Abstract
The significance of DNA hydroxymethylation in replicative senescence of mesenchymal stem cells (MSCs) and aging-related osteoporosis remains unknown. Here, we reveal 5hmC levels positively regulate MSC self-renewal and osteoblast differentiation. Mechanistically, PARP1 recruits TET1 to hydrolyze methylated nucleotides on DNMT1 exons, aiding CTCF in preventing DNMT1 alternative splicing in early MSCs. Additionally, ATM phosphorylates TRIM37 at Th203, promoting its nuclear entry and the monoubiquitination of PARP1, stabilizing the protein. CTCF or TRIM37 knockdown induces replicative senescence of MSCs with loss of full-length DNMT1. Co-treatment with resveratrol (ATM activator) and vitamin C (TET1 activator) rejuvenates late MSCs via the TRIM37/PARP1/DNMT1 pathway and alleviates osteoporosis in aged mice. Gene knockout experiments further reveal the participation of TRIM37 and PARP1 in MSC aging, contributing significantly to bone maintenance and repair in vivo. This study emphasizes the role of DNA hydroxymethylation in stemness, suggesting therapeutic strategies, especially for osteoporosis.
Similar content being viewed by others
Introduction
Age-related osteoporosis presents a significant health challenge, impacting bone density and strength. Understanding the dynamic functions of mesenchymal stem cells or multipotential stromal cells (MSCs) within the bone cavity is crucial for maintaining bone mineral content1. The intricate interplay between aging-related osteoporosis and MSCs in bone homeostasis underscores the need for a comprehensive understanding.
MSCs, capable of self-renewal and differentiation into mesenchymal and non-mesenchymal cells2, exhibit genetic stability through in vitro culture expansion, making cell-based therapies increasingly promising. However, for clinical applications, generating a substantial number of MSCs with the desired phenotype and function relies on the expansion of large cultures. Nevertheless, culture-related modifications, such as in vitro aging, may lead to adverse reactions and unsatisfactory results3. Like other normal human somatic cells, MSCs divide approximately forty times until they are close to the “senescence”, after which they can no longer replicate, and are characterized by enlargement, changes in morphology, and senescence-associated β-galactosidase expression4. The concept of senescence5 was found to correlate with the length of the telomeric region at the end of chromosome6,7. In addition to telomere erosion, the senescence or senescence is caused by DNA damage response (DDR)-dependent and independent activation of cyclin-dependent kinase (Cdk) inhibitors, such as p21Waf1/Cip1 and p16Ink4a8, and other genetic and epigenetic modifications9,10,11. Like many other cells, MSC aging is caused by various factors10. However, the underlying mechanism for the sudden loss of replication after the cell reaches the senescence remains elusive.
Long-term culture causes MSCs to impair ataxia telangiectasia mutated (ATM) function and increase the accumulation of DNA double-strand breaks (DSBs)12, which are the most deleterious forms of DNA damage that cause genomic instability13. Moreover, accumulation of DNA damage during expansion may contribute to the loss of differentiation potential of expanded MSCs14. Epithelial-mesenchymal transition (EMT) regulator, zinc-finger E-box-binding homeobox 1 (ZEB1), which can induce stemness in cancer, promotes DDR and tumor radioresistance15. MSC aging is also associated with reduced radioresistance via the downregulation of DDR16. Deficiencies in DDR limit the function of aging hematopoietic stem cells (HSCs). However, it is unclear whether DDR itself or a specific DDR regulator is responsible for the properties associated with early stem cells, such as self-renewal and multi-differentiation.
Recent findings suggest that epigenetic regulators responsible for modifying histones, such as sirtuin (SIRT)317, SIRT618, and SIRT719, as well as those involved in DNA methylation, including methylcytosine oxidases ten-eleven translocation (TET)120 and TET221, play a vital role in preserving tissue-specific stem cells like HSCs22. TET2 is also required for pre-B cell differentiation to macrophage23. Intriguingly, aging-associated functional decline24 or cell linage-specific functional change and oncogenic potential25 in MSCs are also regulated by epigenetic regulators, including DNA methyltransferase (DNMT)124 and DNMT3B25, which can be reversed by manipulating the epigenetic factors1,26 or culture conditions, such as reduced oxygen tension, to enhance MSC stemness and prevent senescence4,24. Recently, the decline in DNMT1 expression has been linked to the senescence-associated 3D genome reorganization27. The expression of DNMT1 plays a crucial role in MSC aging, being regulated by pluripotency factors and upregulated under hypoxic conditions24.
DNA 5-hydroxymethylcytosine (5hmC) modification, an oxidation product of the TET enzymes28, can potentially lead to DNA demethylation29,30, which is known to be associated with gene transcription. The dynamic changes in 5hmC levels have been well studied during embryonic31 or postnatal tissue-specific development process32 and aging33,34. Moreover, its roles in pluripotency35 and somatic reprogramming36 have been studied. Moreover, 5hmC is essential for the homeostasis of adult stem cells, such as the neural stem cells37 and HSCs38, and its dysregulation, caused by aging37 or mutation in TET enzymes, also predisposes a person to various diseases38. Through the development of a 5hmC tissue map by characterizing the genomic distribution of 5hmC in human tissues, 5hmC was found to be preferentially enriched on tissue-specific gene bodies and enhancers32. In mouse and human stem cells, 5hmC promotes co-transcriptional R-loop formation and regulates specific gene expression required for stem cell proliferation39. However, its role in maintaining the self-renewal and multi-differentiation potential of MSCs as well as the underlying molecular mechanisms remain to be studied.
In the current study, we first found that increased 5hmC levels are required to maintain the early passage characteristics of human bone marrow (BM)-derived MSCs. We further revealed that CTCF-mediated prevention of DNMT1 alternative splicing plays a key role in sustaining the proliferative capacity and differentiation potential of early passage BM-MSCs, which depends on the coordinated actions of TET1 and DNA damage response (DDR) components such as ATM, PARP1, and TRIM37. Mechanistically, PARP1 recruits TET1 to demethylate DNMT1 exons, facilitating CTCF-mediated splicing regulation, while ATM-mediated phosphorylation of TRIM37 promotes PARP1 stabilization. Knockdown of CTCF or TRIM37 induces replicative senescence. Importantly, combined treatment with an ATM activator (resveratrol) and a TET activator (vitamin C) rejuvenates late-passage MSCs, restores osteogenic capacity, and enhances bone regeneration in aged mice. These findings identify a TRIM37/PARP1/DNMT1 regulatory axis critical for MSC stemness and bone homeostasis, suggesting a therapeutic strategy for osteoporosis.
Results
Early human MSCs show higher levels of 5hmC, thereby exhibiting increased proliferation and differentiation potential
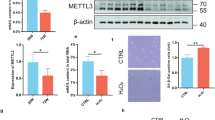
TET1/2 double knockout (DKO) mice had significantly reduced bone mineral density and bone volume/tissue volume ratio compared to the littermate controls40. Moreover, BM-MSCs isolated from TET1/2 DKO mice exhibited impaired bone formation compared to TET1/2 single KO or control mice, but did not exhibit any impaired proliferation and adipogenic differentiation capacities40. Although the 5hmC levels in BM-MSCs of TET1/2 DKO mice were lower than those of the TET1/2 KO or control group40, whether the 5hmC level plays an important role in replicative senescence and biological functions of expanded human BM-MSCs (henceforth referred to as MSCs, if not indicated) was not determined. We hypothesized that 5hmC levels may affect the senescence, proliferation, and differentiation capacities of MSCs. For this purpose, we first compared the 5hmC and 5mC levels between MSCs at early (P2-4) and late (P10-12) passages. Compared with early (passage) MSCs, late (passage) MSCs exhibit multiple hallmarks of cellular senescence, including enlarged and flattened morphology, increased expression of senescence markers (p16Ink4a and p21Waf1/Cip1), and reduced DNMT1 levels. Functionally, they show decreased proliferation and impaired differentiation into osteoblasts and adipocytes. Transcriptomic analysis reveals enrichment of the senescence-associated secretory phenotype (SASP) pathway, accompanied by elevated secretion of IL-1α and CXCL-8. Increased SA-β-gal⁺ staining further confirms the senescent phenotype (Supplementary Fig. 1)4. We observed an increase in the level of 5hmC in early MSCs using dot-blot (Supplementary Fig. 2a) and immunofluorescence (Fig. 1a) assays. In contrast, the level of 5mC was elevated in late MSCs (Supplementary Fig. 2a and Fig. 1a), consistent with findings from a previous study41,42. We next demonstrated that treatment of early MSCs with 2-hydroxyglutarate (2-HG), a TET inhibitor (5 mM), reduced the level of 5hmC without affecting 5mC (Fig. 1b and Supplementary Fig. 2b). Additionally, this treatment suppressed their proliferative capacity (Fig. 1c) and differentiation potential into osteoblasts and adipocytes (Fig. 1d). These data suggest that the 5hmC level plays an important role in the maintenance of replication and biological functions of early MSCs.
a Immunofluorescence shows that early MSCs (P2-4) exhibited increased 5hmC level compared to late MSCs (P10-12). b–d Early MSCs were treated with 2-hydroxyglutarate (2-HG) (5 mM) or vehicle control for 3 d, followed by (b) immunofluorescence assay to determine the 5hmC level, (c) WST-1 assay for growth curve, or (d) the induction of cells for osteogenic and adipogenic differentiation for 14 d. Cells induced for osteogenesis were stained by alizarin red S (ARS) staining, followed by measurement of extracted dye at an optical density (OD) of 550 nm, while the cells induced for adipogenesis were stained with Oil Red O staining, followed by measurement of extracted dye at OD 510 nm. We found that 2-HG treatment decreased the 5hmC level, inhibited cell proliferation, and suppressed the differentiation into osteoblasts and adipocytes. e–h Early MSCs were lentivirally transduced with scrambled (CTR), TET1 (TET1-KD) or TET2 short hairpin RNAs (shRNAs) (TET2-KD), followed by (e) western blotting for the detection of TET1 or TET2 levels and (f) immunofluorescence for the detection of 5hmC level. TET1 KD, rather than TET2 KD, decreased the 5hmC level. Cells without or with TET1 KD were subjected to (g) growth curve analysis by WST-1 assay, or (h) induced for osteogenic and adipogenic differentiation for 14 d. TET1 KD inhibited the cell proliferation, and suppressed the differentiation into osteoblasts and adipocytes. Results are expressed as the mean ± standard deviation (SD) of three independent experiments. (**p < 0.01, ***p < 0.001; ****p < 0.0001; unpaired two-tailed t-test, or ordinary one-way ANOVA) Exact p-values are shown in the Source Data file. Scale bar: 50 μm (a, b, f) and 500 μm (d, h).
TET1 knockdown reduces 5hmC levels, and suppresses the proliferation and differentiation potential of early human MSCs
We further used lentivirus-mediated knockdown (KD) to directly target and investigate the roles of TET1 and TET2 in maintaining early MSC properties (Fig. 1g), which are known to maintain the self-renewal and osteogenic differentiation potential of mouse BM-MSCs40. Our data further provide strong evidence that TET1 is the primary regulator of 5hmC levels in MSCs. KD of TET1, but not TET2, leads to a significant reduction in 5hmC levels, as demonstrated by both immunofluorescence (Fig. 1f) and dot blot analysis (Supplementary Fig. 2c). While dot blot results show that KD of either TET1 or TET2 can reduce 5hmC, the magnitude of reduction is substantially greater following TET1 KD (Supplementary Fig. 2c). Notably, immunofluorescence analysis reveals a more prominent and consistent decrease in nuclear 5hmC levels specifically upon TET1 depletion. These findings indicate that, although TET2 may contribute to 5hmC regulation, TET1 plays a dominant and non-redundant role in maintaining 5hmC levels in MSCs. Moreover, early MSCs with TET1 KD exhibited reduced cell growth (Fig. 1g) and decreased differentiation potential into osteoblasts and adipocytes (Fig. 1h) compared to cells transduced with scrambled (CTR) shRNAs. These findings suggest that TET1 is the primary enzyme responsible for increasing 5hmC levels in early MSCs, and that 5hmC is crucial for MSC growth and differentiation potential.
PARP1/TET1 complex regulates the proliferation and differentiation potential of early human MSCs
PARP1 interacts with target proteins involved in histone modification and DNA methylation43. For instance, it has been reported that PARP1 facilitates the establishment of H3K4me3 marks at the TET1 promoter and directly interacts with TET1, contributing to the progression of T-cell acute lymphoblastic leukemia44. To investigate the role of PARP1 in TET1-mediated elevation of 5hmC levels and the maintenance of early MSC properties, we first demonstrated an endogenous interaction between PARP1 and TET1 in early MSCs using co-immunoprecipitation (Fig. 2a). To confirm that this interaction is direct and independent of PARP1 enzymatic activity or poly(ADP-ribose) (PAR) levels, we treated cells with the PARP inhibitor PJ3445. Notably, the interaction between PARP1 and TET1 remained detectable by both immunoprecipitation and Duolink proximity ligation assay (PLA), even under PARP inhibition (Supplementary Fig. 3). Then, we showed that early MSCs from different individuals showed higher levels of PARP1 compared to late MSCs (Fig. 2b), We further analyzed the RNA-seq of gene expression profiles in early MSCs, late MSCs, and early MSCs with PARP1 KD or TET1 KD. Interestingly, hierarchical clustering analysis revealed that, with the exception of MSCs from donor 3, early MSCs from different individuals clustered together and were distinct from late MSCs. Notably, late MSCs clustered with early MSCs that had PARP1 or TET1 knocked down (Fig. 2c). Pathway analysis of 1026 differentially expressed genes in early MSCs compared to late MSCs revealed several altered pathways that are responsible for the early MSC phenotypes, including increased activity of “differentiation of osteoblast” and “adipogenesis of cells” (Supplementary Fig. 4, Supplementary Data 1). PARP1 KD downregulated 5hmC in early MSCs, while PARP1 overexpression upregulated 5hmC in late MSCs (Fig. 2d). In addition, PARP1 KD increased p16Ink4a and p21Waf1/Cip1 levels (Fig. 2e) and suppressed the proliferation and differentiation potentials in early MSCs (Fig. 2f, g), while PARP1 overexpression decreased p16Ink4a and p21Waf1/Cip1 levels and promoted the proliferation and differentiation potentials in late MSCs (Fig. 2e, h, i). To further confirm the functional relationship between PARP1 and TET1 in maintaining MSC identity, we showed that TET1 KD effectively reduced TET1 protein levels despite robust PARP1 overexpression and reversed the associated decrease in senescence marker expression (Supplementary Fig. 5a). Consistently, PARP1-induced elevation of global 5hmC levels was abolished upon TET1 KD (Supplementary Fig. 5b). Functionally, TET1 KD impaired the proliferative capacity of PARP1-overexpressing (OE) MSCs (Supplementary Fig. 5c) and significantly reduced their differentiation potential into both osteoblasts and adipocytes (Supplementary Fig. 5d). Together, these data indicate that PARP1 interacts with TET1 to increase 5hmC levels and maintain early MSC properties.
a Co-immunoprecipitation assay shows the interaction between PARP1 and TET1 in early MSCs. b Western blotting shows that early MSCs from different donors had elevated PARP1 levels compared to late MSCs. c The dendrogram of hierarchical clustering of the 12 samples shows that TET1-KD or PARP1-KD early-MSCs are closer to late MSCs than to parental early MSCs. The gene expression data is standardized to Z score and displayed as a heatmap. d–i Early MSCs were subjected to lentiviral knockdown of PARP1 (PARP1-KD), while late MSCs were subjected to lentiviral overexpression of PARP1 (PARP1-OE). The cells were used for different assays. (d) PARP1-KD decreased 5hmC levels in early MSCs, while PARP1-OE increased 5hmC levels in late MSCs. e–g Knockdown of PARP1 in early MSCs upregulated the senescence markers (e), and suppressed their proliferation (f) and differentiation potentials (g). e, h, i Overexpression of PARP1 downregulated the senescence markers (e) and enhanced the proliferation (h) and differentiation potentials in late MSCs (i). Results are expressed as the mean ± SD of three independent experiments. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; unpaired two-tailed t-test) Exact p-values are shown in the Source Data file. Scale bar: 50 μm (d) and 500 μm (g, i).
MeDIP and ChIP-seq reveal multiple downstream targets of PARP1/TET1, including the alternative splicing of DNMT1
To identify the downstream targets with increased 5hmC levels of the PARP1/TET1 complex in early MSCs, we performed methyl DNA immunoprecipitation (MeDIP)-seq and hydroxymethyl DNA immunoprecipitation (hMeDIP)-seq analyses in early and late MSCs, and PARP1, and TET1 chromatin immunoprecipitation sequencing (ChIP-seq) analyses in early MSCs. Unfortunately, the hMeDIP-seq data were inconclusive, as the majority of 5hmC-enriched reads across the genome did not show significant enrichment over input controls. One possibility with fewer reads in hMeDIP-seq is that 5hmC in the DNA has been replaced by replication-dependent and independent DNA demethylation cycle46. Enriched DNA segments overlapped in MeDIP-seq (late > early), PARP1 ChIP-seq (ChIP > input), and TET1 ChIP-seq (ChIP > input) were analyzed and the overlapping gene numbers are shown in a Venn diagram (Fig. 3a). The triple overlapping enrichment included 20 genes targeted by the PARP1/TET1 complex (Supplementary Table 1), which showed reduced DNA methylation in early MSCs. The DNMT1 gene, a triple-overlapping gene, is essential for maintaining the self-renewal and undifferentiated state of early human MSCs24. Overexpression of DNMT1 in MSCs cultured under normoxic conditions led to reduced expression of p16Ink4 and p21Waf1/Cip1, increased proliferation rates, higher BrdU incorporation, and enhanced differentiation potential24. PARP1 and TET1 ChIP-seq analysis (Supplementary Fig. 6) and additional ChIP-PCR revealed that the region covering flanking to transcription start site (TSS, Chr19:10305629) of the DNMT1 gene was bound by PARP1 and TET1 in early MSCs (Fig. 3b). These data are similar to a previous study that showed the binding of PARP1 at or near the TSSs of genes positively regulated by PARP147. KD of PARP1 or TET1 reduced DNMT1 full-length protein levels (Fig. 3c). Interestingly, compared with early MSCs, the enriched MeDIP peaks of DNMT1 in the late MSCs were located exons 21, 30, 35, 36, 37, and 39, rather than the TSS site, which is the catalytic domain of DNMT1 (Fig. 3d). Because there is a direct mechanistic association between intragenic DNA methylation and alternative pre-mRNA splicing48, we checked the existence of the DNMT1 isoform with alternative splicing in this region in late MSCs (Fig. 3e) or in early MSCs with PARP1 or TET1 KD (Supplementary Fig. 7a) using 5’-RACE-PCR to amplify generated cDNA first strands. An additional band, significantly smaller than the full-length product, was observed when using primers flanking exons 25 and 38, but not with primers flanking exons 13 and 25 (Fig. 3e, Supplementary Fig. 7a). DNA sequencing of the shorter PCR product showed that it was an alternative splicing of DNMT1, resulting in exon exclusion from exon 30 to exon 37 (Supplementary Fig. 8). Unfortunately, the available antibodies against DNMT1 cannot recognize this isoform. Western blotting of cell lysates from early and late MSCs with or without pretreatment with MG132 also failed to detect this isoform (Supplementary Fig. 9), suggesting that the mRNA or protein isoform was not stable. We identified differential alternative splicing events between different subgroups of MSCs by RNA-Seq and replicated multivariate analysis of transcript splicing (rMATS)49,50. Notably, the number of genes increased in alternative splicing events in late MSCs was slightly higher than that in early MSCs (Supplementary Table 2, Supplementary Data 2), and the number of genes increased in alternative splicing events in early MSCs with PARP1 or TET1 KD was slightly lower than that in early MSCs (Supplementary Table 2, Supplementary Data 3, 4). Among the 20 genes (Supplementary Table 1) targeted by the PARP1/TET1 complex, GUSBP1and PDE4DIP increased in alternative splicing events in late MSCs when compared to early MSCs; GUSBP1and LOC100506990 increased in alternative splicing events in early MSCs with TET1 KD when compared to early MSCs; and LOC100506990 increased in alternative splicing events in early MSCs with PARP1 KD when compared to early MSCs. These data indicate that the PARP1/TET1 complex-mediated loss of intragenic DNA methylation in DNMT1 prevents alternative splicing of DNMT1 in early MSCs.
a Venn diagram showing overlap of genes enriched in MeDIP analysis (Late > Early), PARP1 ChIP-seq analysis (Early > Late) and TET1 ChIP-seq analysis (Early > Late). Numbers indicate the number of overlapping genes or regions. b ChIP-polymerase chain reaction (PCR) showing the binding or PARP1 and TET1 at the transcription-start site (TSS) of DNMT1 in early MSCs. c Early MSCs were lentivirally transduced with scrambled (CTR), PARP1 (PARP1-KD) or TET1 short hairpin RNAs (shRNAs) (TET1-KD), followed by western blotting showed that the knockdown of PARP1 or TET1 decreased the levels of the DNMT1 full-length protein. d Representative MeDIP peaks shown in early and late MSCs for genes encoding DNMT1. The high methylation region of late MSCs is located in exons 30 to 37, which is the catalytic domain of DNMT1. e 5’-rapid amplification of cDNA ends (RACE)-PCR revealed the increase of alternative splicing forms of DNMT1 in late MSCs. Arrow indicates the PCR product of the alternative splicing isoform. f CTCF ChIP real-time PCR showing the binding of CTCF in exon30 and exon37 in early and late MSCs. H19 serving as the positive control. g Early MSCs were lentivirally transduced with scrambled (CTR) and CTCF shRNAs (#1” and “#2” refer to two different shRNA constructs), followed by western blotting, which showed that the knockdown of CTCF decreased the DNMT1 full-length protein levels. h Realtime PCR showed that the knockdown of CTCF increased the levels of the alternative splicing isoform of DNMT1 in early MSCs. Late MSCs served as the control. i Plasmids carrying full-length (Full) or alternative splicing isoform (AS) of DNMT1 were transfected into 293 T cells, followed by immunoprecipitation of exogenous DNMT1 to determine the in vitro DNMT1 activity. Compared to Full, AS reduced the DNMT1 activity. Moreover, the addition of AS reduced the DNMT1 activity of Full in a dose-dependent manner. j Dot-blot assay shows that the 5mC level in plasmids carrying Full or AS of DNMT1 transfected 293 T cells. Addition of AS reduced the 5mC level in 293 T cells. Quantitation of dot-blot assay (j lower panel). k–m Early MSCs were inserted in lentiviral overexpression control vectors (CV) or vectors carrying AS DNMT1, followed by (k) western blotting analysis of senescence marker levels (AS M.W. size: around 130 kDa), (l) growth curve analysis, and (m) differentiation potential analysis. Cells were induced for osteogenic and adipogenic differentiation for 14 d, followed by ARS and Oil Red staining, respectively. Extracts of dyes were subjected to optic measurement. mright Quantification of optical measurements. Overexpression of AS-DNMT1 in MSCs inhibited their proliferation and differentiation potential, and increased the senescence marker levels. Results are expressed as the mean ± SD of three independent experiments. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, unpaired two-tailed t-test, or ordinary one-way ANOVA). Exact p-values are shown in the Source Data file. Scale bar: 500 μm.
Involvement of CTCF in PARP1/TET1 complex-mediated loss of intragenic DNA methylation and prevention of DNMT1 alternative splicing
Since PARP1 interacts with CTCF51, a key insulator in intragenic chromatin looping and exon inclusion via RNA polymerase II pausing48,52, we hypothesized that CTCF facilitates the inclusion of unmethylated exons during transcription elongation in early MSCs. Similar to previous studies51, the immunoprecipitation and PLA assay revealed the interaction of the PARP1/TET1 complex with CTCF (Supplementary Fig. 3). Prediction of CTCF binding sites in 20 triple overlapping enrichment genes by CTCFBSDB 2.0 software53 revealed that most of the CTCF binding sites were restricted to intragenic regions (Supplementary Table 3). Moreover, CTCF-binding sites were found in exons 21, 27, 30, 37, 39, and 41 (Supplementary Table 4) of the DNMT gene, which were largely overlapped with the exons hypermethylated in late MSCs. Subsequent quantitative real-time PCR of CTCF-ChIP showed that, compared with that of early MSCs, the binding of CTCF in exons 30 and 37 of DNMT1 in late MSCs and TET1 or PARP1 KD in early MSCs was reduced, while no difference was observed between early and late MSCs in the H19 gene, the positive control region (Fig. 3f, Supplementary Fig. 7b). Furthermore, KD of CTCF in early MSCs reduced the protein level of full-length DNMT1 (Fig. 3g) and the mRNA level of DNMT1 in exon 34, but not in exon 13 (Fig. 3h), suggesting that CTCF forms intragenic chromatin loops including exon 34 and its KD induces alternative splicing. These data indicate the involvement of CTCF in PARP1/TET1 complex-mediated loss of intragenic DNA methylation and prevention of DNMT1 alternative splicing in early MSCs.
DNMT1 alternative spliced form promotes human MSC senescence and reduces stemness
To examine the biological function of the DNMT1 alternative spliced form with a truncated catalytic domain, we constructed a plasmid encoding full-length (Full-DNMT1) or alternative splicing form of DNMT1 (AS-DNMT1), which was then overexpressed in 293 T cells. We then isolated and purified exogenous DNMT1 from 293 T cells. The in vitro DNMT1 activity of Full-DNMT1 was 3.5 times that of AS-DNMT1 (Fig. 3i). In addition, AS-DNMT1 reduced the activity of Full-DNMT1 in a dose-dependent manner, both in vitro (Fig. 3i) and in vivo, as evidenced by the loss of 5mC levels (Fig. 3j). This suggests that AS-DNMT1 not only reduces the activity of DNMT1, but also competes to inhibit the activity of full-length DNMT1. Early MSCs were lentivirally transduced with a control vector (CV) or a vector containing AS-DNMT1, followed by western blotting analysis for the expression of senescence markers and functional analysis of proliferation and multi-differentiation potential. Compared with CV, overexpression of AS-DNMT1 increased the expression of a short-form DNMT1 ( ~ 135 kDa) and the levels of senescence markers, p16Ink4a and p21Waf1/Cip1 (Fig. 3k), but inhibited cell growth (Fig. 3l) and differentiation potential into osteoblasts and adipocytes (Fig. 3m). These results indicate that DNMT1 with alternative splicing in the catalytic domain not only loses DNMT1 activity, but also inhibits the activity of full-length DNMT1, thereby promoting MSC senescence and reducing stemness.
T203 phosphorylation is required for TRIM37 to stabilize PARP1 via K337 mono-ubiquitination
To identify the upstream proteins of PARP1, we compared phosphorylated proteins that interacted with PARP1 in early and late MSCs using immunoprecipitation (IP)-tandem mass spectrometry (MS/MS) analysis (Supplementary Table 5). We first demonstrated that phosphor-TRIM37T203 was one of the enriched phosphopeptides associated with endogenous PARP1 in early MSCs (Fig. 4a) and confirmed the protein-protein interaction between exogenous PARP1 and TRIM37 in 293 T cells co-transfected with plasmids encoding Flag-TRIM37 and HA-PARP1 (Fig. 4b). Because TRIM37 is an E3 ubiquitin-protein ligase that induces mono-ubiquitination of nuclear proteins, such as H2A54 and NEMO46, and results in H2A stabilization and nuclear export of NEMO in cancer cells, we then examined whether TRIM37 ubiquitinates PARP1. IP-MS/MS analysis revealed that Lys337 (K337) and Lys438 (K438) were the possible TRIM37-mono-ubiquitinated residues in PARP1 (Supplementary Table 6). Overexpression of Flag-TRIM37, His-ubiqutin, HA-PARP1/wild-type (WT), or mutants in 293 T cells followed by immunoprecipitation with antibodies against HA revealed that TRIM37 interacted with and ubiquitinated PARP1 at K337, rather than with K438 (Fig. 4c). Moreover, K337 mono-ubiquitination caused by TRIM37 stabilized PARP1 (Fig. 4c). Furthermore, the function of T203 phosphorylation of TRIM37 was examined using 293 T cells co-transfected with Flag-TRIM37/WT or T203A (non-phosphorylatable) inactive mutant or T203E (phosphomimetic) constitutively active mutant, His-ubiqutin, and HA-PARP1. When compared with cells transfected with WT Flag-TRIM37, TRIM37 binding, ubiquitination, and stabilization of PARP1 were significantly decreased in cells transfected with the T203A mutant (Fig. 4d) and a significant increase in cells transfected with the T203E mutant (Fig. 4e) in immunoprecipitation assay. Furthermore, lentiviral-mediated KD of TRIM37 was performed to confirm that PARP1 acts downstream of TRIM37. TRIM37 KD using two independent shRNAs targeting TRIM37 resulted in decreased protein levels of PARP1 and full-length DNMT1, along with increased expression of senescence markers (Fig. 4f). This was accompanied by reduced cell proliferation (Fig. 4g) and impaired differentiation into both osteoblasts and adipocytes (Fig. 4h) compared to cells transduced with control shRNAs. To determine the functional importance of TRIM37 and its phosphorylation at T203, we performed rescue experiments in TRIM37-KD MSCs using shRNA-resistant constructs encoding wild-type (WT), T203A, or T203E TRIM37. Re-expression of WT or T203E TRIM37 restored PARP1 protein levels, reduced senescence marker expression (Supplementary Fig. 10a), and rescued both proliferation (Supplementary Fig. 10b) and differentiation capacities into osteoblasts and adipocytes (Supplementary Fig. 10c). In contrast, re-expression of the T203A mutant failed to restore these functions (Supplementary Fig. 10). These results indicate that phosphorylation at T203 is critical for TRIM37-mediated regulation of PARP1 expression and MSC function.
a Immunoprecipitation of PARP1 and the identification of phosphorylated proteins by tandem mass spectrometry in early MSCs. Interpretation of MS2 spectra revealed enriched phosphopeptides associated with PARP1. Phosphor-TRIM37T203 is indicated. Detailed information is in Supplementary Table 3. b Co-immunoprecipitation assay showed that the exogenous TRIM37 binds to PARP1 in 293 T cells cotransfected with the plasmids of Flag-TRIM37 and HA-PARP1. c Western blotting for the protein levels of immunoprecipitated HA-PARP1 in 293 T cells cotransfected with plasmids of Flag-TRIM37, His-ubiqutin (His-Ub), and HA-PARP1/wild-type (WT) or indicated mutants. The overexpression of HA-PARP1 with K337R mutation, rather than with K438R mutation, significantly reduced the immunoprecipitated HA-PARP1 and His-Ub protein levels compared to WT HA-PARP1. d, e Western blotting for the expression of Flag-TRIM37 bound to immunoprecipitated PHA-ARP1 in 293 T cells cotransfected with plasmids of HA-PARP1, His-Ub, and Flag-TRIM37/WT or indicated mutant. d The overexpression of Flag-TRIM37 with T203A mutation reduced Flag-TRIM37 binding to and ubiquitination of HA-PARP1 compared to WT Flag-TRIM37. eThe overexpression of Flag-TRIM37 with T203E mutation increased Flag-TRIM37 bind–ing to, and the ubiquitination and stabilization of HA-PARP1 compared to WT Flag-TRIM37. f–h Early MSCs were lentivirally transduced with scrambled (CTR) and TRIM37 shRNAs (TRIM37-KD #1 & #2), followed by (f) western blotting for the detection of PARP1, DNMT1, and senescence marker levels, (g) WST-1 assay for growth curve analysis, and (h) induction for osteogenic and adipogenic differentiation for 14 d. TRIM37 knockdown decreased the PARP1 and DNMT1 full-length protein levels, increased the senescence marker levels, inhibited the cell proliferation, and suppressed the differentiation of cells into osteoblasts and adipocytes. Results are expressed as the mean ± SD of three independent experiments. (*p < 0.05; **p < 0.01; ****p < 0.0001; ordinary one-way or two-way ANOVA). Exact p-values are shown in the Source Data file. Scale bar: 500 μm.
ATM upregulates TRIM37 protein levels and nuclear interaction with PARP1 via T203 phosphorylation
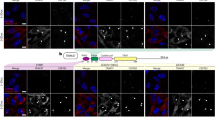
According to a previous study showing that ATM promotes nuclear translocation of TRIM37 by phosphorylation55, we examined the role of ATM in the nuclear localization of the TRIM37-PARP1 complex. Early MSCs had higher levels of Ser1981 phosphor-ATM (p-ATM), total ATM, and TRIM37 than late MSCs (Fig. 5a). Treatment of early MSCs with ATM inhibitor KU-55933 reduced TRIM37, PARP1 and full length DNMT1 levels (Fig. 5b), while treatment of middle-passage (p7-9) MSCs with ATM activator resveratrol increased TRIM37, PARP1 and full length DNMT1 levels (Fig. 5c). In esophageal cancer cells, activated ATM kinase directly interacts with and phosphorylates TRIM37 in the cytoplasm, which induces translocation of TRIM37 into the nucleus55. Therefore, we checked the location of p-ATM, TRIM37, and PARP1 using immunofluorescence (Fig. 5d, e) and analyzed their association using the PLA assay (Fig. 5f, g). Compared with late MSCs, early MSCs increased the nuclear localization of p-ATM, TRIM37, and PARP1 (Fig. 5d, e). PLA revealed an increased association of TRIM37 with p-ATM and PARP1 (mainly in the nucleus) in early MSCs compared with late MSCs (Fig. 5f, Upper panel). Quantification data further revealed that these proteins have increased nuclear binding in early MSCs compared to late MSCs (Fig. 5f, Lower panel). Moreover, treatment with the ATM activator resveratrol enhanced the nuclear binding of TRIM37 with p-ATM and PARP1 in middle-passage MSCs (Fig. 5g). Interestingly, overexpression of a dominant-negative T203A TRIM37 mutant in 293 T cells inhibited the nuclear translocation of exogenous TRIM37, while overexpression of a constitutively active T203E TRIM37 mutant promoted nuclear translocation of exogenous TRIM37 (Fig. 5h). These results indicate that ATM activation promotes the formation of the TRIM37/PARP1 complex and its nuclear translocation via phosphorylation of TRIM37 at T203.
a Western blotting showed that the total and phosphor-ATM (p-ATM) and TRIM37 levels increased in early MSCs. b, c Western blotting showed that ATM inhibition (b) with KU55933 (10 μM) for 2 h in early MSCs decreased the TRIM37, PARP1, and DNMT1 levels, while ATM activation (c) with resveratrol (5 μM) for 2 d in middle passage (p7-9) MSCs increased the TRIM37, PARP1 and DNMT1 levels. d, e Immunofluorescence showing increased nuclear localization of TRIM37 with (d) p-ATM or (e) PARP1 in early MSCs. f, upper panel Duolink proximity ligation assay (PLA) showed that the nuclear association of TRIM37 with p-ATM and PARP1 was increased in early MSCs compared to late MSCs. f, lower panel Quantification of the percentage of PLA nuclear-positive cells in early or late MSCs. g, upper panel Duolink PLA assay showed that ATM activation with resveratrol induced the nuclear association of TRIM37 with p-ATM and PARP1 in middle passage MSCs. g, lower panel Quantification of the percentages of PLA nuclear-positive cells in the vehicle control or resveratrol-treated MSCs. h The 293 T cells were overexpressed with wildtype (WT), T203A or T203E mutant Flag-TRIM37. Immunofluorescence with anti-Flag antibodies showed that the phosphorylation at T203 is required for the nuclear localization of TRIM37. Arrows indicate cells with nuclear translocation of specific proteins. Results are expressed as the mean ± SD of three independent experiments. (*p < 0.05; **p < 0.01; ****p < 0.0001; unpaired two-tailed t-test, or ordinary one-way ANOVA). Exact p-values are shown in the Source Data file. Scale bar: 20 μm.
ATM activator combined with TET activator maintains DNMT1 in full length and promotes MSC stemness
According to the above findings, the PARP1/TET1 complex maintains the 5hmC level of DNMT1, while the ATM/TRIM37 complex regulates the stability and nuclear localization of PARP1, thereby further promoting the proliferation and differentiation of early MSCs. We hypothesized that the combination of an ATM activator, resveratrol, and a TET activator, vitamin C, may improve the stemness of in vitro expanded MSCs. Compared with vehicle control and any single treatment, the combined treatment of MSCs increased growth rates in the logarithmic phase and the cumulative population-doubling level not only in medium supplemented with 10% fetal bovine serum (FBS) but also in serum-free medium containing 2% human platelet lysate (hPL) (Fig. 6a). Furthermore, compared with vehicle control and any single treatment, the combination treatment with resveratrol and vitamin C in medium containing 10% FBS and 2% hPL enhanced the differentiation potential of MSCs into osteoblasts (Fig. 6b) and adipocytes (Fig. 6c). As expected, combinational treatment with resveratrol and vitamin C also increased the DNMT1 full-length protein level and reduced senescence marker levels when MSCs were expanded to late passages (Fig. 6d). Together, these data suggest that the ATM/TRIM37 complex and PARP1/TET1 complex upregulate DNMT1 full-length protein levels, which is essential for preventing replicative senescence and maintaining early MSC properties.
Passage 4 MSCs were seeded at a density of 1000 cells/cm2 and expanded in a medium containing 10% fetal bovine serum (FBS) or 2% human platelet concentrate (hPL), supplemented with or without the ATM activator, resveratrol (RSV, 0.1 μM), or/and the TET activator, vitamin C (vit C, 250 μM), and subcultured for 7 d. The cells were reseeded at the same density in the same medium and subcultured on the same day. a Cumulative population doubling level (PDL) was calculated up to passage 16. b, c Cells at passage 11 (FBS) and 9 (hPL) were subjected to differentiation potential analysis. Cells were induced for (b) osteogenic and (c) adipogenic differentiation for 14 d, followed by ARS and Oil Red staining. The extracts of the dyes were subjected to optical measurements. b, right panel; c, right panel Quantification of optical measurements. The combination group, in a medium containing 10% FBS and 2% hPL, showed increased cumulative PDL and differentiation potential into osteoblasts and adipocytes. d Western blotting showed that the combination group exhibited an increase in DNMT1 full-length protein levels and a decrease in the senescence marker levels. Results are expressed as the mean ± SD of three independent experiments. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ordinary one-way ANOVA). Exact p-values are shown in the Source Data file. Scale bar: 500 μm.
The involvement of TRIM37 and PARP1 in MSC aging and function in vivo
To demonstrate the involvement of TRIM37 and PARP1 in MSC aging in vivo, BM-MSCs were isolated from B6 mice and minimally expanded and cell lysates were subjected to western blotting. BM-MSCs from mice less than 8 weeks increased in the levels of TRIM37, PARP1, PAR, total DNMT1, and decreased in the levels of senescence markers, p16 Ink4a and p21Waf1/Cip1, than those from mice around 6 to 10 months (Supplementary Fig. 11a, b). To further demonstrate the involvement of TRIM37 and PARP1 in MSC function in vivo, BM-MSCs isolated from 8 weeks of both WT and TRIM37- and PARP1-deficient mice were subjected to comparisons of senescence marker expression, growth capacity and differentiation potentials. Compared with BM-MSCs from WT mice, BM-MSCs from TRIM37- or PARP1-deficient mice decreased in the levels of TRIM37, PARP1, and total DNMT1, increased in the expression of senescence markers, such as p16Ink4a, p21Waf1/Cip1 (Supplementary Fig. 11c, d) and senescence β-galactosidase staining (Supplementary Fig. 11e, f), and decreased in proliferation capacity (Supplementary Fig. 11g, h) and differentiation potentials into osteoblasts and adipocytes (Supplementary Fig. 11i, j). These data suggest the involvement of TRIM37 and PARP1 in MSC aging and function in vivo.
TRIM37 and PARP1 affect MSC ability to maintain bone homeostasis and contribute to in vivo bone repair
To link MSC function with bone formation and bone mineral content maintenance, we assessed distal femur bone parameters in mice at 12 weeks and over 53 weeks of age. Old mice demonstrated a decrease in BV/TV, BMD, Trabecular No., and an increase in Trabecular separation (Tb. Sp) compared to their counterparts at a younger age (Fig. 7b). Notably, young Trim37-deficient mice exhibited results similar to the changes associated with aging when compared to young WT mice (Fig. 7c). To further elucidate the roles of TRIM37 and PARP1 in new bone ingrowth in adults following injury, a single cortical drill hole was created in the medial side of the right femoral diaphysis in WT, TRIM37-deficient, and PARP1-deficient mice at 8 weeks of age. This procedure was followed by an intraperitoneal injection of calcein three weeks later and single-photon (Fig. 7a, d, e) or two-photon fluorescence imaging four weeks post-injury (Supplementary Fig. 12). In the control sham group, no fluorescence was observed in the right femurs of WT and genetically modified mice (Supplementary Fig. 12). However, continuous fluorescence throughout the entire drill hole was evident in WT young mice that underwent drill-hole surgery (Fig. 7d, e, Supplementary Fig. 12). In contrast, in aged mice and those lacking TRIM37 or PARP1, diminished and non-continuous fluorescence was observed near the drill-hole area (Fig. 7d, e, Supplementary Fig. 12). These findings collectively indicate the potential role of TRIM37 and PARP1 in the aging process of MSCs, influencing their ability to maintain bone homeostasis and contribute to in vivo bone repair.
a Flowchart illustrating the experimental procedures. b,c, Micro-CT analysis conducted in the distal femur of mice at different stages: young (12 weeks), old (57 weeks), wild type (WT, 12 weeks), and TRIM37-deficient (TRIM37 KO, 12 weeks). Panel (b) depicts the morphological characteristics of trabecular bone in old mice, while panel (c) focuses on the comparison between WT and TRIM37 KO mice. (Lower panels) Quantification of micro-CT data on bone volume fraction (Bone volume (BV) / Total volume, %), bone mineral density (mg HA/ml), trabecular number (1/mm), and trabecular spacing (mm) (p values represent in number on top of each assessment as determined with unpaired t-test). d, e At 8 weeks of age, the medial cortex of the right femoral shafts was drilled in young versus old mice (d) and in WT versus TRIM37 KO mice (e). Three weeks later, calcein injection was performed, followed by fluorescence microscopy analysis of new bone formation another 1 week later. Representative fluorescent images from more than three independent experiments were captured using a single-photon microscope. Rectangular areas are magnified in the right. Arrow in (e) highlights the observed pinhole. Scale bar, 500 μm. n ≥ 3 biologically independent mice. Results are expressed as the mean ± SD of independent experiments. (*p < 0.05; **p < 0.01). Exact p-values are shown in the Source Data file.
The combination of an ATM activator and a TET activator promotes in vivo bone repair and exhibits anti-osteoporosis effects
Since ATM activator combined with TET activator maintains DNMT1 in full length and promotes MSC stemness, we therefore hypothesized that ATM activator combined with TET activator might rescue MSC osteogenic potential in vivo. We assessed left distal femur bone and fat parameters in 83-week-old female mice that had received daily treatment with resveratrol, vitamin C, or a combination of both starting at 75 weeks of age (Fig. 8a). Compared to the control and each of the single-treatment groups, the combination treatment group exhibited increased bone mineral density (BMD), bone volume fraction (BV/TV), trabecular number, and trabecular thickness, along with reduced bone surface-to-volume ratio (BS/BV) and trabecular separation (Tb. Sp) (Fig. 8b). However, fat content remained unchanged across all treatment groups (Supplementary Fig. 13). Following the creation of a single cortical drill hole on the medial side of the right femoral diaphysis at 79 weeks of age, calcein was administered intraperitoneally three weeks later, and single-photon fluorescence imaging was performed four weeks post-injury (Fig. 8c). Notably, the combination treatment group exhibited increased calcein fluorescence at the defect site in the right femoral shaft compared to the control and each single-treatment group, indicating enhanced bone repair capacity (Fig. 8c). Together, these findings suggest that the ATM/TRIM37 and PARP1/TET1 complexes cooperatively upregulate full-length DNMT1, which is critical for preventing replicative senescence, preserving early MSC characteristics in vitro, and promoting bone homeostasis, in vivo repair, and anti-osteoporotic effects.
a Flowchart illustrating the experimental procedures. b, c Micro-CT analysis conducted in the distal femur of female mice at different stages: control (CTR, 83 weeks), ATM activator, resveratrol (RSV, 10 mg/kg, daily), or/and the TET activator, vitamin C (vit C, 3.3 g/L in drinking water, daily) for 8 weeks. Panel a depicts the morphological characteristics of trabecular bone of combinational group in old female mice. (Lower panels) Quantification of micro-CT data on bone mineral density (mg HA/ml), bone volume fraction (Bone volume (BV) / Total volume, %), bone-specific surface (bone surface (BS)/BV), trabecular spacing (mm) trabecular number (1/mm), and trabecular thickness (Tb.Th). b At 79 weeks of age, the medial cortex of the right femoral shafts was drilled in old female mice. Three weeks later, calcein injection was performed, followed by fluorescence microscopy analysis of new bone formation another 1 week later. Representative fluorescent images from three independent experiments were captured using a single-photon microscope. Rectangular areas are magnified in the lower panels. Scale bar, 200 μm. Quantification of mean fluorescent intensity by Image J software was in lower panel. n = 3 biologically independent mice. Results are expressed as the mean ± SD of independent experiments. (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ordinary one-way ANOVA). Exact p-values are shown in the Source Data file.
Discussion
TET and 5hmC have been suggested to participate in the maintenance of pluripotency at regulatory elements in embryonic stem cells35 and somatic cell reprogramming36 and are required for HSC function and homeostasis38, and its dysregulation caused by mutations in TET enzymes also predisposes diseases38. However, information on their roles in expanded MSCs is limited. In the current study, we found that TET1-dependent increases in 5hmC levels are required for the maintenance of early human MSC properties, including proliferation and multi-differentiation potential. This study also supports a recent report that demonstrates that TET proteins play a critical role in maintaining mouse BM MSCs and bone homeostasis through demethylation of P2rX7 to control exosome and miRNA release40. It should be noted that the levels of 5hmC in human adipose stem cells56 were higher in age-advanced than in young individuals and correlated with population doubling time56, suggesting that the roles of 5hmC levels in aging and replication capacity of MSCs are text-dependent.
The decline in global 5hmC levels observed in aged MSCs is associated with reduced proliferation and diminished osteogenic/adipogenic differentiation potential. Mechanistically, PARP1 recruits TET1 to hydrolyze methylated nucleotides on DNMT1 exons, assisting CTCF in preventing DNMT1 alternative splicing. Notably, DNMT1 plays a crucial role in maintaining MSC stem cell properties24. Recent research supports these findings, revealing decreased chromatin accessibility and lower histone acetylation in aged MSCs1. The root cause is impaired mitochondrial acetyl-CoA export due to lower citrate carrier levels1. Both histone acetylation and elevated 5hmC levels, leading to DNA demethylation, contribute to chromatid access, playing a critical role in regulating MSC properties. This underscores the significance of chromatid accessibility in maintaining stemness and suggests potential therapeutic strategies for conditions such as osteoporosis. Additionally, it establishes an age-dependent connection between chromatin, DNA modification, and cell fate, mediated by citrate carrier or DDR, and epigenetic modulation factors.
PARP1 and TET2 are required for the early establishment of histone modifications that typify an activated chromatin state at pluripotency loci, whereas PARP1 induction further promotes accessibility to the Oct4 reprogramming factor36. We also demonstrated that both PARP1 and TET1 are required for early MSC properties. KD of either PARP1 or TET1 in early MSCs induces a switch of the transcription profile into late MSCs with a loss in proliferation and multi-differentiation potential and an increase in the expression of senescence markers. In early MSCs, PARP1 binds to the TSS sites, thereby promoting accessibility of PARP1-bound TET1 to the gene body of DNMT1, where TET1 generates 5hmC, thereby reducing DNA methylation, allowing CTCF to access the exons and prevent alternative splicing. Taken together, these findings suggest that PARP1 and TET proteins not only contribute to an epigenetic program that directs subsequent transcriptional induction at pluripotency loci during somatic cell reprogramming36, but also play an important role in the maintenance of adult stem cell function and homeostasis.
DDR, including core DNA repair and associated pathways, is essential for maintaining the integrity of the cell genome, and its destruction is one of the hallmarks of cancer57. Traditionally, defects in genome stability mechanisms have been used to treat cancer through radiotherapy or genotoxic chemotherapy. Recently, DDR protein components have been identified as promising candidates for targeted cancer therapy58. For example, olaparib (Lynparza™; AZD2281) is a potent PARP inhibitor used to treat tumors with BRCA1 or BRCA2 mutations59. It has recently been approved and represents the first drug based on this principle, exploiting an underlying cause of tumor formation that also represents an Achilles’ heel. The DDR proteins against which anti-cancer drugs were tested in vitro or in vivo for efficacy include PARP1, ATM, and DNMT158. Prolonged DDR signaling, convergent p53 activation, and subsequent cell cycle arrest trigger replicative senescence, while inhibition of DDR signaling allows senescent cells to re-enter the cell cycle60. However, whether these DDR proteins also play an important role in preventing the aging of senescent cells, and whether related mechanisms can be considered as opportunities for anti-aging treatments is rarely discussed60. PARP1 and ATM recognize and bind to both single-and double-strand DNA breaks in response to different DNA damage triggers61. Early embryonic lethality in PARP1 and ATM double-mutant mice suggests a functional synergy between these two DDR proteins in cell proliferation during embryonic development62. Compared with late MSCs, we found that early MSCs increased PARP1 protein levels, which was mediated by activated ATM that phosphorylates TRIM37 and induces translocation into the nucleus, where TRIM37 monoubiquitinates and stabilizes PARP1. Lentiviral transduction of shRNAs against TRIM37 and CTCF in early MSCs induces senescence and the loss of proliferation and multi-differentiation. These data suggest that DDR proteins, such as PARP1, ATM, and DNMT1, and related mechanisms can be considered novel therapeutic opportunities for anti-aging.
TRIM37 is an E3 ubiquitin ligase that has been reported to T196 and S801 phosphorylated sites by ATM kinase55, but no other functional phosphorylated site has been reported. We found that mutating the TRIM37 T203 site to alanine or glutamate regulates TRIM37/PARP1 interaction and nuclear translocation. Furthermore, TRIM37-directed PARP1 monoubiqutination as well as histone H2A monoubiquitination54 enforces changes in gene expression and DNA repair that render multiple tumors resistant to chemotherapy that trigger ATM signaling to transcriptionally upregulate TRIM3763,64,65. On the other hand, TRIM37 is highly expressed in triple-negative breast cancer (TNBC) under chemotherapeutic stress63, as well as in MCF7 breast cancer and neuroblastoma, where it inhibits PLK4 self-assembly to the centrosome, stabilizes the centrosomal protein CEP19266,67, and promotes peroxisomal matrix protein import by stabilizing the peroxisomal receptor PEX568. Together, these data suggest that different phosphorylation sites of TRIM37 are required for its different E3 ubiquitin ligase functions in different stem cells or cancer cells.
Our findings provide mechanistic insight into how TRIM37 phosphorylation at T203 modulates its subcellular localization and function in MSCs. We show that T203 phosphorylation enhances TRIM37 nuclear localization and promotes short-term MSC proliferation, with the phosphomimetic T203E mutant exhibiting the greatest effect. Interestingly, this phosphorylation appears dispensable for TRIM37-mediated PARP1 stabilization and differentiation potential, as both wild-type and T203E TRIM37 restored these functions in TRIM37 KD MSCs. These results suggest that TRIM37’s nuclear relocalization and its functional outputs may be governed by distinct regulatory mechanisms. Specifically, T203 phosphorylation is critical for proliferation-related activities, while PARP1 stabilization and differentiation may depend on other regions or modifications of TRIM37. This functional uncoupling underscores the complexity of TRIM37’s role in MSC biology and raises the possibility that additional post-translational modifications or co-factors modulate its specificity in proliferation versus differentiation contexts. Future studies dissecting these pathways may provide further clarity on how TRIM37 integrates multiple signals to regulate stem cell fate and bone homeostasis.
For future expansion of clinical-grade MSCs for clinical use, we demonstrate that simultaneous treatment of MSCs in FBS-containing and defined serum-free medium with an ATM activator, such as hydroxychloroquine or resveratrol and a TET activator, such as vitamin C69, inhibits replicative senescence and maintains early MSC properties. Since MSCs isolated from BM have many clinical applications, and senescence-associated epigenomic changes, such as reduced 5hmC levels, identified in this study, could be applied to MSCs derived from other sources, and may be associated with related functional defects. Therefore, expansion of MSCs in growth medium containing ATM and TET activators should be considered in future clinical settings.
In our study, we assessed bone parameters and long bone repair capacity in young and old mice, extending this assessment to WT and TRIM37-deficient mice. Notably, young TRIM37-deficient mice exhibited results similar to the changes associated with aging when compared to young WT mice. Through these animal studies, we have successfully established a crucial link between in vitro MSC senescence, in vivo bone formation, and the maintenance of bone mineral content. The pivotal roles played by TRIM37 and PARP1 in these processes underscore their significance. Our findings present an exciting prospect for clinical applications, suggesting that targeted interventions aimed at TRIM37 and PARP1 could offer promising strategies to mitigate osteoporosis and enhance bone repair.
This study analyzed MSCs from three human donors who were not strictly age- or gender-matched, which may contribute to inter-donor variability. Despite this, key findings—such as transcriptomic shifts in TET1- and PARP1-KD MSCs resembling late-passage MSCs—were consistent across donors. Notably, Donor 1 samples, including early, late, and TET1- and PARP1-KD MSCs, were sequenced in the same flow cell, minimizing batch effects and reinforcing the biological relevance of the clustering patterns. While Donor 3 displayed a distinct gene expression profile, likely related to older age, this variation highlights the natural heterogeneity in human MSCs. Nevertheless, future studies with larger, age- and gender-matched donor cohorts will be essential to validate and extend these results.
In conclusion, our study establishes a crucial link between in vitro MSC senescence and in vivo bone formation, unraveling the potential involvement of TRIM37 and PARP1 in the aging of MSCs and their capacity to facilitate bone repair. These findings not only deepen our understanding of MSC dynamics but also pave the way for innovative therapeutic strategies, particularly in the realm of age-related osteoporosis. The stabilization of PARP1 by TRIM37 introduces a promising avenue for targeted interventions, offering hope for mitigating osteoporosis and advancing regenerative medicine.
Methods
Cell culture and reagent treatment
BM aspirates were collected from patients who received an orthopedic surgery in Taipei Veterans General Hospital with the approval of Institute of Review Board (#2012-05-007 A). Informed consent was obtained from all subjects. Details regarding the donor information are shown in Supplementary Table 7. The protocols of MSC isolation and expansion were modified from previously described methods25. In brief, mononuclear cells were isolated from heparinized BM by density gradient centrifugation using Ficoll-Hypaque (Sigma-Aldrich, St. Louis, MO) of a density of 1.077 g/L at 400 x g, followed by seeding with complete medium [CM: Dulbecco’s Modified Eagle Medium (DMEM; Gibco-BRL, Gaithersburg, MD), supplemented with 10% fetal bovine serum (FBS, Gibco-BRL), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine (Invitrogen, Carlsbad, CA)] at 37 °C under 5% CO2 atmosphere. Upon reaching semi-confluence, MSCs were retrieved and reseeded at an initial density of 4 × 103 cells/cm2 and the culture medium was changed twice per week. Mycoplasma contamination was checked every 1 month. Cells with mycoplasma contamination were discarded. For treatment with 2-hydroxyglutarate (2-HG, HY-100542, MedChem Express, Monmouth Junction, NJ), cells were cultured with 5 mM 2-HG in CM. For treatment with KU55933 (SML1109, Sigma-Aldrich, St. Louis, MO), cells were cultured with 10 μM KU55933 in CM. For treatment with resveratrol (R5010, Sigma-Aldrich), cells were treated with 5 μM resveratrol in CM. For treatment with vitamin C (A4544, Sigma-Aldrich), cells were cultured with 50 μg/ml vitamin C in CM. Cells were harvested and stained with Trypan Blue (Sigma-Aldrich) for counting cell numbers at each passage. Cumulative population doubling level (PDL) was calculated using (logN–logN0)/log2, where N = cell number harvested during passaging and N0 = cell number at initial seeding.
Senescence-associated β-Galactosidase (SA-β-gal) staining
SA-β-gal staining was performed using a β-galactosidase staining kit (BioVision, Milpitas, CA) according to the manufacturer’s instructions. Briefly, cells were washed with PBS and fixed with 3.7% formaldehyde for 5 min at room temperature. After washing, the cells were incubated with β-gal staining solution (1 mg/mL, 5-bromo-4-chloro-3-indolyl-b-galactoside) overnight at 37 °C. SA-β-gal-positive cells were observed and acquired at room temperature using an inverted microscope (Nikon Eclipse TE2000, Tokyo, Japan).
Proliferation assay
For proliferation assays, cells were plated in triplicate in 96-well tissue culture plates with growth media. At each time point, cells were seeded at a density of 1000 cells/well in 96-well plate, followed by recover and assay with a water-soluble tetrazolium salt, WST-1, 4-[3-(4iodophenyl)−2-(4-nitrophenyl)−2H-5-tetrazolio]−1,3-benzene disulfonate), kit (Cayman Chemical Company, Ann Arbor, MI) at indicated time periods. Absorbance was measured at a wavelength of 450 nm.
Cell differentiation protocols
Cell differentiation was performed as previously described25. Before the initiation of differentiation, cells were seeded in CM at a density of 104 cells/cm2. For differentiation into osteoblasts and adipocytes, cells were induced in osteogenic induction medium [OIM: CM supplemented with 10−8 M dexamethasone, 50 µg/ml ascorbic acid-2 phosphate, 10 mM β-glycerophosphate (Sigma-Aldrich)] and adipogenic induction medium [AIM: CM supplemented with 50 mg/ml ascorbate-2 phosphate (Sigma), 10−7 M dexamethasone (Sigma-Aldrich), 50 mg/ml indomethacin (Sigma-Aldrich), and 10 mg/ml insulin (Sigma-Aldrich)], respectively. After induction in defined induction medium for 14 days, cells in OIM and AIM were stained with Alizarin red staining (ARS) and Oil Red O, respectively, followed by extraction and measurement of O.D. values of ARS at 550 nm and Oil Red O at 510 nm.
Quantitative analysis of 5hmC levels using dot blot
The procedure of dot-blot was modified from previously described method70. Briefly, genomic DNA samples were prepared in 2 N NaOH and 10 mM Tris-HCl, pH 8.5. The samples were spotted onto Hybond-N+ nylon membrane (GE Healthcare, Chicago, Illinois) using a 96-well dot-blot apparatus (Bio-Rad, Hercules, CA), baked in 80 °C for 30 min, blocked with 5% skim milk for 1 h at room temperature, and incubated with anti-5-hydroxymethylcytosine antibody (39791, Active Motif, Carlsbad, CA) or anti-5-methylcytosine antibody (BI-MECY-0500, Eurogentec, Seraing, Belgium) at 4 °C overnight, followed by incubation with species-specific HRP-conjugated secondary antibody, and dot signal was visualized with the ECL Plus chemiluminescence assay kit (GE Healthcare, Chicago, Illinois).
Genomic DNA preparation
Early-passage (P2-P3) and late-passage (P10-P11) MSCs were treated with RNase cocktail (Roche Diagnosis, Basel, Switzerland) for 5 min and subsequently incubated in 0.5% SDS and Proteinase K (100 ng/ul) for 2 h at 55 °C. Genomic DNA was extracted with phenol:chloroform: isoamyl alcohol (25:24:1) and then chloroform using phase lock gel, followed by ethanol/NaCl precipitation overnight at −20˚C and two washes with 70% ethanol. The DNA pellets were resuspended in 30μl nuclease-free MilliQ water, and the supernatant was transferred to low-binding tubes. Microplate Spectrophotometer (Multiskan™ GO, Thermo Fisher Scientific, Waltham, MA), and μDrop™Plate (Thermo Fisher Scientific) were used to quantify DNA concentration.
RNA-seq and transcriptomic data analysis
RNA-seq experiments and data analysis were performed as previously described25. In brief, total RNA was isolated from three donors with the RNeasy Micro Kit (QIAGEN, Hilden, Germany). Illumina TruSeq RNA sample Prep kit (Illumina, San Diego, CA) was used to prepare RNA-Seq library. RNA-Seq experiments of early MSCs, late MSCs, TET-KD early-MSCs and PARP1-KD early-MSCs were performed for all three donors. More than 30 million (mean ± standard deviation= 36,146,115 ± 2,891,116) 100 bp paired-end reads for each RNA-Seq sample were generated using an Illumina HiSeq 2000 sequencer in National Center for Genome Medicine, Academia Sinica, Taiwan. Sequencing bases with low quality (Phred + 33 quality score <20) were trimmed using the TrimGalore program (Babraham Bioinformatics, Cambridge, UK) from the 5’- and 3’- ends of reads. After trimming, reads shorter than 50 bp were discarded. Then, low complexity reads (repeat sequences) were removed using the prinSeq program. The human genome assembly GRCh37/hg19 and NCBI RefSeq Genes were downloaded from the University of California Santa Cruz (UCSC) Genome Browser for genome mapping and gene annotation. Genome mapping was performed using Tophat (v2.0.10) with Bowtie (v.2.0.6.0) and Samtools (v. 0.1.18.0). Cufflinks with GFF was used for transcript identification and counting. Quantification was done at gene-level. Normalization of gene expression level in individual sample was done by transforming count number to Fragments Per Kilobase of transcript per Million mapped read (FPKM). A total of 13326 genes with FPKM > 0 in at least half of the samples and with an average FPKM ≥ 1. were selected for unsupervised hierarchical clustering of the genes and samples. Z-score was calculated for each gene across samples and presented in the heat map. Paired sample t-test was performed to compare the early MSCs to the late MSCs. Due to the small sample size, very few genes reached the genome-wide significance. Therefore, genes with differential expression in the early MSCs compared to the late MSCs were selected according to fold change. Differentially expressed genes should fulfilled the following two criteria: (1) mean fold change larger than or equal to 2 or smaller than or equal to −2, and (2) the three pairs of MSCs showed the same direction of fold change. A total of 1026 genes were identified. Activation or suppression of biological pathways by the altered expression of the 1026 genes was predicted using Ingenuity Pathway Analysis (IPA) (QIAGEN Inc) and quantified using the Activation Z-score metric. The enrichment of altered genes in biological pathways was calculated using Fisher Exact test with IPA. Only the pathways with P-value smaller than 0.05 for all the three donors are considered significant. RNA-seq raw data are accessible at NCBI GEO with accession number GSE178804. All relevant data are available from the authors with restrictions.
Alternative splicing analysis using rMATS
Differential alternative splicing (AS) events were identified between the two sample groups using rMATS version 4.1.1 (https://csibioinfo.nus.edu.sg/csingsportal/login/home.php)50, which detects five major types of AS events from RNA-Seq data (GSE178804) with replicates71. In each analysis, the experimental groups (late, PARP1KD or TET1KD MSCs) were compared to the reference group (early MSCs) to identify differentially spliced events with an associated change in the percent spliced in (ΔPSI) of these events. We computed P values of splicing events with a cutoff <0.05 and then collected the splicing events with a |ΔPSI| of > 0.2. When the ΔPSI is a positive value, the number of AS events in the experimental group is more than the reference group.
5mC/5hmC DIP and high-throughput sequencing analysis
Aliquots of purified genomic DNA from early and late MSCs were first sonicated to ∼250 bp using Bioruptor (Diagenode, Seraing, Belgium), then end-repaired and ligated to Illumina PE adaptors using NEB Next DNA Library Prep Master Mix Set (New England Biolabs, Beverly, MA). In DIP assays, 10 μg of the adaptor-ligated genomic DNA was used as input, and 5 μl of anti-5-methylcytosine antibody (BI-MECY-0500, Eurogentec, Seraing, Belgium) or 5 μl of 5hmC antibody (39791, Active Motif, Carlsbad, CA) was added to immunoprecipitate modified DNA. Each of the immunoprecipitated DNA was amplified with forward primer and reverse primers in a 50 μl PCR reaction with 0.2× Sybr-Green I and 1× Phusion High-fidelity PCR Master Mix (New England Biolabs, Beverly, MA). PCR products that contained barcodes were sequenced after the standard first read using N2IndSeq primer. Raw data are accessible at NCBI GEO with accession number GSE178805. All relevant data are available from the authors with restrictions.
TrimGalore program (Babraham Bioinformatics, Cambridge, UK) was applied to clean the raw sequences to increase the quality of sequences for downstream analysis. After trimming, the sequence length of each read ranged from 36 bases to 101 bases. Interestingly, per sequence GC content of input control samples were generally all right, but the samples with methylated DNA immunoprecipitation (MeDIP) were tagged either warning or failed. This result suggested that the IP worked since the GC content was biased due to enrichment of methylated DNA fragments. The read sequences were aligned to human genome reference GRCh37/hg19 using Bowtie (v.2.0.10). Any read with more than two mapped ends were filtered out. The bam. files of MeDIP samples and input control samples were imported into PARTEK software for identification of methylated DNA peaks enriched in late passages of MSC. Data from late MSCs input was used as baseline to identify peaks enriched in samples with methylated DNA immunoprecipitation treatment. Firstly, sequencing peaks of each sample were detected by setting maximum fragment size at 200 bases with a widow size of 100 bases. Overlapping DNA fragments were merged. The peak cut-off FDR was set at 0.01. Then enriched peaks in late MSCs were identified by comparing sequencing peaks detected in late MSCs to paired early MSCs. DNA peaks whose q value is smaller than or equals to 0.001 and scaled fold change is larger than or equals to 4 were selected. The overrepresented peaks in late MSCs were annotated according to gene sections. Promoter region was defined as 5000 bp upstream of transcription start site (TSS, the first nucleotide of an mRNA according to RefSeq database) or 3000 bp downstream of TSS. By performing finding the nearest genomic features, 7633 genes were found to be hypermethylated at late-passage compared to early -passage of T7 MSC line.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde. For 5hmC and 5mC detection, cells were treated with 2 N HCl for 30 min at 37 °C, incubated with an anti-5hmC or anti-5mC monoclonal antibody, followed by incubation with a species-specific DyLightTM 488-conjugated secondary antibody (GTX213111-04, GeneTex, Inc., Irvine, CA), and counterstained with DAPI. For TRIM37 immunofluorescence, we used a mouse antibody against TRIM37 (sc-515044, Santa Cruz Biotechnology, Santa Cruz, CA) and reacted with a corresponding species-specific DyLightTM 488-conjugated secondary antibody. For pSer1981-ATM, PARP1 and FLAG-tag immunofluorescence, we used rabbit antibodies against pSer1981-ATM (GTX132146, GeneTex), PARP1 (#9532, Cell Signaling Technology, Danvers, MA) and FLAG-tag (GTX115043, GeneTex) and reacted with corresponding species-specific DyLightTM 594-conjugated secondary antibodies (GTX213110-05, GeneTex). Cells were counterstained with DAPI. Immunofluorescence was observed with a Leica TCS SP8 (Wetzlar, Germany) confocal system.
Proximity ligation assay
Cells were fixed with 4% formaldehyde in PBS for 10 min. Cells were then permeabilized and blocked against nonspecific binding with 0.1% Triton X-100 and 10% BSA in PBS for 1 h at room temperature. Primary antibodies, included anti-TRIM37, anti-pSer1981-ATM and anti-PARP1 antibodies, were then incubated with cells in 1% BSA in PBS at the indicated concentrations for 2 h at room temperature. Following the manufacturer-recommended procedure of Duolink® In Situ Red Starter Kit Mouse/Rabbit Kit (DUO92101-1 KT, Sigma-Aldrich), cells were counterstained with DAPI and in situ protein-protein association was analyzed with a Leica TCS SP8 confocal system.
Lentiviral vector production and cell infection
The expression plasmids and the bacterial clones for PARP1 shRNA (TRCN0000007932), TET1 shRNA (TRCN0000075026), TET2 shRNA (TRCN0000144344), CTCF shRNA (TRCN0000218498 and TRCN0000230191), and TRIM37 shRNA [TRCN0000221060, TRCN0000370523 and TRCN0000370461 (3’UTR)] were provided by the RNAi core of the National Science Council in Taiwan. Lentiviral transfer vector pLAS2w.Ppuro carrying PARP1 (NM_001618.4) was used to generate stable PARP1 expression cells and lentiviral transfer vector (LV038, Applied Biological Materials Inc.) carrying wild-type TRIM37 or mutants (T203A or T203E) was used to generate stable TRIM37 expression cells. Lentiviral production was performed by transfection of 293 T cells using Lipofectamine 3000 (L3000001, Invitrogen, Carlsbad, CA). Supernatants were collected 48 h after transfection and then were filtered. Subconfluent cells were infected with lentivirus in the presence of 100 μg/ml protamine sulfate (P4020, Sigma-Aldrich). At 24 h post-infection, we use puromycin (2 μg/ml) to select for infected cells for 48 h.
Western blotting
Cell extracts were prepared with M-PER (Pierce, Rockford, IL) or Subcellular Protein Fractionation Kit (Thermo Scientific, Waltham, MA) plus protease inhibitor cocktail (Halt™; Pierce, Rockford, IL) and protein concentrations were determined using the BCA assay (Pierce, Rockford, IL). Aliquots of protein lysates were separated on SDS–8 or 15% polyacrylamide gels and transferred to PVDF membrane filters, which were blocked with 5% blotting grade milk (Bio-Rad, Hercules, CA) in TBST (20 mM Tris-HCl [pH 7.6], 137 mM NaCl, 0.1% Tween 20) for 1 h. The filters were then incubated 1 h at room temperature with a 1:1000 dilution in TBST of antibodies against TET1 (GTX627420, GeneTex), TET2 (ab94580, Abcam, Cambridge, UK), PARP1 (#9532, Cell Signaling Technology), DNMT1 (#5032, Cell Signaling Technology, Danvers, MA), pSer1981-ATM (GTX132146, Gene Tex), TRIM37 (GTX114565, GeneTex), p16Ink4a (#92803, Cell Signaling Technology, Danvers, MA), p21Waf1/Cip1 (sc-6246, Santa Cruz Biotechnology), CTCF (GTX65926), HA tag (GTX115044), His tag (GTX628914), and Flag tag (GTX115043, GeneTex), reacted with corresponding secondary antibodies, and detected using a chemiluminescence assay (Millipore, Billerica, MA). The uncropped scans of the most important blots are shown in the Source Data file.
Co-immunoprecipitation assay
Cell extracts were prepared with Pierce IP lysis buffer (Thermo Scientific, Waltham, MA) plus protease inhibitor cocktail (Halt™; Pierce, Rockford, IL) and protein concentrations were determined using the BCA assay (Pierce, Rockford, IL). For TET1 and PARP1, the protein complexes were isolated by Dynabeads Protein G Immunoprecipitation Kit (Thermo Scientific, Waltham, MA). Anti-TET1 (GTX627420, GeneTex) or anti-PARP1 (#9532, Cell Signaling Technology, Danvers, MA) antibodies were crosslinked with protein G-magnetic beads by DSP (Proteochem, Inc., Denver, Colo.). The cell extracts were incubated with antibody-cross-linked protein G-magnetic beads for 30 min at room temperature and were collected by magnetic stand. For HA tag, cell extracts were incubated with Pierce Anti-HA Magnetic Beads (Thermo Scientific, Waltham, MA) for 30 min at room temperature and were collected by magnetic stand. The sample components were electrophoretically separated on SDS-PAGE for western blot analysis.
Phosphopeptides analysis by mass spectrometry
The TRIM37 or PARP1 phosphopeptides in anti-PARP1 immunoprecipitation from Early MSCs were performed in-gel digestion and detected on a nano-flow high-performance liquid chromatography system connected to an Orbitrap Q Exactive mass spectrometer. Each peptide fraction was autosampled and separated on a 15 cm analytical column (75 μm inner diameter) packed with 3 μm C18 beads with a 2 h gradient ranging from 5%–40% acetonitrile in 0.5% acetic acid at a flow rate of 250 nl/min. The Q Exactive mass spectrometer was operated in data-dependent acquisition mode, and all samples were analyzed with a “sensitive” acquisition method. All raw data analysis was performed with MaxQuant version 1.3.0.5 supported by the Andromeda peptide search engine.
Plasmid construction and cell infection
The pcDNA3-HA-PARP1, pcDNA3.1-Flag-TRIM37 and pcDNA3-His-Ubiquitin were gifts from Dr. Sheau-Yann Shieh72, Dr. Ping-Hui Tseng, and Heng-Hsiung Wu73, respectively. Human DNMT1-His (NM_001379.4) and alternative splicing DNMT1-His (3203-4521 deletion) constructs were generated by cloning specific coding sequences into the pcDNA3.1/V5-His vector (K4800-01, Invitrogen, Life Technologies, CA). Mutation of K337 or K438 on pcDNA3-HA-PARP1 and mutation of T203 on pcDNA3.1-Flag-TRIM37 were generated by QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) using the following primers (Supplementary Table 8). Transient infection was performed by transfection of 293 T human cell line using Lipofectamine 3000 (L3000001, Invitrogen, Carlsbad, CA). For primary MSCs, transient infection was performed by Neon transfection system (Invitrogen, Life Technologies, Carlsbad, CA).
DNMT1 activity assay
Full-length and alternative splicing form DNMT1 proteins generated in 293 T cells were purified by Ni Sepharose 6 Fast Flow (GE Healthcare, Chicago, Illinois). The DNMT1 activity was determined with the EpiQuikTM DNA Methyltransferase Activity/Inhibition Assay Kit (Epigentek Inc., New York, NY) with a microplate reader at 450 nm. DNMT1 activity was calculated as OD/h/μg of protein.
Chromatin immunoprecipitation (ChIP) assay
To demonstrate the binding of PARP1, TET1 and CTCF protein to genomic DNA, the ChIP assay was performed using a commercial kit (Pierce, Rockford, IL) according to the manufacturer’s protocol with minor adjustments. The MSCs were grown to confluence, crosslinked with 1% formaldehyde for 8 min at room temperature, washed in ice-cold PBS containing protease inhibitors, then lysed on ice for 10 min in M-PER Extraction Reagent (Thermo Scientific, Waltham, MA) with phosphatase and protease inhibitors. DNA-protein complexes were sonicated to 200 and 500 base pairs. One aliquot of the soluble chromatin was stored at 20 °C for use as input DNA, and the remainder was diluted 10 times in Pierce IP buffer (Thermo Scientific, Waltham, MA) containing phosphatase and protease inhibitors, and incubated overnight (4 °C) with anti-PARP1 (#9532, Cell Signaling Technology, Danvers, MA), anti-TET1 (GTX627420, GeneTex) or anti-CTCF (GTX65930, GeneTex) antibodies. DNA-protein complexes were eluted with IP elution buffer. Cross-linking was reversed by incubation at 65 °C for overnight. Proteins were removed with proteinase K, and DNA was extracted with phenol/chloroform, redissolved, and PCR-amplified with specific primers for DNMT1 exon 30, 34 and 37 (Supplementary Table 9).
ChIP-seq and data analysis
The immunoprecipitated DNA and input DNA were also subjected to next-generation sequencing using an Illumina HiSeq 2000 sequencer. The ChIP assay was performed using the Millipore chromatin immunoprecipitation (ChIP) assay kit (17-295, Millipore). The library preparation and sequencing procedure was performed following the manufacture’s instruction. After base-calling, trimming, and quality control, the high-quality reads (Phred + 33 quality score > 20) were aligned to the human genome reference (GRCh37/hg19) using Bowtie (v.2.0.6.0) and Samtools (v. 0.1.18.0). Bam files of T7E_INPUT, T7L_INPUT, T7_PARP1_ChIP, and T7_TET1_ChIP were imported into Partek software. The alignments per read of each sample past quality assurance and quality control. Sequencing peaks of each sample were detected by setting the maximum fragment size at 350bp with a window size of 100bp. Overlapping fragments were merged. The peak cut-off FDR was set at 0.001. Enriched PARP1 or TET1 binding sites were identified by comparing sequencing peaks detected in T7E_PARP1_ChIP or T7E_TET1_ChIP to T7E_INPUT. DNA segments with qvalue smaller or equals to 0.01 and scaled fold change larger than or equals to 2 were selected. Additional filter criterion was non-overlapping between the selected segments with peaks detected in T7L_INPUT. A total of 249 peaks were specified to PARP1-ChIP and 960 peaks to TET1-ChIP. RefSeq transcripts database was used for annotation of the identified peak fragments. Promoter region was defined as 2000 bp upstream of transcription start site (TSS) or 1000 bp downstream of TSS. By performing finding the nearest genomic features, the 249 PARP1 binding peaks were co-localized with 117 genes and the 960 TET1 binding peaks were co-localized with 466 genes. The binding of PARP1 and TET1 to the promoter region, including TSS of the DNMT gene was enriched in early MSCs.
5’-Race PCR
Total RNA was extracted from early and late MSC using a RNeasy Micro Kit (QIAGEN) and was used for the first-strand cDNA synthesis by a SMARTer RACE 5’/3’Kit (Takara Bio., Shiga, Japan). The first-strand reaction products in Tricine-EDTA buffer were stored at −80 °C as the PCR templates. According to the cDNA sequences of DNMT1 (NM_001379.4), we designed gene-specific primers in Supplementary Table 9.
Animals and mouse BM MSC isolation
All animal experiments were conducted in accordance with the guidelines of the Animal Care Committee of China Medical University and the protocol was approved by Animal Care Committee of China Medical University (CMUIACUC-2019-374-1 and CMUIACUC- 2021-374-1). All mice used in this study were on the C57BL/6 background. Male mice were used exclusively to avoid the influence of sex hormones on bone parameter assessment, except for experiments in Fig. 8 and Supplementary Fig. 13, in which female mice were used because osteoporosis models commonly employ females. The age of the animals used in each experiment is indicated in the corresponding figure legends. The strains used were as follows: C57BL/6-PARP1-deficient mice (129S-Parp1tm1Zqw/J)74 and their wild-type littermates (C57BL/6 background) obtained from The Jackson Laboratory (Bar Harbor, ME); and C57BL/6-Trim37em1(flox)Smoc and C57BL/6-Tg(Pgk1-RFP, -cre/ERT2)3Narl (RMRC13179) mice, which were kindly provided by Prof. Ping-Hui Tseng (National Yang Ming Chiao Tung University) and purchased from the National Laboratory Animal Center (Taipei, Taiwan). TRIM37-deficient mice were generated by cross-breeding C57BL/6-Trim37em1(flox)Smoc with C57BL/6-Tg(Pgk1-RFP, -cre/ERT2)3Narl mice. Mice were maintained under specific pathogen-free (SPF) conditions at 22 ± 2 °C with a 12 h light/dark cycle and ad libitum access to food and water. Health and welfare were monitored daily by trained animal care staff, and euthanasia was performed by CO₂ inhalation followed by cervical dislocation, in accordance with institutional and international guidelines.
For mouse BM MSC isolation, intact femurs and humeri were dissected carefully from the mice. The bones were cut at both ends, then a 26-gauge needle was inserted at the exposed end of the bones and the marrow plug was flushed out by 1% BSA in PBS. Cell suspension was collected through a 70-mm filter mesh to remove any cell clumps. MSCs were isolated and expanded with protocols described previously75. In brief, BM cells were cultured in 100-mm culture dishes with complete alpha MEM medium (with 16.6% FBS) and incubated at 37 °C with 5% CO2 in a humidified chamber. After 12 h, the nonadherent cells were removed on the surface of the dish by changing the medium. Thereafter, this step was repeated every 24 h for up to 72 h of initial culture. Within 4–8 days, the culture becomes more confluent and distinct fibroblastic cells reaches 65–70% confluence within 2 weeks. After 2 weeks of initiating culture, the culture was trypsinized with 0.05% trypsin/1 mM ethylenediaminetetraacetic acid for 2 min at 37 °C. The trypsin was neutralized by adding 5 ml of complete medium, and culture in a 100-mm dish. The culture medium was changed every 3 days. Typically, cell confluence is achieved in 7 days.
Single-photon or two-photon evaluation of new bone growth in right femoral shaft drilling hole
TRIM37 deficiency was induced in mice at 6 weeks of age through intraperitoneal injection of 2 mg of tamoxifen (diluted in 100 μl of 20 mg/ml corn oil from Sigma-Aldrich) administered daily for a consecutive 5-day period. At 8 weeks of age, both young WT mice and mice deficient in TRIM37 or PARP1, as well as older mice (53 weeks), were anesthetized using 2.5–5% isoflurane in the presence of oxygen. A longitudinal incision, measuring 1 cm in size, was performed in the mid-portion of the right medial thigh of the mouse. Soft tissue retraction was accomplished using small retractors. A single medial cortical drill hole, created with a 1-mm pin, was carefully executed to avoid damage to the opposite cortex. Subsequently, the drill hole was rinsed with PBS, and the wound was closed using sutures. The animal was left move freely in the cage. Intraperitoneal injection of calcein (20 mg/kg) was performed 3 weeks later. Fluorescence microscopic evaluation was performed 4 weeks later. The femur bone was cleaned thoroughly, immersed in PBS and sophisticatedly attached on 6 cm dish for single-photon or two-photon microscopic observation.
For single-photon microscopic observation, the microscope system equipped with a Nikon AXR inverted microscope using a 2X objective (PLAN APO λD 2x OFN25) was used. For two-photon microscopic observation, the microscope system was operated using a near-infrared femtosecond laser (Mira 900, Coherent) at the central wavelength of 810 nm, 76 MHz pulse repetition rate, and 200 fs pulse width for imaging. The laser power was controlled to 20 mW that is sufficient to produce second harmonic generation (SHG) and two-photon excitation fluorescence (TPEF), and also prevented photodamage during continuous illumination. Thus, the wavelength of SHG from collagen fibers is 405 nm, while the TPEF from collagen, elastin, FAD, and NADH is approximately ranging from 450 to 650 nm. All images were obtained by a laser scanning unit (Fluoview 300, Olympus), a pair of two objective lenses for both lasers focusing and collection of photons (UPlanSApo 20×/0.75, Olympus), and two photomultiplier tubes, respectively, for SHG and TPEF detection (R3896, Hamamatsu). SHG and TPEF were filtered from the intense excitation laser background by a combination of band-pass filter (FF01-405/10, Semrock) and color glass (BG39, Schott, Germany). And, then splitted by a dichroic mirror (FF435-Di01, Semrock) and forward detected. Note that we used a cube polarizing beam-splitter (GT10-B, Thorlabs) combined with a half (AHWP05M-980, Thorlabs) and a quarter (AQWP05M-980, Thorlabs) waveplates to demonstrate LP and CP imaging, respectively. Only the extinction ratio of linear polarization larger than 50:1 and the ellipticity of circular polarization (Imax/Imin) less than 1.1 after the focusing objective lens can then be used for the following two-photon imaging. The acquired images were mainly processed with Image J software. The collagen structure reconstructed through the second harmonic generation images showed porous collagen fiber inter-connected structures (white color) with fluorescent calcein (green area).
Measurement of bone and BM fat parameters by micro-CT
Mouse femurs were dissected and harvested, and subsequently, they were fixed in 10% neutral buffered formalin for a period of 72 h. Femurs were scanned at an isotropic voxel size of 4 μm (80 kV, 450 μA and 2000 ms integration time) by Extra-Ultra-High Resolution micro-CT system (U-CTXUHR, MILabs, Utrecht, Netherlands). Images were reconstructed and analyzed using MILabs Reconstruction 13.0 and Imalytics Preclinical software 3.1.0.7 (Gremse-IT GmbH, Aachen, Germany), respectively. Trabecular bone regions of distal femurs were selected with reference to the growth plate (0.3–1.3 mm from growth plate). Bone mineral density, trabecular bone volume to tissue volume ratio, trabecular number, and increased trabecular spacing was determined based on calibration with phantoms of Imalytics Preclinical software, which were scanned under the same conditions as the bone samples. The calculated thresholds were bone (white) >800, BM fat (yellow) −100 to +80, and other tissues 80–800, and fat volume measured by Imalytics Preclinical software.
Statistical analysis and reproducibility
Data are presented as mean ± SD. Statistical comparisons were performed by unpaired two-tailed Student’s t-test, Tukey’s multiple comparisons test of one-way analysis of variance (ordinary one-way ANOVA), or two-way ANOVA and p values<0.05 were considered significant. All calculations were performed using GraphPad Prism 8.0. All in vivo data are representative of at least 3 independent experiments as indicated.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus database (GEO) under accession GSE178804. The 5mC/5hmC DIP and high-throughput sequencing data are available in the GEO repository database under accession code GSE178805. All other data supporting the findings of this study are available from the corresponding author upon request. Source data are provided with this paper.
References
Pouikli, A. et al. Chromatin remodeling due to degradation of citrate carrier impairs osteogenesis of aged mesenchymal stem cells. Nat. Aging 1, 810–825 (2021).
Prockop, D. J. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 276, 71–74 (1997).
Pittenger, M. F. et al. Mesenchymal stem cell perspective: cell biology to clinical progress. NPJ Regen. Med 4, 22 (2019).
Tsai, C. C. et al. Hypoxia inhibits senescence and maintains mesenchymal stem cell properties through down-regulation of E2A-p21 by HIF-TWIST. Blood 117, 459–469 (2011).
Hayflick, L. & Moorhead, P. S. The serial cultivation of human diploid cell strains. Exp. Cell Res 25, 585–621 (1961).
Shay, J. W. & Wright, W. E. Hayflick, his limit, and cellular ageing. Nat. Rev. Mol. Cell Biol. 1, 72–76 (2000).
Okamoto, K. & Seimiya, H. Revisiting Telomere Shortening in Cancer. Cells 8, https://doi.org/10.3390/cells8020107 (2019).
van Deursen, J. M. The role of senescent cells in ageing. Nature 509, 439–446 (2014).
Fernandez, A. F. et al. H3K4me1 marks DNA regions hypomethylated during aging in human stem and differentiated cells. Genome Res 25, 27–40 (2015).
Khademi-Shirvan, M., Ghorbaninejad, M., Hosseini, S. & Baghaban Eslaminejad, M. The importance of stem cell senescence in regenerative medicine. Adv. Exp. Med Biol. 1288, 87–102 (2020).
Rando, T. A. & Chang, H. Y. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell 148, 46–57 (2012).
Hladik, D. et al. Long-term culture of mesenchymal stem cells impairs ATM-dependent recognition of DNA breaks and increases genetic instability. Stem Cell Res Ther. 10, 218 (2019).
Khanna, K. K. & Jackson, S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet 27, 247–254 (2001).
Alves, H. et al. A link between the accumulation of DNA damage and loss of multi-potency of human mesenchymal stromal cells. J. Cell Mol. Med 14, 2729–2738 (2010).
Zhang, P. et al. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 16, 864–875 (2014).
Wu, P. K. et al. Early passage mesenchymal stem cells display decreased radiosensitivity and increased DNA repair activity. Stem Cells Transl. Med 6, 1504–1514 (2017).
Brown, K. et al. SIRT3 reverses aging-associated degeneration. Cell Rep. 3, 319–327 (2013).
Wang, H. et al. SIRT6 controls hematopoietic stem cell homeostasis through epigenetic regulation of Wnt signaling. Cell Stem Cell 18, 495–507 (2016).
Mohrin, M. et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347, 1374–1377 (2015).
Rudenko, A. et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79, 1109–1122 (2013).
Fuster, J. J. et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017).
Chen, D. & Kerr, C. The epigenetics of stem cell aging comes of age. Trends Cell Biol. 29, 563–568 (2019).
Wang, D. et al. Active DNA demethylation promotes cell fate specification and the DNA damage response. Science 378, 983–989 (2022).
Tsai, C. C., Su, P. F., Huang, Y. F., Yew, T. L. & Hung, S. C. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol. Cell 47, 169–182 (2012).
Shen, W. C. et al. Methylation and PTEN activation in dental pulp mesenchymal stem cells promotes osteogenesis and reduces oncogenesis. Nat. Commun. 10, 2226 (2019).
Sui, B. D., Zheng, C. X., Li, M., Jin, Y. & Hu, C. H. Epigenetic regulation of mesenchymal stem cell homeostasis. Trends Cell Biol. 30, 97–116 (2020).
Sati, S. et al. 4D genome rewiring during oncogene-induced and replicative senescence. Mol. Cell 78, 522–538 e529 (2020).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011).
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011).
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011).
Cui, X. L. et al. A human tissue map of 5-hydroxymethylcytosines exhibits tissue specificity through gene and enhancer modulation. Nat. Commun. 11, 6161 (2020).
Beerman, I. & Rossi, D. J. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell 16, 613–625 (2015).
Szulwach, K. E. et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 14, 1607–1616 (2011).
Pastor, W. A. et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473, 394–397 (2011).
Doege, C. A. et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488, 652–655 (2012).
Song, C. X. et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 29, 68–72 (2011).
Busque, L. et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet 44, 1179–1181 (2012).
Sabino, J. C. et al. Epigenetic reprogramming by TET enzymes impacts co-transcriptional R-loops. Elife 11, https://doi.org/10.7554/eLife.69476 (2022).
Yang, R. et al. Tet1 and Tet2 maintain mesenchymal stem cell homeostasis via demethylation of the P2rX7 promoter. Nat. Commun. 9, 2143 (2018).
Choi, M. R. et al. Genome-scale DNA methylation pattern profiling of human bone marrow mesenchymal stem cells in long-term culture. Exp. Mol. Med 44, 503–512 (2012).
Ames, B. N. & Gold, L. S. Carcinogenesis mechanisms: the debate continues. Science 252, 902–904 (1991).
Ciccarone, F., Zampieri, M. & Caiafa, P. PARP1 orchestrates epigenetic events setting up chromatin domains. Semin Cell Dev. Biol. 63, 123–134 (2017).
Bamezai, S. et al. TET1 promotes growth of T-cell acute lymphoblastic leukemia and can be antagonized via PARP inhibition. Leukemia 35, 389–403 (2021).
Abdelkarim, G. E. et al. Protective effects of PJ34, a novel, potent inhibitor of poly(ADP-ribose) polymerase (PARP) in in vitro and in vivo models of stroke. Int J. Mol. Med 7, 255–260 (2001).
Hahn, M. A., Szabo, P. E. & Pfeifer, G. P. 5-Hydroxymethylcytosine: a stable or transient DNA modification? Genomics 104, 314–323 (2014).
Krishnakumar, R. & Kraus, W. L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749 (2010).
Shukla, S. et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 (2011).
Shen, S. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. USA 111, E5593–E5601 (2014).
An, O. et al. CSI NGS Portal: An Online Platform for Automated NGS Data Analysis and Sharing. Int. J. Mol. Sci. 21, https://doi.org/10.3390/ijms21113828 (2020).
Zampieri, M. et al. ADP-ribose polymers localized on Ctcf-Parp1-Dnmt1 complex prevent methylation of Ctcf target sites. Biochem J. 441, 645–652 (2012).
Ruiz-Velasco, M. et al. CTCF-mediated chromatin loops between promoter and gene body regulate alternative splicing across individuals. Cell Syst. 5, 628–637.e626 (2017).
Ziebarth, J. D., Bhattacharya, A. & Cui, Y. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res 41, D188–D194 (2013).
Bhatnagar, S. et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature 516, 116–120 (2014).
Wu, G. et al. An ATM/TRIM37/NEMO axis counteracts genotoxicity by activating nuclear-to-Cytoplasmic NF-kappaB Signaling. Cancer Res 78, 6399–6412 (2018).
Borkowska, J. et al. Alterations in 5hmC level and genomic distribution in aging-related epigenetic drift in human adipose stem cells. Epigenomics 12, 423–437 (2020).
Lord, C. J. & Ashworth, A. The DNA damage response and cancer therapy. Nature 481, 287–294 (2012).
Pearl, L. H., Schierz, A. C., Ward, S. E., Al-Lazikani, B. & Pearl, F. M. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer 15, 166–180 (2015).
O’Connor, M. J. Targeting the DNA damage response in cancer. Mol. Cell 60, 547–560 (2015).
Di Micco, R., Krizhanovsky, V., Baker, D. & d’Adda di Fagagna, F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021).
Aguilar-Quesada, R. et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 8, 29 (2007).
Menisser-de Murcia, J., Mark, M., Wendling, O., Wynshaw-Boris, A. & de Murcia, G. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol. Cell Biol. 21, 1828–1832 (2001).
Przanowski, P. et al. Oncogenic TRIM37 Links Chemoresistance and Metastatic Fate in Triple-Negative Breast Cancer. Cancer Res 80, 4791–4804 (2020).
Fu, T. et al. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-kappaB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer 24, 45–59 (2021).
Tan, G. et al. TRIM37 overexpression is associated with chemoresistance in hepatocellular carcinoma via activating the AKT signaling pathway. Int J. Clin. Oncol. 26, 532–542 (2021).
Yeow, Z. Y. et al. Targeting TRIM37-driven centrosome dysfunction in 17q23-amplified breast cancer. Nature 585, 447–452 (2020).
Meitinger, F. et al. TRIM37 controls cancer-specific vulnerability to PLK4 inhibition. Nature 585, 440–446 (2020).
Wang, W., Xia, Z. J., Farre, J. C. & Subramani, S. TRIM37, a novel E3 ligase for PEX5-mediated peroxisomal matrix protein import. J. Cell Biol. 216, 2843–2858 (2017).
Agathocleous, M. et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481 (2017).
Blaschke, K. et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 (2013).
Chou, L. Y., Ho, C. T. & Hung, S. C. Paracrine Senescence of Mesenchymal Stromal Cells Involves Inflammatory Cytokines and the NF-kappaB Pathway. Cells 11, https://doi.org/10.3390/cells11203324 (2022).
Hsu, P. C., Gopinath, R. K., Hsueh, Y. A. & Shieh, S. Y. CHK2-mediated regulation of PARP1 in oxidative DNA damage response. Oncogene 38, 1166–1182 (2019).
Wu, H. H. et al. Characterization of initial key steps of IL-17 receptor B oncogenic signaling for targeted therapy of pancreatic cancer. Sci Transl Med 13, https://doi.org/10.1126/scitranslmed.abc2823 (2021).
Wang, Z. Q. et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9, 509–520 (1995).
Yen, Y. T., Chien, M., Wu, P. Y. & Hung, S. C. PP2A in LepR+ mesenchymal stem cells contributes to embryonic and postnatal endochondral ossification through Runx2 dephosphorylation. Commun. Biol. 4, 658 (2021).
Acknowledgements
Grants supported by National Science and Technology Council (MOST 111-2326-B-039-001; NSCT 109-2314-B-039 −009 -MY3; S.C.H receiving them). This work was also financially supported by the “Drug Development Centre, China Medical University” from the Featured Areas Research Centre Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. The funding sources are not involved in the design of the study, in the collection, analysis and interpretation of data, in the writing of the report, and in the decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
Conceptualization, S.C.H.; Data Curation, C.T.H., W.C.C., and L.H.L.; Formal Analysis, C.T.H., and L.H.L; Validation, C.T.H. and W.C.C.; Investigation, C.T.H., L.H.L. and S.C.H.; Visualization, C.T.H., and S.C.H.; Methodology, L.H.L., W.C.C., Y.H.C and H.H.W.; Writing – Original Draft, C.T.H. and S.C.H.; Writing – Review & Editing, all authors; Supervision, S.C.H.; Funding Acquisition, S.C.H.; Project Administration, S.C.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ho, CT., Li, LH., Chang, WC. et al. TRIM37–PARP1–TET1 axis maintains stemness and prevents osteoporosis by inhibiting DNMT1 alternative splicing via 5hmC regulation. Nat Commun 16, 11420 (2025). https://doi.org/10.1038/s41467-025-66281-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66281-y