd1RC/RC mice. In addition, we found that high dose PN14 treatment also achieved higher DNA base-editing efficiency than the same dose treated at PN28 and analyzed in 3-month Pkd1RC/RC tissues; DNA base-editing efficiency was 11.6 ± 1.5% (PN14) vs. 7.1 ± 3.0% (PN28) in kidneys, 21.3 ± 3.2% (PN14) vs. 17.4 ± 4.7% (PN28) in hearts, and 40.1 ± 8.7% (PN14) vs. 30 ± 8.2% (PN28) in livers (Fig. 1i). Treatment with non-targeting sgRNA did not convert the “T” in TGC to “C” in Pkd1RC/RC kidneys and treatment with AAV-ABE9 also did not convert the codon around the editing window in wide type kidneys (Supplementary Fig. 4a, b). These results suggest that a single dose injection of a broadly expressed ABE9-encoding AAV delays cyst growth through the partial correction of the RC variant to restore the expression of Pkd1 gene and the mature protein form in the kidneys.
Treatment with Cbh-ABE9-dual-AAV9 decreases heart hypertrophy in Pkd1 RC/RC mice
ADPKD gene mutations not only drive cyst progression in the kidney but also result in extrarenal complications, including left ventricular hypertrophy (LVH)20, which has also been shown in Pkd1RC/RC mice21. We found that one injection of Cbh-ABE9-dual-AAV9 particles, total 1 × 1011 or 1 × 1012 vg also decreased cardiac hypertrophy as seen by a decrease of heart weight/body weight (HW/BW) ratios in three-month-old Pkd1RC/RC mice compared to vehicle treated age matched control mice. Again, early injection at PN14 with a high dosage of 1 × 1012 vg had a stronger effect on decreasing heart hypertrophy compared to injection at PN28 with the low dosage in those mice (Fig. 2a, b, Supplementary Fig. 5a). We detected a doublet of PC1 around 470 kDa in three-month-old wild type hearts, in which the PC1-NTR was decreased in vehicle treated age matched Pkd1RC/RC hearts compared to WT hearts and was partially increased in Cbh-ABE9-dual-AAV9 treated age matched Pkd1RC/RC hearts as examined by Western blot analysis (Fig. 2c, Supplementary Fig. 5b). In addition, the expression of Pkd1 mRNA was also increased in Cbh-ABE9-dual-AAV9 treated Pkd1RC/RC hearts compared to vehicle treated controls as examined by qRT-PCR analysis (Fig. 2d).
a Representative histological images of hearts from Pkd1RC/RC mice injected with total 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14 and PN28. Scale bar, 1 mm. b Treatment with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14 and PN28 decreased HW/BW ratios in three-month-old Pkd1RC/RC mice. (n = 10 independent replicates, equal numbers of male and female mice, the colors represent female (pink) and male mice (blue)). c Western blot analysis of PC1 protein in three-month-old wild-type and age-matched Pkd1RC/RC hearts treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 or vehicle at PN14. Data were analyzed from three experiments. d qRT-PCR analysis of relative Pkd1 mRNA expression in three-month Pkd1RC/RC hearts treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 or vehicle at PN14 (n = 6 independent replicates). The expression of Pkd1 mRNA in age matched wild type hearts was used as a control. Bars indicate means values ± s.d; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 or as indicated in the figure, using two-tailed t-test. Source data are provided as a Source Data file.
A long-term duration effect of Cbh-ABE9-dual-AAV9 treatment in Pkd1 RC/RC mice
To evaluate the long-term effectiveness of injected Cbh-ABE9-dual-AAV9 particles in vivo, we found that one-time injection of Cbh-ABE9-dual-AAV9 particles of 1 × 1011 vg (n = 10) or 1 × 1012 vg (n = 10, equal numbers of male and female mice) delayed cyst growth in six-month-old Pkd1RC/RC mice as seen by a decrease of cystic index and KW/BW ratio as well as BUN and serum creatinine levels (Fig. 3a–d, Supplementary Fig. 5c, 5d), and treatment with high dose of Cbh-ABE9-dual-AAV9 particles decreased HW/BW ratios in Pkd1RC/RC mice compared to vehicle treated controls (n = 10) (Fig. 3e, f, Supplementary Fig. 5c). An increase of PC1-NTR product was detected in Cbh-ABE9-dual-AAV9 particle treated Pkd1RC/RC kidneys and hearts compared to that in vehicle treated controls (Fig. 3g, h, Supplementary Fig. 5e, 5f). These results suggest that there is a long-term or lasting effect of Cbh-ABE9-dual-AAV9 particles in vivo. Furthermore, DNA base-editing outcomes were still high in organs from 6-month Pkd1RC/RC mice when treated with the high dose of Cbh-ABE9-dual-AAV9 at PN14; 12.1 ± 0.5% in kidneys, 22.4 ± 1.4 % in hearts and 38.2 ± 3.9% in livers, but these were not significant differences compared to 3-month Pkd1RC/RC mice when treated with the high dose of Cbh-ABE9-dual-AAV9 at PN14 (Fig. 3i). These results suggest that DNA base-editing on the correction of the RC variant may continue from 3 months to 6 months in vivo. Also, there is a possibility that edited cells may have a survival advantage over uncorrected cells in those organs, increasing the prevalence of edited alleles over time, which is consistent with the known persistence of AAV-ABE in mammals11.
a Representative histological images of six-month old kidneys from Pkd1RC/RC mice treated with 1 × 1011 or 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14. Scale bar, 1 mm. b Percent cystic area relative to total kidney area of kidneys from Pkd1RC/RC mice treated with 1 × 1011 or 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14 (n = 10 independent replicates). c, d Treatment with 1 × 1011 or 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14 decreased KW/BW ratios (c) and BUN levels (d) in six-month old Pkd1RC/RC mice. (n = 10, equal numbers of male and female mice, the colors represent female (pink) and male mice (blue)). e Representative images of hearts from six-month old Pkd1RC/RC mice injected with 1 × 1011 or 1 × 1012 of Cbh-ABE9-dual-AAV9 at PN14. Scale bar, 1 mm. f Treatment with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14 decreased HW/BW ratios in six-month-old Pkd1RC/RC mice compared to vehicle-treated controls (n = 10 independent replicates). g, h Western blot analysis of PC1 protein in six-month-old wide type and age-matched Pkd1RC/RC kidneys and heart treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 or vehicle at PN14 (n = 3 independent replicates). i the editing efficiency (%) of the targeted pathogenic variant R3277C in kidneys, hearts and livers from three and six-month-old Pkd1RC/RC mice injected with a single dose of dual-AAV9-ABE9 (1 × 1012 vg) at PN14 (n = 3 independent replicates). Values and error bars reflect mean ± SD from indicated counts of independent experiments, ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 or as indicated in the figure, using two-tailed t-test. Source data are provided as a Source Data file.
Assessment of bystander editing in Cbh-ABE9-dual-AAV9 treated Pkd1 RC/RC mice
As amino acid off-target substitutions could be deleterious, we assessed the frequency of the editing of bystander mutations of adenosine residues at protospacer positions 1 and 6 and the introduction of indels in base-edited Pkd1RC/RC mice using genomic DNA (gDNA) and RT–PCR-amplified Pkd1 mRNAs from kidneys in three-month Pkd1RC/RC mice treated with Cbh-ABE9-dual-AAV9 particles (1 × 1012 vg) at PN14 (Supplementary Fig. 6a–e). The result showed levels of Pkd1 cDNA editing efficiency in kidney is 28.7 ± 3.6% (Supplementary Fig. 6c), in heart is 40.89 ± 4.7% (Supplementary Fig. 6d), and in liver is 56.96 ± 2.8% (Supplementary Fig. 6e). We did not detect the bystander edit around the potential editing site in gDNA and RNA. In addition, no significant differences were observed in the frequency of A-to-I RNA nucleotide changes in AAV-ABE9 treated mice (Supplementary Fig. 6f). Furthermore, to evaluate the Cas9-dependent off-target effect, we predict potential off-target sites using CRISPOR, a tool ranks the sgRNA with off-target effects and on-target activity, and then conducted targeted amplicon deep sequencing (Amplicon-seq). There were no detectable off-target editing above background levels at the top candidate sites analyzed (Supplementary Fig. 6g), supporting the high specificity and safety of the Cbh-ABE9-dual-AVV9 base editing system in vivo.
Establish kidney specific promoter mediated ABE9-AAV delivery system
To test whether base editing can be used to specifically target the genome in kidneys and to convert the RC mutation in the Pkd1 gene in kidney only, we generated a kidney specific promoter mediated dual AAV base editor by replacing the broadly expressed Cbh promoter as well as the cytomegalovirus enhancer and hybrid intron in the established Cbh-ABE9-dual-AAV9 vectors with a kidney specific promoter mediated cadherin-16 promoter (Ksp), Ksp-ABE9-dual-AAV9 (Fig. 4a). Before testing the effect of Ksp-ABE9-dual-AAV9 in vivo, we first determined the distribution and expression of AAV-Ksp-EGFP particles. We found that the AAV-Ksp-EGFP particles entered and the GFP auto-fluorescent signal was observed only in kidney and liver but not in other organs, such as heart (Supplementary Fig. 7a, b). The expression of GFP was also detected by immunofluorescence staining with GFP antibody, which co-localized with DBA (about 39% in total DBA positive cells), LTL (about 36% in total LTL positive cells), and NCC (about 6% in total NCC positive cells) in kidneys treated with AAV-Ksp-EGFP (Supplementary Fig. 7c). The average percentage of GFP positive cells was 27% among those cells in AAV9-Ksp-EGFP treated kidneys. These results indicate that the transduction of AAV-Ksp-EGFP particles is expressed in specific renal tubular cells in the kidney, which is consistent with prior reports showing that AAV9 can efficiently target renal tubules, even though those reports did not quantify the percentages of GFP-positive cells in kidneys22,23,24,25. In addition, we found that GFP is expressed in kidneys injected with AAV9-Cbh-EGFP and AAV9-Ksp-EGFP but not in kidneys injected with empty AAV vectors as examined with immunohistochemistry (IHC) staining (Supplementary Fig. 7d, 7e), confirming the specificity of AAV-mediated GFP expression in kidneys.
a Schematics of kidney specific promoter mediated dual AAV vectors (not to scale) that encode an amino-terminal fragment (N-term) and carboxyl-terminal fragment (C-term) that form base editors after protein splicing. b Representative histological images of kidneys from Pkd1RC/RC mice injected with 1 × 1012 vg of Ksp-ABE9-dual-AAV9 at PN14. Scale bar, 1 mm. c Percent cystic area relative to total kidney area of kidneys from Pkd1RC/RC mice treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 as well as vehicle at PN14 (n = 10 independent replicates). d–f Treatment with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 at PN14 decreased KW/BW ratios (d), BUN (e), and serum creatinine levels (f) in three-month-old Pkd1RC/RC mice. (n = 10, equal numbers of male and female mice, the colors represent female (pink) and male mice (blue)). g Representative hearts and histological images of hearts from three-month-old Pkd1RC/RC mice injected with 1 × 1012 vg of Ksp-ABE9-dual-AAV9 and Cbh-ABE9-dual-AAV9 at PN14. Scale bar, 1 mm. h Treatment with 1 × 1012 vg of Ksp-ABE9-dual-AAV9 has no effect on HW/BW ratios in three-month-old Pkd1RC/RC mice (n = 10). ns, not significant. i The comparison of the editing efficiency (%) of the targeted pathogenic variant R3277C in kidneys and hearts as analyzed with high-throughput sequencing of gDNA in these tissues from three-month-old Pkd1RC/RC mice injected with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 at PN14 (n = 3 independent replicates). j–l Western blot analysis of PC1 protein in three-month-old wide type and age-matched Pkd1RC/RC and wide type kidney (j), livers (k), and heart (l) treated with 1 × 1012 vg of Ksp-ABE9-dual-AAV9 or vehicle at PN14 (n = 3 independent replicates). Values and error bars reflect mean ± SD from indicated counts of independent experiments, ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 or as indicated in the figure, using two-tailed t-test. Source data are provided as a Source Data file.
Treatment with Ksp-ABE9-dual-AAV9 particles delays cyst growth in Pkd1 RC/RC mice
To test the effect of Ksp-ABE9-dual-AAV9 particles in vivo, we found that treatment with Ksp-ABE9-dual-AAV9 particles (1 × 1012 vg) at PN14 delayed cyst growth as seen by a decrease of cystic index and KW/BW ratio as well as BUN and serum creatinine levels in 3-month Pkd1RC/RC mice compared to vehicle treated age matched mice (n = 10) (Fig. 4b–f, Supplementary Fig. 8a). In contrast, this treatment did not normalize HW/BW ratio and cardiac hypertrophy in those mice (n = 10) (Fig. 4g, h, Supplementary Fig. 8b). Treatment with high dosage of 1 × 1012 vg Ksp-ABE9-dual-AAV9 particles had no significant effect on the KW/BW ratio, BUN and serum creatinine levels as well as HW/BW ratio in WT mice (n = 10) (Fig. 4d–f and Fig. 4h).
We further found that DNA base-editing was 23.4 ± 0.4% in 3-month Ksp-ABE9-dual-AAV9 treated kidneys, which was higher than the DNA base-editing efficiency in kidneys in age matched Pkd1RC/RC mice treated with the broadly expressed Cbh-ABE9-dual-AAV9 particles (Fig. 4i). The on-target DNA base-editing was very low in heart tissues in 3-month Pkd1RC/RC mice treated with Ksp-ABE9-dual-AAV9 particles (1 × 1012 vg) at PN14, consistent with the Ksp promoter’s kidney-specific expression (Fig. 4i). Treatment with Ksp-ABE9-dual-AAV9 could partially rescue the PC1-NTR product in kidneys (Fig. 4j, Supplementary Fig. 8c) and livers (Fig. 4k, Supplementary Fig. 8d) but not that in hearts compared to that in vehicle treated controls (Fig. 4l, Supplementary Fig. 8e).
Treatment with Cbh -ABE9-dual-AAV9 and Ksp -ABE9-dual-AAV9 particles corrects mutations in primary renal tubular cells isolated from Pkd1 RC/RC mice
To determine the editing efficiency in primary renal tubular cells, we isolated the renal tubules and then tubular cells from Pkd1RC/RC kidneys injected with Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 particles. We found that the editing efficiency in renal tubular cells isolated from, 1) Cbh-ABE9-dual-AAV9 particle injected Pkd1RC/RC kidneys is about 7.21% ± 0.3, similar to that in the whole kidneys (Supplementary Fig. 9a), and 2) Ksp-ABE9-dual-AAV9 injected Pkd1RC/RC kidneys was 34.2% ± 8.8, which is higher than that in the whole kidneys (Supplementary Fig. 9a), possibly due to the fact that the Ksp promoter is expressed exclusively in tubular epithelial cells in the kidney but not in other cells26.
It has been reported that chromatin accessibility differs substantially between C57BL/6 J and 129S1/SvImJ mice, contributing to distinct regulatory landscapes and potentially affecting gene editing outcomes27. By treating the C57BL/6 (B6) background Pkd1RC/RC mice with Cbh- and Ksp-ABE9-dual-AAV9 particles, we found that the editing efficiency was approximately 4.9% and 20.4% (Supplementary Fig. 9b) in the kidneys of the B6 background Pkd1RC/RC mice treated with Cbh-ABE9-dual-AAV9 and Ksp-ABE9-AAV particles, respectively. These values are lower than those observed in 129 Pkd1RC/RC mice (7.7% for Cbh-ABE9-AAV and 23.4% for Ksp-ABE9-AAV), supporting the notion that mouse genetic background can affect base editing outcomes.
Treatment with Cbh -ABE9-dual-AAV9 rescues the liver function and did not induce liver toxicity and immune infiltration in Pkd1 RC/RC mice
In ADPKD, liver cysts are not a primary or early feature of the disease, and liver function is generally not significantly affected despite the presence of liver cysts28. To evaluate the effect of base editors in the liver, we first found that treatment with Cbh-ABE9-dual-AAV9 led to an increase of Pkd1 mRNA in 3- and 6-month-old livers compared to vehicle treated age matched controls, even though it was still lower than that in wild type livers (Fig. 5a, b), and high dose treatment achieved a better effect on the expression of Pkd1 mRNA in livers (Fig. 5a, b). We also found that the level of PC1-NTR was decreased in 3- and 6-month-old vehicle treated Pkd1RC/RC livers, which could be restored in Cbh-ABE9-dual-AAV9 treated age matched Pkd1RC/RC livers (Fig. 5c, d, Supplementary Fig. 9c, Supplementary Fig. 9d).
a, b qRT-PCR analysis of relative Pkd1 mRNA expression in livers of three-month-old (a) and six-month-old (b) mice treated with 1 × 1011 or 1 × 1012 of Cbh-ABE9-dual-AAV9 and vehicle at PN14. The expression of Pkd1 mRNA in age matched wild type livers was used as a control (n = 6 independent replicates). c, d Western blot analysis of PC1 protein in livers of three (c) and six-month-old (d) wide type and age-matched Pkd1RC/RC mice treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 or vehicle at PN14 (n = 3 independent replicates). e, f H&E histology sections of three and six-month-old livers from Pkd1RC/RC mice treated with 1 × 1011 or 1 × 1012 vg of Cbh-ABE9-dual-AAV9 at PN14, as well as WT and vehicle control (n = 6 independent replicates). Scale bar, 50 μm. Values and error bars reflect mean ± SD from indicated counts of independent experiments, ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001 or as indicated in the figure, using two-tailed t-test. Source data are provided as a Source Data file.
With H&E staining, we found that there was no massive hepatocyte necrosis in Pkd1RC/RC livers, and only mild structural disorganization and liver cyst were observed in 3- and 6-month-old Pkd1RC/RC livers (Fig. 5e, f), and treatment with Cbh-ABE9-dual-AAV particles ameliorated liver cyst in 3- and 6-month-old Pkd1RC/RC livers, suggesting a protective effect of Cbh-ABE9-AAV on hepatocellular injury in Pkd1RC/RC mice (Fig. 5e, f). In liver function tests, alanine aminotransferase (ALT), aspartate aminotransferase (AST), and alkaline phosphatase (ALP) are key enzymes that can indicate liver health, and the elevated levels of ALT, AST, and ALP often suggest liver cell damage29. We found that the levels of ALT, AST and ALP were increased in 3-month old Pkd1RC/RC livers compared to age-matched wild type livers, while treatment with Cbh-ABE9-dual-AAV9 could decrease the level of ALT, AST, and ALP in Pkd1RC/RC livers comparable to wild type livers compared to Ksp-ABE9-AAV treated Pkd1RC/RC (Supplementary Fig. 10a–c), suggesting that treatment with Pkd1 base editors could rescue liver function in Pkd1RC/RC mice.
To evaluate the liver toxicity of base editors, we detected the levels of white blood cell (WBC), red blood cell (RBC), platelet in blood of wild type and Pkd1RC/Null mice at PN20 as well as in blood of 3-month-old wild-type and Pkd1RC/RC mice treated with Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 particles. We found that treatment with high dose Cbh-ABE9-dual-AAV9 or Ksp-ABE9-dual-AAV9 has no significant effect on the level of WBC, RBC, platelets in the blood from wild-type and age-matched Pkd1RC/null and Pkd1RC/RC mice. These results suggested that the used doses of Ksp-ABE9-dual-AAV9 and Cbh-ABE9-dual-AAV9 did not cause an obvious immune response in mice (Supplementary Fig. 10d–i). C-reactive protein (CRP) is primarily produced by the liver in response to inflammation, and as an acute-phase reactant, its levels rise in the blood when there is inflammation in the body30. We found that treatment with high dose Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 particles decreased the level of CRP in blood, and IL-6 and TNF-α in livers in Pkd1RC/RC mice compared to age matched control mice treated with vehicle, whereas those treatments had no obvious effect on these parameters in wild type mice compared to age matched WT mice treated with vehicle (Supplementary Fig. 10j–l).
To further evaluate the hepatic immune infiltration in Pkd1RC/RC livers with the treatment of Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 particles, we performed the immunofluorescence staining with CD45 (a pan-leukocyte marker), CD3 (T cell marker), CD4 (helper T cell marker) as well as F4/80 antibodies. We found that the numbers of CD45, CD3 + , CD4 + T cells and macrophage were increased in Pkd1RC/RC livers compared to age-matched wild-type controls, indicating enhanced immune infiltration, whereas treatment with either Cbh-ABE9-dual-AAV9 or Ksp-ABE9-dual-AAV9 decreased the levels of CD45, CD3 + , CD4 + T cells and macrophage cells in 3-month-old Pkd1RC/RC livers, and had no significant effect on those in livers of wild-type controls (Supplementary Figs. 11 and 12). Because the doses of AAV that we used in this study is within the safety range as reported in previous studies11,31,32, these results further indicate that either Cbh-ABE9-AAV or Ksp-ABE9-AAV treatment leads to the decrease of immune infiltration and early signs of hepatocyte stress in Pkd1RC/RC livers.
Treatment with ABE9-dual-AAV9 particles extends lifespan of Pkd1 RC/null mice
By crossing Pkd1RC/RC mice with Pkd1null/+ mice, we generated more rapidly progressing Pkd1RC/null mice, which only survive up to about 25 days13. We evaluated the effect of the ABE9-dual-AAV9 treatment on the lifespan of these Pkd1RC/null mice by injection of 1 × 1012 vg broadly expressed Cbh-ABE9-dual-AAV9 and kidney specific promoter mediated Ksp-ABE9-dual-AAV9 particles at PN 7 (see details in Methods). We found that treatment with ABE9-dual-AAV9 particles did not affect the body weight of Pkd1RC/null mice (n = 10) compared to vehicle treated controls (n = 10) (Fig. 6a). We further found that the broadly expressed Cbh-ABE9-dual-AAV9 treated Pkd1RC/null mice (n = 10) lived to a mean age of 33.0 days, while vehicle treated mice (n = 10) died of PKD at a mean age of 23.0 days (p < 0.0001) (Fig. 6b). Importantly, we found that treatment with Ksp-ABE9-dual-AAV9 further increased the lifespan of Pkd1RC/null mice (n = 10) to a mean age of 37 days (p < 0.0001) (Fig. 6b). These PN7-injected Pkd1RC/null mice exhibited normal behavior and vitality at ages well beyond the lifespan of vehicle treated Pkd1RC/null mice. We also saw a significant decrease of cystic index, KW/BW ratio and BUN level, as well as serum creatinine levels (Supplementary Fig. 13a and 13b) in Pkd1RC/null mice collected at 20 days, after treatment with Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 (Fig. 6c–f). In addition, treatment with Ksp-ABE9-dual-AAV9 increased editing efficiency in kidneys compared to that in Cbh-ABE9-dual-AAV9 treated kidneys (Fig. 6g). We further found that treatment with Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 particles increased 1) the PC1-NTR level in Pkd1RC/Null kidneys (Fig. 6h, Supplementary Fig. 13c) and 2) Pkd1 mRNA and protein in PN20 Pkd1RC/+ and Pkd1RC/null kidneys compared to that in vehicle treated controls (Supplementary Fig. 13d).
a Statistic analysis of body weights and KW/BW ratios of Pkd1RC/null mice treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9, Ksp-ABE9-dual-AAV9 and vehicle (n = 10), p = 0.43 of body weight calculated by one-way ANOVA test of age matched mice. The KW/BW ratio was calculated at the endpoint of Pkd1RC/null mice died at different days. b The survival rate of Pkd1RC/null mice treated with Cbh-ABE9-dual-AAV9 (1 × 1012 vg) (n = 10) lived to a mean age of 33.4 ± 1.6 days (p < 0.0001) compared with vehicle treatment (n = 10) (22 ± 3 days), and Pkd1RC/null mice treated with Ksp-ABE9-dual-AAV9 (1 × 1012 vg) lived to a mean age of 37.1 ± 5.9 days (n = 10) (p < 0.05) compared with Cbh-ABE9-dual-AAV9 treatment. c–f Representative kidneys (c), cystic index (d), KW/BW ratios (e) and BUN levels (f) in 20-days-old Pkd1RC/null mice treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 as well as vehicle (n = 10, equal numbers of male and female mice in each group). The colors in the graphs represent female (pink) and male mice (blue). Scale bar, 1 mm. g The comparison of the editing efficiency (%) of the targeted pathogenic variant R3277C in kidneys from Pkd1RC/null mice treated with 1 × 1012 vg of Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 at PN20 (n = 3 independent replicates). h PC1 protein in kidneys from PN20 Pkd1RC/null mice treated with Cbh-ABE9-dual-AAV9,Ksp-ABE9-dual-AAV9 and vehicle (n = 3 independent replicates). Values and error bars reflect mean ± SD from indicated counts of independent experiments, ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 or as indicated in the figure, using two-tailed t-test. Source data are provided as a Source Data file.
Treatment with AAV9-ABE9 reduces the recruitment/accumulation of macrophages at interstitial and pericystic regions and decreases cyst cell proliferation
It has been reported that an abundance of M2-like macrophages can be recruited to interstitial and pericystic regions in ADPKD mouse kidneys to promote cystic renal cell proliferation and cyst growth33,34,35. We found that one time injection of broadly expressed Cbh-ABE9-dual-AAV9 particles of 1 × 1011 and 1 × 1012 vg at either PN14 or PN28 decreased the accumulation/recruitment of macrophages to pericystic and interstitial sites (Supplementary Fig. 14a) and cyst lining epithelial cell proliferation (Supplementary Fig. 14b) in three-month-old Pkd1RC/RC kidneys compared to vehicle treated age matched Pkd1RC/RC mice. In addition, treatment with broadly expressed Cbh-ABE9-dual-AAV9 particles of 1 × 1011 and 1 × 1012 vg at PN14 also decreased the accumulation/recruitment of macrophages and cyst lining epithelial cell proliferation in 6-month Pkd1RC/RC kidneys compared to age-matched vehicle-treated Pkd1RC/RC kidneys (Supplementary Fig. 15a, b). Furthermore, one-dose injection of Ksp-ABE9-dual-AAV9 particles of 1 × 1012 vg at PN14 also decreased the accumulation/recruitment of macrophages and cyst lining epithelial cell proliferation in three-month-old Pkd1RC/RC kidneys compared to vehicle treated age matched Pkd1RC/RC mice (Supplementary Fig. 16a, b).
Treatment with Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 decreases fibrosis in Pkd1 RC/RC mice
Renal fibrosis is a hallmark of ADPKD36. As the progression of disease, renal fibrosis increases and kidney function declines37. Treatment with high dose of either Cbh-ABE9-dual-AAV9 or Ksp-ABE9-dual-AAV9 particles decreased renal fibrosis in 3- and 6-month-old Pkd1RC/RC kidneys compared to that in age matched vehicle treated Pkd1RC/RC kidneys (Supplementary Fig. 17a and 17b). Myocardial fibrosis is also prominently increased in mouse models of ADPKD38,39. We found that treatment with Cbh-ABE9-dual-AAV9 particles significantly decreased fibrosis in 3- and 6-month-old Pkd1RC/RC hearts compared to that in hearts from vehicle treated Pkd1RC/RC mice (Supplementary Fig. 17c and 17d). We also found that treatment with Cbh-ABE9-dual-AAV9 decreased heart and renal fibrosis in Pkd1RC/Null mice compared to that in vehicle treated control mice (Supplementary Fig. 17e), whereas treatment with Ksp-ABE9-dual-AAV9 decreased renal fibrosis only but had no effect on heart fibrosis compared to vehicle treated and Cbh-ABE9-dual-AAV9 treated kidneys and hearts (Supplementary Fig. 17f). Furthermore, we found that treatment with Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 significantly decreased fibrosis in 3-month and 6-month-old Pkd1RC/RC and PN20 Pkd1RC/null livers compared to that in vehicle treated age matched livers (Supplementary Fig. 18a–18c). Together, these results indicate that Cbh-ABE9-dual-AAV9 and Ksp-ABE9-dual-AAV9 treatments decrease organ fibrosis in Pkd1RC/RC mice.
Discussion
This is the proof-of-principle study to correct a pathogenic single-nucleotide variant in an ADPKD mouse model with the developed broadly expressed and kidney specific promoter mediated CRISPR/ABE9 base editing systems. The efficiency and duration of kidney specific promoter-mediated AVV-ABE9 base editor to correct mutations in ADPKD kidneys should provide a guide for using a similar strategy to convert mutations in other kidney diseases.
Correcting pathogenic alleles is a longstanding challenge in medicine, especially those alleles that cause devastating kidney diseases. With base editing technique, we may directly correct point mutations that drive many disorders without requiring double-strand DNA breaks10,40,41. Recently, the ABE7 and ABE8e systems have been developed and tested in Hutchinson-Gilford progeria syndrome and cardiac disease11,12,42. However, it has been reported that ABE7 and ABE8e base editing achieves no obvious editing efficiency in kidneys11,43. In addition, ABE8e has a broad editing window (positions A3–A9) and a known off-target cytosine editing44. For that the target mutation in Pkd1RC/RC mice lies precisely at position A5 within the protospacer (Supplementary Fig. 6a), and ABE9 has been specifically engineered to achieve high precision editing at A5 with minimized bystander editing at adjacent adenines and cytosines14, thus, we adapted the ABE9 system to generate broad and kidney specific promoter mediated base editors. With kidney specific promoter mediated base editor, we intend to, 1) determine whether base editor can work in specific organ, such as kidney, 2) minimize off-target editing and potential toxicity in non-renal tissues, and 3) explore whether restricting base editing activity to renal epithelial cells can enhance therapeutic precision compared to universal base editor. Importantly, our results support that kidney specific promoter mediated base editor works better than the universal base editor in correcting point mutation in kidney. In addition, our results that kidney specific promoter mediated base editor does not express in the heart support the potential to target specific organs with organ specific base editor in the clinic. We should point out that ADPKD is a systemic disease with extrarenal manifestations, such as cardiac hypertrophy45 and liver cysts46, thus, the application of universal base editor which can correct mutation in organs other than kidney may be beneficial in the context of systemic manifestations of ADPKD.
The AAV-ABE9 system mainly installed desired single A-to-G conversion in the mouse genome with very minimal RNA and undetectable Cas9-independent DNA off-target effects11,14,47, indicating preferential editing of the target nucleotide on the R3277C allele to decrease cyst growth and rescue heart abnormalities. The results that AAV-ABE9 system does not induced the significant DNA and RNA off-target effect as well as bystander editing in ADPKD mouse model suggest that genome-wide off-target DNA editing should not be a problem in AAV-ABE9-treated mice as being reported in literatures47,48,49.
Extensive efforts have been focused on AAV-mediated gene therapy in the kidney50,51. Recent studies have shown that adeno-associated virus 2/9 mediated gene therapy prevents the progression of disease in genetic mouse models, such as nephrotic syndrome and chronic kidney disease, and have demonstrated a good transduction of AAV2/9 in kidney cells in mouse kidneys in vivo22,52,53. The entry and expression of AAV particles had also been confirmed by the constructed AAV9 vectors carrying EGFP under Cbh and Ksp promoters in collecting ducts and proximal tubules in wild type kidneys (Supplementary Fig. 1 and Supplementary Fig. 7). In addition, we observed the expression of GFP in cyst lining epithelial cells when Pkd1RC/RC mice was treated with AAV9-Cbh-EGFP and AAV9-Ksp-EGFP particles (Supplementary Fig. 19a). We further found that the expression of GFP under Ksp promoter was a little bit more in kidneys than that in kidneys under the promoter of Cbh (Supplementary Fig. 19b). This result suggests that Ksp promoter works more efficiently than Cbh promoter in kidneys, which may also explain the results that treatment with Ksp-ABE9-dual-AAV9 particles have better effect on delaying cyst growth than the treatment with Chh-ABE9-dual-AAV9 particles, and this strategy may also generate a better therapeutic outcome in other kidney diseases.
AAVs are favored for their safety, low immunogenicity, and ability to provide long-lasting gene expressions, which is particularly useful for targeting non-dividing cells, such as those in the heart, kidney and liver. A number of theories have been proposed to explain the relatively low immunogenicity of AAV. First, the poor transduction of professional antigen-presenting cells (APCs) by most AAV serotypes may only lead to a minimal increase of major histocompatibility complex (MHC) proteins in target cell types. Second, there is a balance between immune tolerance and the clearance of AAV-delivered components (e.g., Cas9) after AAVs treatment. High dosage of AAV is easy to activate T cells to induce the immune clearance of AAV, resulting in the loss of efficacy, but low dose of AAV is more likely to be neutralized by anti-AAV antibodies54. In some contexts, moderate doses may promote immune tolerance, rather than immune activation, particularly in immune-modulatory organs, such as the liver. Certain degree of base editing vector uptake/accumulation, and the expression of base editor is inevitable in livers, even with tissue-enriched promoters55. This is a known limitation of AAV9 delivery and does not necessarily reflect promoter leakiness but rather a vector biodistribution.
It has been reported that AAV vector-mediated expression of the transgene is generally maintained in vivo for at least 6 months56. In essence, the initial burst of AAV gene delivery, possibly resulting in a higher copy number, and the stability of episomal genomes in non-dividing cells and the potential for low-level integration, can collectively contribute to the observed long-term maintenance of transgene expression despite a decline in the overall AAV copy number over time57. As such, copy number and transgene expression are not always directly proportional. With the junction-spanning RT-qPCR analysis to target the spliced 5′-UTR–GFP junction in the mature mRNA, GFP transcripts were detected in kidneys, livers, and hearts treated with Cbh- and Ksp-driven GFP-AAV9 particles, respectively (Supplementary Fig. 20a and 20b), supporting a genuine viral transduction and the transcription of transgene. We also observed the expression of Cas9 protein and mRNA in hearts, kidneys and livers and in base editor injected 3-month and 6-month -old Pkd1RC/RC mice (Supplementary Fig. 20c–20f) and a decline in AAV vector copy number and Cas9 protein expression over time in the heat, kidney and liver (Supplementary Fig. 20g and h), whereas the base editing efficiency remained stable, which is consistent with previous report that even short-term expression of the editor can result in permanent genetic correction58. These results suggest that the subsequent declines in AAV copy number and Cas9 expression do not negate those already established genomic changes, which supports a “hit-and-go” editing model, in which the initial expression of Cas9 introduces the desired genomic changes and then is cleared gradually, thereby minimizing long-term Cas9 exposure and associated risks. This concept is crucial for the development of safe and effective long-term gene therapies using CRISPR-Cas9 delivered via AAV vectors.
The results that Pkd1 mRNA gets stabilized or increased upon gene-editing suggest that mutation of PC1 (R to C) may negatively affect Pkd1 mRNA stability in Pkd1 mutant renal epithelial cells and Pkd1RC/RC kidneys. In addition, a possible scenario to address that non-corrected cells do not take over the corrected ones as time goes by is that Pkd1 mutation mediated the increase of cell proliferation may not be a dominant factor to promote cyst growth or expansion, and other factors, such as fluid secretion into the cyst, the recruitment of macrophages and the activation of fibroblasts in the interstitial, etc. are also contribute to the progression of ADPKD.
The injection of AAV9-ABE9 base editor significantly improves kidney function, lifespan, and vitality in ADPKD mice suggest that this strategy is promising to correct mutations and improve renal function and lifespan in patients with ADPKD. However, several limitations must be clarified. First, prolonged expression of Cas9 and the deaminase may increase the risk of off-target editing and elicit immune responses, particularly in larger animals and humans59,60. Second, the use of high AAV doses required for dual-vector delivery to efficiently target kidney raises safety concerns in both preclinical and clinical settings61. Future efforts are necessary to develop more compact base editor systems that can be packaged into a single AAV, and non-viral delivery methods, such as lipid nanoparticles (LNPs) that are testing in sickle cell disease for human gene editing applications62. In sum, our current study demonstrates therapeutic potential and kidney accessibility of base editor, laying the groundwork for further optimization of the delivery technologies/methods for its clinic potential.
Methods
Ethics statement
All animal protocols were approved by and conducted in accordance with Laboratory Animal Resources of the Mayo Clinic and Institutional Animal Care and Use Committee regulations.
AAV vector cloning
The ABE9 was cloned into dual-vector split-intein base editor AAV genome plasmids (Addgene plasmids 137176 and 137177) as described in ref. 8. In brief, the ABE9 fragments were amplified by PCR, and then cloned into the N-AAV vector using isothermal assembly in NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs (NEB), E2621S). The protospacer sequence targeting the pathogenic variant R3277C was cloned into the sgRNA cassette in AAV genome vectors as previously described in ref. 11. In brief, AAV genome vectors were digested with BsmBI and ligated with pre-annealed oligos encoding the spacer sequence. SgRNA Forward oligo: aaaccgtagccgctttactTGCgtC. Reverse oligo: caccGacGCAagtaaagcggctacg. Non-target sgRNA Oligo-scramble-F: caccGTGTAGTTCGACCATTCGTG, scramble-R: aaacCACGAATGGTCGAACTACAC. For the vehicle AAV, we used the Cas9-ABE9 vectors with the non-targeted sgRNA and injection with 1 × 1012 vg/ mice.
To generate the Ksp-AAV vectors, we replaced the ubiquitously expressing synthetic Cbh promoter with the AAV9 vectors with the kidney-specific cadherin 16 promoter (Ksp) promoter (1.3 kb), which was amplified from mouse genomic DNA with the following primers, including forward oligo: ATGGTACCagcttgctctgccatg and reverse oligo: CGACCGGTgcaaatttggcttagg.
AAV production
AAV production was performed as previous reported8. Briefly, HEK293T/17 cells (ATCC, cat: CRL-11268) were plate in 10 cm dishes with DMEM/10% FBS without antibiotic (Thermo Fisher Scientific 157150). Each plate was transfected with 5 μg AAV genome, 5 μg pAdDeltaF6 and 5 μg pAAV2/9n plasmid. PEI 40k transfect regent mix with plasmids and incubate in 37 °C for 15 min. Three days after transfection, cells were scraped and used the AAVpro purification kit (All serotypes, Takara) to purify the AAV particles. Then the concentration of AAV particles was detected by AAVpro® Titration Kit (for Real Time PCR) Ver.2 (Takara).
Mouse strains and treatments
All mice were housed and maintained under pathogen-free conditions. The housing conditions were: 12 h dark/light cycle (lights on at 8:00 AM), 22 °C temperature, and 30%–70% humidity. 129 S/6 Pkd1RC/RC mice were treated with AAV particles by intravenous injection at postnatal day 14 and day 28, and then those mice were euthanized at three-month-old and six-month-old for further analysis. To evaluate the effect of gene editing on the lifespan of ADPKD mice, we generated Pkd1null/RC mice by crossing Pkd1null/+ mice with Pkd1RC/RC. Pkd1null/RC mice and Pkd1null/RC mice were treated with AAV particles by intravenous injection at postnatal day 7 (retro-orbital injection).
High-throughput sequencing of in vivo samples
Fresh tissues (5-10 mg) were digested with 600 lysis buffers (10 mM Tris-HCl, pH 7.5, 0.05% SDS, 25 μg/ml proteinase K (NEB)). Lysing tissues were incubated at 55 °C overnight. Isolated DNA was amplified as described for the genomic DNA samples using mouse Pkd1 specific primers (F: 5-CAGTTCCAGCGGCTCCTC-3, R: 5-GTCCAGCAAACCTCACAGGA-3). The sgRNA library readout was performed using two steps of PCR, where the first PCR includes enough genomic DNA to preserve full library complexity, and the second PCR adds appropriate sequencing adapters to the products from the first PCR The sgRNA library for each sample (plasmid, genomic DNA from cells and tissues) was amplified and prepared for Illumina sequencing using the following two-step PCR procedure. All PCR was performed using Phusion Flash High Fidelity Master Mix (Thermo). For PCR#1, the thermocycling parameters were: 98 oC for 30 s, 18–24 cycles of (98 oC for 1 s, 62 oC for 5 s, 72 oC for 35 s), and 72 oC for 1 minute. In each PCR#1 reaction, we used 3 μg of gDNA. Then the PCR products were harvested and send to GENEWIZ (Amplicon EZ; GENEWIZ Germany GmbH) for a second PCR and deep sequencing. As for the editing assay at the RNA level, cDNA was synthesized using 1 μg of RNA, and reverse transcription was performed11. As for the editing assay at the RNA level, cDNA was synthesized using 1 μg of RNA, and reverse transcription was performed. PCR amplification was performed using 50 ng of cDNA, and the PCR program was performed at 95 °C for 5 min, followed by 39 cycles of 94 °C for 30 s and 60 °C for 30 s at a slope of 2 °C/s. Raw sequencing data are available at the NCBI Sequence Read Archive (SRA) under the Bioproject accession no PRJNA1157189. Amplicon sequencing results were analyzed using CRISPResso2.
AAV copy number detection
DNA was extracted from kidneys and livers tissues using the QIAGEN DNeasy Blood and Tissue kit and resuspended in AE buffer. The AAV copy number was detected by using AAVpro® Titration Kit (for Real Time PCR) Ver.2 (Takara, Cat 6233). Brifely, qPCR reactions were prepared in 10 μL reaction volume with AAV9 primers, TB Green Premix, positive control. Thermocycling parameters were as follows: initial denaturation at 95 °C for 2 min, followed by 35 cycles of denaturation at 95 °C for 5 s and elongation at 60 °C for 30 s. The standard curve was created with positive AAV9 vector and diluted to an appropriate range for primer efficiency to estimate vector copies per reaction.
RNA-sequencing and analysis of transcriptome-wide off-target RNA base editing
Mouse kidneys were collected and homogenized in Trizol Reagent (Life Technologies, USA) with TissueLyzer (QIAGEN, Inc., Valencia, CA) for the RNA extraction. The construction and sequencing of the RNA-seq library were performed by CD Genomics (USA) on an Illumina PE150 platform. Analysis of transcriptome-wide off-target RNA base editing was performed as previously described63. Data were processed as previously described and normalized to the total number of reads per million exon kilobases (RPKM). Reads were aligned to the GRCm38 mouse genome using Spliced Transcripts Alignment to a Reference (STAR). RNA edits in ABE9 compared with the negative controls were filtered to include only loci with ten or more reads and greater than 0% reads containing an alternate allele. In brief, REDItools version 1.312 was used to quantify the average percentage of A-to-I editing among all sequenced adenosines in each sample. Any adenines with a read depth less than 10 or a read quality score below 30 were removed from the analysis. The transcriptome-wide A-to-I editing frequency was calculated as: (number of reads in which an adenosine was called as a guanosine) divided by (total number of reads covering all analyzed adenosines).
Tissue histology and Immunohistochemistry
The sections of the kidney, liver, and heart were fixed in 4% paraformaldehyde (pH 7.4), and stained with hematoxylin and eosin (H&E) and Masson trichrome staining methods. For the Immunohistochemistry (IHC) staining, a monoclonal mouse anti-GFP antibody (Santa Cruz, Dallas, TX, USA, sc-9996, 1:500), biotinylated secondary antibody (Santa Cruz Biotechnology Inc., 1:500), and DAB substrate system were used. Kidney sections were counterstained by hematoxylin. For the Masson trichrome staining, collagen fibers in kidney sections were stained with Masson trichrome. Images were analyzed using a NIKON ECLIPSE 80i microscope.
Primary renal tubular isolation
Fresh renal tissue (approximately 10 mg) was placed in a sterile Petri dish and soaked with PBS to remove blood. The outer cortex was cut from the medulla to form pieces about 1 mm3 and then washed three times. The tissue was then digested in 1 mg/ml collagenase type IV with 0.1% DNase I in fetal bovine serum (FBS) at 37 °C for 30 min in a shaking water bath. The digestion was then stopped by adding an equal volume of ice-cold PBS/10% FBS. The digestion was stopped by adding an equal volume of ice-cold PBS containing 10% FBS. The cell suspension was first passed through a 100 μm sterile cell strainer to remove undigested tissue chunks and large structures such as glomeruli. The filtrate was collected and centrifuged at 4 °C at 200 g for 5 min. The cell pellet was gently resuspended in cold PBS and layered onto a discontinuous Percoll gradient (25%–50%), followed by centrifugation at 3000 × g for 30 min at 4 °C. The layer of renal tubular epithelial cells was harvested, then diluted in ice-cold PBS and centrifuged at 200 × g at 4 °C for 5 min to remove any contaminated Percoll. After centrifugation, the supernatant was removed carefully, and the pellets were resuspended in culture medium for further analyses.
Immunofluorescence (IF) staining
The sections of the kidney and liver were fixed in 4% paraformaldehyde (pH 7.4) and stained with IF staining with a pan-macrophage marker, F4/80. After antigen retrieval, tissue sections were incubated with a rat anti-mouse F4/80 antibody (14-4801-82; eBioscience Inc.; 1:100 dilution) overnight, and then were incubated with Fluro-555 anti-rat IgG secondary antibody and mounted in Prolong Gold Anti-fade reagent with DAPI (Invitrogen). For Ki67 staining, a rabbit anti-Ki67 antibody (ab15580; Abcam) and Alexa Fluor 488 anti-rabbit IgG secondary antibody were used. For DBA, LTL, NCC and NKCC2 staining, the fixed kidney sections was stained with DBA Rhodamine (RL-1032-2, Vector Laboratories, 1:200), LTL Texas Red (21761195-1, bioPLUS™, 1:200), NCC (GeneTex, GTX41969, 1:1000), NKCC2 (Proteintech, 18970-1-AP, 1:1000) and mouse monoclonal GFP antibody (Santa cruz, sc-9996, 1:1000). For liver immune infiltration staining, the mouse FITC-CD45 antibody (Biolegend, Cat: 103107), PE-CD3 antibody (Biolegend, Cat: 100205) and PE-CD4 antibody (Biolegend, Cat: 100407) were used. Images were analyzed using a Nikon TI2E microscope.
Quantitative reverse-transcription PCR
The RNA extraction and reverse transcription (RT) were performed as previously described in ref. 64. Total RNA was extracted using the RNeasy Plus Mini Kit (QIAGEN). Total RNA (1 μg) was used for RT reactions in a 20 μl reaction to synthesize cDNA using an iScript cDNA Synthesis Kit (Bio-Rad). RNA expression profiles were analyzed by real-time PCR using iTaq SYBR Green Supermix with ROX (Bio-Rad) in an iCycler iQ Real-Time PCR Detection System. The complete reactions were subjected to the following program of thermal cycling: 40 cycles of 10 seconds at 95 °C and 20 seconds at 60 °C. A melting curve was run after the PCR cycles, followed by a cooling step. Each sample was run in triplicate in each experiment, and each experiment was repeated 3 times. Expression levels of target genes were normalized to the expression level of actin. Mouse Pkd1 primer used as: Forward 5’- TCAATGGGTCTGTGTCAAGG-3’, Reward: 5’-CCAGGGAGCATAGGAACATC-3’. Cas9 primer used as Forward 5’: TCTGAGACACCTGGCACAAG-3’, Reward: 5’-: CTGGGCACCTTGTACTCGTC-3’. Mouse TNF-α Forward 5′-GCCTCTTCTCATTCCTGCTTG-3′, Reward 5′-CTGATGAGAGGGAGGCCATT-3′, Mouse IL-6 Forward 5′-ACGGCCTTCCCTACTTCACA-3′, Reward 5′-CATTTCCACGATTTCCCAGA-3′.
Western blot
Western blotting analysis was performed on 3%–8% TA gels (Invitrogen) for 4.5 hours at 150 V at 4 °C. Gels were transferred to 0.45 μm nitrocellulose membranes at 40 V overnight at 4 °C. The blots were probed with anti-PC1 antibody (7e12), Cas9 Antibody (Santa Cruz Biotechnology Inc, sc-517386), and then with HRP-conjugated secondary antibodies (Southern Biotech, 1:1000).
Measurement of cyst area
The cyst volume was quantified in the whole kidney after H&E staining using Image-Pro Plus v5 software (Media Cybernetics). The cyst area was calculated as (cyst area/total area) × 100. Three sections from both kidneys were analyzed for each mouse.
Quantitative BUN determination
Serum samples were diluted 5-fold in distilled water prior to assay. Next, 5 μl water (blank), 5 μl standard (50 mg/dl) and 5 μl sample were transferred in triplicate into wells of a clear-bottom 96-well plate. Approximately 200 μl of working reagent was added and tapped lightly to mix, and the samples were incubated 20 min at room temperature. Optical density was read at 520 nm.
Analysis of serum creatinine and C-reactive protein (CRP)
Creatinine levels were measured with the QuantiChrom Creatinine Assay Kit from BioAssay Systems (Hayward, CA). The CRP levels were analyzed with the Mouse C-Reactive Protein/CRP ELISA Kit (Protentech, Cat No. KE10085). Serum was obtained by centrifuging the blood samples at 4000 × g/min for 15 min following 2 h resting period. The collected serum samples were subjected to ELISA, and serum levels of creatinine and CRP were quantified in accordance with the manufacturer’s instructions.
Analysis of liver aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP)
Liver tissues were cut into about 1 mm3 blocks with a sterile knife and washed twice with PBS and lysate to grind tissue homogenates on ice. After centrifuging for 5 minutes at 5000 × g then collect the supernatant and detect total protein concentration by bicinchoninic acid assay (BCA assay). The levels of AST, ALT, and ALP in the liver supernatant were measured by AST (ELK Biotechnology, Cat: ELK1778), ALT (Biocompare, Cat: EM0829), and ALP (ebiohippo, Cat: BHE10810355). ELISA kits according to the manufacturer’s instructions. The absorbance (450 nm) of each sample was detected on a standard automatic microplate reader.
Analysis of complete blood count (CBC) parameters
The whole blood was collected within an hour before the analysis of CBC, including white blood cell (WBC), red blood cell (RBC), platelets. The Piccolo XpressTM Chemistry Analyzer was utilized according to manufacturer instructions65 along with the Abaxis VetScan HM5 Analyzer for veterinary hematology in Mayo Clinic.
Biodistribution of injected EGFP-AAV
To assess the biodistribution of cystic cell-derived AAV-Ksp-EGFP and AAV-Cbh-EGFP particle, 1.5-month-old wide type and Pkd1RC/RC mice were treated with a dose of 1 × 1012 vg AAV-Ksp-EGFP and AAV-Cbh-EGFP by intravenous injection. The AAV-Ksp-EGFP and AAV-Cbh-EGFP treated wide type and Pkd1RC/RC mice were sacrificed 2 weeks post-administration, and then the organs (kidney, heart, liver, lung, and spleen) were harvested and analyzed with vivo imaging system (IVIS PerklinElmer, MA, USA). In addition, fresh frozen sections of whole kidney, liver, and heart were examined under fluorescence microscopy, where GFP auto-fluorescence confirmed the presence of AAV-derived signals in these organs.
Junction-spanning RT-qPCR analysis
To analyze the expression of GFP transcripts in kidneys, the primers were designed to target the spliced 5′-UTR–GFP junction in the mature mRNA: Forward 5′-ATCACTCTCGGCATGGACGA-3’, and Reward 5′- AAGCAGCGTATCCACATAGC-3’. The RNA extraction and reverse transcription (RT) were performed as previous described in ref.64. RT-qPCR was conducted using the following program: an initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s and annealing/extension at 60 °C for 20 s. A melting curve analysis was performed at the end of amplification, followed by a cooling step. The experiment was repeated three times in AAV-Ksp-EGFP and AAV-Cbh-EGFP groups vs. vehicle controls. GFP transcript abundance was normalized to the expression of actin.
Statistics & reproducibility
All statistical data are presented as mean ± SEM. All statistical analyses were performed using SPSS Statistics 22 software. P-values were calculated by two-tailed unpaired Student’s t-test and one-way ANOVA; and a P-value less than 0.05 was considered significant. The Investigators were blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The High-throughput DNA sequencing data generated in this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession code PRJNA1157189. There are no restrictions on data availability associated with any part of this work. Source data are provided with this paper.
References
Agborbesong, E., Li, L. X., Li, L. & Li, X. Molecular mechanisms of epigenetic regulation, inflammation, and cell death in ADPKD. Front. Mol. Biosci. 9, 922428 (2022).
Torres, V. E. & Harris, P. C. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int. 76, 149–168 (2009).
Chebib, F. T. & Torres, V. E. Autosomal dominant polycystic kidney disease: core curriculum 2016. Am. J. Kidney Dis. 67, 792–810 (2016).
Ecder, T. & Schrier, R. W. Cardiovascular abnormalities in autosomal-dominant polycystic kidney disease. Nat. Rev. Nephrol. 5, 221–228 (2009).
Cornec-Le Gall, E. et al. Type of PKD1 mutation influences renal outcome in ADPKD. J. Am. Soc. Nephrology: JASN 24, 1006 (2013).
Zhou, J. X. et al. DNA methyltransferase 1 (DNMT1) promotes cyst growth and epigenetic age acceleration in autosomal dominant polycystic kidney disease. Kidney Int. 106, 258–272 (2024).
Mao, X. et al. Cellular senescence and its association with aldose reductase promote cyst growth in autosomal dominant polycystic kidney disease. Kidney Int. 108, 811–826 (2025).
Levy, J. M. et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 4, 97–110 (2020).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N. M. et al. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Koblan, L. W. et al. In vivo base editing rescues Hutchinson–Gilford progeria syndrome in mice. Nature 589, 608–614 (2021).
Reichart, D. et al. Efficient in vivo genome editing prevents hypertrophic cardiomyopathy in mice. Nat. Med. 29, 412–421 (2023).
Hopp, K. et al. Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J. Clin. Investig. 122, 4257–4273 (2012).
Chen, L. et al. Engineering a precise adenine base editor with minimal bystander editing. Nat. Chem. Biol. 19, 101–110 (2023).
Tan, Y. et al. Rationally engineered Staphylococcus aureus Cas9 nucleases with high genome-wide specificity. Proc. Natl Acad. Sci. 116, 20969–20976 (2019).
Nishimasu, H. et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361, 1259–1262 (2018).
Miller, S. M. et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 38, 471–481 (2020).
Piontek, K., Menezes, L. F., Garcia-Gonzalez, M. A., Huso, D. L. & Germino, G. G. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat. Med. 13, 1490–1495 (2007).
Gainullin, V. G., Hopp, K., Ward, C. J., Hommerding, C. J. & Harris, P. C. Polycystin-1 maturation requires polycystin-2 in a dose-dependent manner. J. Clin. Investig. 125, 607–620 (2015).
Chapman, A. B. et al. Left ventricular hypertrophy in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 8, 1292–1297 (1997).
Atwood, D. J. et al. Increased mTOR and suppressed autophagic flux in the heart of a hypomorphic Pkd1 mouse model of autosomal dominant polycystic kidney disease. Cell. Signal. 74, 109730 (2020).
Ding, W. Y. et al. Adeno-associated virus gene therapy prevents progression of kidney disease in genetic models of nephrotic syndrome. Sci. Transl. Med. 15, eabc8226 (2023).
Furusho, T. et al. Enhancing gene transfer to renal tubules and podocytes by context-dependent selection of AAV capsids. Nat. Commun. 15, 10728 (2024).
Wu, G. et al. Adeno-associated virus–based gene therapy treats inflammatory kidney disease in mice. J. Clin. Investig. 134, e174722 (2024).
Gupta, N., Zhang, K., Sabbisetti, V., Shu, J. & Morizane, R. AAV for gene therapy drives a nephrotoxic response via NFκB in kidney organoids. Signal Transduct. Target. Ther. 10, 252 (2025).
Igarashi, P. et al. Ksp-cadherin gene promoter. II. Kidney-specific activity in transgenic mice. Am. J. Physiol.-Renal Physiol. 277, F599–610 (1999).
Mononen, J. et al. Genetic variation is a key determinant of chromatin accessibility and drives differences in the regulatory landscape of C57BL/6J and 129S1/SvImJ mice. Nucleic Acids Res. 52, 2904–2923 (2024).
Arroyo, J. et al. The genetic background significantly impacts the severity of kidney cystic disease in the Pkd1RC/RC mouse model of autosomal dominant polycystic kidney disease. Kidney Int. 99, 1392–1407 (2021).
Lala, V., Zubair, M. & Minter, D. Liver function tests. StatPearls (2023).
Sproston, N. R. & Ashworth, J. J. Role of C-reactive protein at sites of inflammation and infection. Front. Immunol. 9, 754 (2018).
Chai, A. C. et al. Base editing correction of hypertrophic cardiomyopathy in human cardiomyocytes and humanized mice. Nat. Med. 29, 401–411 (2023).
Gopalappa, R. et al. In vivo adenine base editing rescues adrenoleukodystrophy in a humanized mouse model. Mol. Ther. 32, 2190–2206 (2024).
Swenson-Fields, K. I. et al. Macrophages promote polycystic kidney disease progression. Kidney Int 83, 855–864 (2013).
Karihaloo, A. et al. Macrophages promote cyst growth in polycystic kidney disease. J. Am. Soc. Nephrology: JASN 22, 1809–1814 (2011).
Chen, L. et al. Macrophage migration inhibitory factor promotes cyst growth in polycystic kidney disease. J. Clin. Investig. 125, 2399–2412 (2015).
Li, L. X. et al. Cross talk between lysine methyltransferase Smyd2 and TGF-β-Smad3 signaling promotes renal fibrosis in autosomal dominant polycystic kidney disease. Am. J. Physiol.-Ren. Physiol. 323, F227–F242 (2022).
Zhou, J.X. et al. CD74 Promotes Cyst Growth and Renal Fibrosis in Autosomal Dominant Polycystic Kidney Disease. Cells 13 (2024).
Sagar, P. S. & Rangan, G. K. Cardiovascular Manifestations and Management in Autosomal Dominant Polycystic Kidney Disease. Kidney International Reports, (2023).
Spinelli, L., Giugliano, G. & Esposito, G. Cardiac involvement in autosomal dominant polycystic kidney disease. Cardiogenetics 11, 39–49 (2021).
Xu, L. et al. Efficient precise in vivo base editing in adult dystrophic mice. Nat. Commun. 12, 3719 (2021).
Jeong, Y. K., Song, B. & Bae, S. Current status and challenges of DNA base editing tools. Mol. Ther. 28, 1938–1952 (2020).
Lebek, S. et al. Ablation of CaMKIIδ oxidation by CRISPR-Cas9 base editing as a therapy for cardiac disease. Science 379, 179–185 (2023).
Qi, M. et al. In vivo base editing of Scn5a rescues type 3 long QT syndrome in mice. Circulation 149, 317–329 (2024).
Tu, T. et al. A precise and efficient adenine base editor. Mol. Ther. 30, 2933–2941 (2022).
Kuo, I. Y. & Chapman, A. B. Polycystins, ADPKD, and cardiovascular disease. Kidney Int. Rep. 5, 396–406 (2020).
Judge, P. K. et al. Biliary tract and liver complications in polycystic kidney disease. J. Am. Soc. Nephrol. 28, 2738–2748 (2017).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Xin, H., Wan, T. & Ping, Y. Off-Targeting of Base Editors: BE3 but not ABE induces substantial off-target single nucleotide variants. Signal Transduct. Target. Ther. 4, 9 (2019).
Konkalmatt, P.R. et al. Renal rescue of dopamine D2 receptor function reverses renal injury and high blood pressure. JCI insight 1 (2016).
Rubin, J. D., Nguyen, T. V., Allen, K. L., Ayasoufi, K. & Barry, M. A. Comparison of gene delivery to the kidney by adenovirus, adeno-associated virus, and lentiviral vectors after intravenous and direct kidney injections. Hum. gene Ther. 30, 1559–1571 (2019).
Furusho, T. et al. Enhancing gene transfer to renal tubules and podocytes by context-dependent selection of AAV capsids. bioRxiv, 2023.2007. 2028.548760 (2023).
Daneshgar, N. et al. Metabolic derangement in polycystic kidney disease mouse models is ameliorated by mitochondrial-targeted antioxidants. Commun. Biol. 4, 1200 (2021).
Schulz, M. et al. Binding and neutralizing anti-AAV antibodies: Detection and implications for rAAV-mediated gene therapy. Mol. Ther. 31, 616–630 (2023).
Schuster, D. J. et al. Biodistribution of adeno-associated virus serotype 9 (AAV9) vector after intrathecal and intravenous delivery in mouse. Front. Neuroanat. 8, 42 (2014).
Hollidge, B. S. et al. Kinetics and durability of transgene expression after intrastriatal injection of AAV9 vectors. Front. Neurol. 13, 1051559 (2022).
Muhuri, M., Levy, D. I., Schulz, M., McCarty, D. & Gao, G. Durability of transgene expression after rAAV gene therapy. Mol. Ther. 30, 1364–1380 (2022).
Schindeler, A. et al. In vivo precision base editing to rescue mouse models of disease. Mol. Ther. Nucleic Acids. 36, 102622 (2025).
Chehelgerdi, M. et al. Comprehensive review of CRISPR-based gene editing: mechanisms, challenges, and applications in cancer therapy. Mol. cancer 23, 9 (2024).
Han, H. A., Pang, J. K. S. & Soh, B.-S. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J. Mol. Med. 98, 615–632 (2020).
Stone, D., Aubert, M. & Jerome, K. R. Adeno-associated virus vectors and neurotoxicity—lessons from preclinical and human studies. Gene Ther. 32, 60–73 (2025).
Zarghamian, P., Klermund, J. & Cathomen, T. Clinical genome editing to treat sickle cell disease—a brief update. Front. Med. 9, 1065377 (2023).
Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020).
Zhou, X. et al. Sirtuin 1 inhibition delays cyst formation in autosomal-dominant polycystic kidney disease. J. Clin. Investig. 123, 3084–3098 (2013).
Kottom, T. J. et al. Preclinical and Toxicology Assessment of ALW-II-41-27, an Inhibitor of the Eph Receptor A2 (EphA2). Drugs RD 24, 425–434 (2024).
Acknowledgements
X.L. acknowledges support from NIH R01 DK129241 and NIH R01 DK126662. J.X.Z. acknowledges support from NIH K01 DK107729, and PKD Foundation Research Grant. P.C.H. acknowledges support from NIH R01 DK058816 and NIH R01 DK059597. X.L. and P.C.H. acknowledge support from Department of Defense (DOD) Focused Program Award, PR221810. A.S.C. acknowledges support from American Heart Association (AHA), 24POST1242218.
Author information
Authors and Affiliations
Contributions
A.S.C. performed most experiments and data analysis. L.X.L. and J.X.Z. performed some of the experiments and data analysis. P.C.H. and J.P.C. assisted in data analysis and manuscript preparation. X.L. supervised the whole project, data analysis, and manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, A.S., Li, L.X., Zhou, J.X. et al. In vivo base editing rescues ADPKD in a humanized mouse model. Nat Commun 16, 11212 (2025). https://doi.org/10.1038/s41467-025-65997-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65997-1