Abstract
The NeoRHEA was a single-arm phase 2 study that included patients with estrogen receptor positive / human epidermal factor receptor 2 negative early breast cancer that received 4 cycles of neoadjuvant palbociclib and endocrine therapy. The primary outcome was baseline biomarkers of treatment resistance and secondary outcome was post-treatment transcriptional and epigenetic changes of tumor, immune and stromal cells. E2F targets and G2M checkpoint proliferation-related genes gene sets were enriched in baseline samples from resistant patients., Downregulation of E2F targets and G2M checkpoint post treatment was observed in tumor, endothelial and T cells. Gene Set Enrichment Analyses (GSEA) based on genes residing in the differentially accessible peaks revealed similar effects,. Moreover, decreases in CD8 + CD103+ tissue-resident memory cell marker genes were observed post-treatment and validated by multiplex immunohistochemistry. Our data reveal that treatment with palbociclib and endocrine therapy diminishes adaptive anti-tumor immunity by decreasing T cell proliferation and the presence of tissue-resident memory T cells NCT03065621.
Similar content being viewed by others
Introduction
Breast cancer is the most common malignancy diagnosed in women worldwide, with the estrogen receptor-positive (ER)/HER2-negative subtype being the most prevalent, encompassing about 70% of all patients. Most cases are detected at an early stage, offering the potential for cure through locoregional and systemic treatments. However, up to 20% of patients relapse within 10 years after first diagnosis, either locally or at distant sites, with the latter incurable. Consequently, research efforts are focused on developing new treatment strategies to reduce the risk of recurrence in this group of patients1.
CDK4/6 inhibitors have become a cornerstone in breast cancer treatment. In the metastatic setting, three CDK4/6 inhibitors (palbociclib, ribociclib, and abemaciclib) are approved in combination with endocrine therapy as a standard first-line treatment and a possible option in later lines2. In the early-stage setting, abemaciclib has been approved as an adjuvant therapy for patients with node-positive, ER + /HER2-, high-risk breast cancer, based on the results of the MonarchE trial3. Recently, ribociclib has been approved in stage II-III high-risk ER + /HER2- early breast cancer based on the results of the phase 3 NATALEE trial (Slamon et al., 2024)4.
Conversely, two trials evaluating the addition of adjuvant palbociclib did not show a benefit over standard endocrine therapy. The PALLAS trial5 investigated the addition of palbociclib for two years in the adjuvant setting but found no improvement in invasive disease-free survival (iDFS) compared to standard endocrine therapy. Similarly, the PENELOPE-B trial6, which tested the addition of one- year palbociclib to endocrine therapy in patients with residual disease after neoadjuvant chemotherapy, did not show a benefit in iDFS, the primary endpoint. CDK4/6 inhibitors have also been tested in the neoadjuvant setting for patients with ER + /HER2- breast cancer. Despite some differences in study designs and patient populations, all of them demonstrated low rates of pathological complete response (pCR) but an effective shutdown of tumor cell proliferation7.
Preclinical data suggest that CDK4/6i increases tumor immunogenicity and thereby promote cytotoxic T cell-mediated killing of tumor cells via two mechanisms: 1. suppressing the proliferation of immunosuppressive regulatory T cells targeting cytotoxic T cell activities, and 2. enhancing tumor antigen presentation8. In the NeoMonarch trial9, gene expression analysis using RNA-seq revealed upregulation of inflammatory and T cell-related pathways by gene set enrichment analysis (GSEA) in a limited subset of samples from patients treated with abemaciclib plus endocrine therapy. Marked upregulation of IFN-gamma, APC antigen presentation and PD-1 expression at the end of treatment were observed, although only among subjects who received the lead-in abemaciclib plus anastrozole (n = 10) but not the other arms.
A study of single tumor cells from ER + /HER2- early breast cancer patients treated with ribociclib10 found that resistance to endocrine therapy combined with CDK4/6 inhibitors in ER + /HER2- early breast cancer is primarily due to alterations in cell cycle regulation and survival pathways. Specifically, the tumor cells showed increased expression of genes involved in cell cycle progression, such as E2F targets, and decreased expression of apoptotic genes. In addition, there were transcriptional changes associated with enhanced cell survival, suggesting that tumor cells adapt to therapy by promoting proliferation and evading cell death mechanisms, contributing to drug resistance.
Research has been performed on ER + /HER2- early breast cancer to study the effect of endocrine therapy and palbociclib on gene expression programs of bulk tumors and individual tumor cells, but not of stromal or immune cells. In the NeoRHEA trial, changes in the transcriptional programs and chromatin accessibility induced by endocrine therapy combined with palbociclib were evaluated for tumor, stromal and immune cells in ER + /HER2- early breast cancer.
Results
NeoRHEA clinical study results
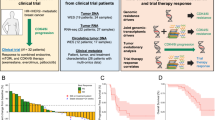
NeoRHEA was an open-label, single-arm, phase 2 study that included patients with ER + /HER2- early breast cancer who were candidates for neoadjuvant therapy. Patients received 4 cycles of palbociclib 125 mg (each cycle of palbociclib involved treatment from D1 to D21, followed by one week of rest) combined with endocrine therapy. Out of 97 evaluable patients, 66 (68%) were postmenopausal, 62 (64%), 74 (76%) and 64 (66%) had histology grade 2, clinical stage T2 and clinical stage N0 breast cancer, respectively (Fig. 1a). PAM50 subtypes were inferred from RNA-seq data of the baseline tumor biopsy with luminal A being the most common subtype. Baseline clinico-pathological characteristics are summarized in Fig. 1a and Supplementary Table 1. Ultrasound (US) response (complete/partial) was observed in 53 (55%) of 97 patients (Fig. 1b). Using centrally-reviewed, semi-automated evaluation of Ki67% in baseline and surgical tumor samples, we observed a significant decrease of Ki67% post-treatment (from 17.4, 0–90 at baseline to 1.6, 0–73 at surgery, p < 0.01, Fig. 1c) with 54% of patients presenting complete cell cycle arrest (CCCA) at surgery (Fig. 1b). We observed no significant differences in the percentage of stromal Tumor Infiltrating Lymphocytes (sTILs %) post-treatment (from 0, 0–50 at baseline to 0, 0–90 at surgery, p = 0.14, Fig. 1d).
a Pie plots of baseline clinico-pathological characteristics of all evaluable (n = 97) patients. b Pie plots of ultrasound response post-treatment and Complete Cell Cycle arrest (CCCA) at surgery. c Ki67 percent (%) assessed by immunohistochemistry (IHC) in baseline and surgery tumor samples. d Tumor Infiltrating Lymphocytes (TILs) percent (%) assessed in H&E slides at baseline and surgery tumor samples. For (c, d), each sample is represented by one point; patients with samples in both timepoints (n = 58 patients for Ki67 and n = 76 for TILs) relate to a line, and points are colored according to the presence or not of CCCA. Patients with samples in at least one timepoint: n = 88 for Ki67 and n = 97 for TILs. Box plots show the median (horizontal line), interquartile range (box = 25th to 75th percentile), and whiskers extending to the smallest and largest values within 1.5 × QR from the lower and upper quartiles, respectively; points beyond the whiskers represent outliers. Two-sided unpaired Wilcoxon was used, no adjustments were made for multiple comparisons. NA not available data. cT clinical tumor status, cN clinical nodal status. Source data are provided as supplementary Table 1.
Baseline and post-treatment transcriptional profiles of treatment resistance
Baseline biomarkers of treatment resistance, defined as the absence of a US response (primary objective of the study) or no CCCA at surgery, were investigated. Baseline clinicopathological characteristics were not associated with the absence of a US response (Supplementary Table 2a) or CCCA, except for patients with luminal B tumors (inferred subtype from PAM50 gene expression) that were associated with an absence of CCCA (Supplementary Fig. 1a and Supplementary Table 2b). Comparing baseline transcriptomic profiles between patients with and without US response or CCCA found that only a few genes were differentially expressed (Supplementary Tables 3, 4 and Supplementary Fig. 1c, d). GSEA using Hallmark gene sets revealed that E2F targets and G2M checkpoint gene sets were significantly enriched in baseline samples from patients without CCCA, while the early estrogen response gene set was significantly enriched in baseline samples from patients with US response and CCCA (Supplementary Table 5 and Supplementary Fig. 1e, f). These findings should be regarded as “hypothesis-generating” with the potential to open up new research directions, requiring external and independent validation. No difference was observed in ESR1mRNA expression at baseline nor at the time of surgery in patient subgroups based on CCCA (Supplementary Fig. 2).
Post-treatment changes of bulk tumor transcriptional profiles
Transcriptional changes following treatment with palbociclib in combination with endocrine therapy were explored by comparing baseline (pre- treatment, n = 78) and surgical (post-treatment, n = 49) tumor samples from 78 patients using bulk RNA sequencing and differential gene expression (DGE) analyses (Supplementary Tables 3-4). Genes related to the Activator Protein 1 (AP-1) transcription factor family, such as JUNB, FOS and FOSB, were significantly up- regulated whereas genes related to proliferation, such as MKI67 and AURKA, were significantly downregulated post-treatment (Fig. 2a). E2F targets, G2M checkpoint and early or late estrogen response gene sets were downregulated, whereas TNFa genes signaling via the NFkB pathway were upregulated post-treatment (Supplementary Table 5). Thus, proliferation and estrogen receptor signaling genes decrease, whereas those involved in inflammatory responses are upregulated post-treatment. (Fig. 2b). DGE and GSEA analyses were performed on patients grouped according to CCCA or US response (Supplementary Tables 4, 5). Interestingly, the data for all patients (Fig. 2b) were mainly driven by patients with CCCA (Fig. 2c).
a Volcano plot of the differential gene expression analysis performed on baseline and surgery samples. Each dot represents a gene. The Y-axis displays the significance of this test (log10 FDR) while the X-axis denotes the fold change of each gene (positive being upregulated and negative downregulated). Genes colored in red are significantly upregulated (FDR ≤ 0.01, log2 Fold change ≥ 0.5) while genes in blue are significantly downregulated (FDR ≤ 0.01, log2 Fold change ≤ 0.5). b Gene Set Enrichment Analysis (GSEA) barplot leveraging data from differential gene expression between baseline and surgery samples from all patients (n = 78). The top 10 most significantly enriched Hallmark gene sets are displayed. The X-axis denotes the normalized enrichment score (NES) of each of the gene sets shown on the Y-axis. c, GSEA barplot leveraging data from differential gene expression between baseline and surgery samples from patients with Complete Cell Cycle arrest (CCCA) at surgery (n = 62). Source data are provided as supplementary Tables 4, 5.
Post-treatment changes of transcriptional profiles in the tumor, stromal and immune cell populations
While changes in gene expression induced by endocrine therapy and palbociclib have been described for tumor cells, there is a paucity of data for stromal and immune cells. Single-nuclei RNA- and ATAC-sequencing (snRNA-seq & ATAC-seq) of baseline (pre-treatment, n = 30) and surgical (post-treatment, n = 33) tumor samples from 37 patients were used to address this question. snRNA-seq transcriptional profiles for 226,097 cells were obtained after rigorous filtering of low-quality nuclei (Supplementary Fig. 3). Counts were processed via the Seurat pipeline so that the PCAs were computed and applied to the shared nearest neighbor and Louvain algorithm for defining nuclei clusters. Once the clusters were defined, major cell types were assigned to each cluster based on marker gene expression across all clusters, which were visualized using the Uniform Manifold Approximation and Projection (UMAP) for dimension reduction (Fig. 3a). Tumor cells were distinguished from normal epithelial cells after profiling the gene copy number aberrations for all cells (Fig. 3b). A total of 144,198 tumor cells, 46,756 stromal cells, 19,438 immune cells and 15,705 normal epithelial cells were identified. Epithelial cells were distinguished by expression of ESR1 as well as ANKRD30A and KRT19. Stromal cells were identified by expression of PECAM1, FAP and ACTA2 and immune cells by expression of CD68, MS4A1 (CD20) and PTPRC (CD45). Cellular subpopulations were further categorized as endothelial (PECAM1, n = 7844), fibroblast (FAP, n = 23,395) or myofibroblast (ACTA2, n = 15,517) stromal cells and myeloid (ITGAX, CD68 n = 9788), T cell (CD3, n = 6837) or B cell (MS4A1, n = 2813) immune cells (Fig. 3c, d). The distribution of the cells in the UMAP for each patient and according to the timepoint is shown in Supplementary Fig. 4. Global analysis of all cells revealed 0.42% upregulated and 0.27%. The percentage of upregulated and downregulated genes for individual analyses of tumor, stromal and immune cell populations are presented in Supplementary Table 6.
a UMAP representation of nuclei colored by cell type. Nuclei are colored according to the assigned cell type. b Copy number aberrations per nuclei; X-axis represents genomic regions from chromosome 1 to chromosome 22, while each row of the Y-axis represents a specific nucleus, colored bars denote the cell type label. Tumor cells display abundant copy number aberrations compared to other cell types. c Barplot of cell type proportions (Y-axis) per patient (X-axis). d Expression of marker genes of tumor cells and normal epithelial cells (ESR1, ANKRD30A, KRT19), stromal cells (PECAM1, FAP, ACTA2, CD68), and immune cells (MS4A1, PTPRC). Source data are deposited in GEO.
Focusing on the E2F target gene set (targets of endocrine therapy and palbociclib), a significant downregulation in tumor and immune but not stromal cells post-treatment was observed (Fig. 4a, Supplementary Fig. 5, and Supplementary Table 8). When examining stroma and immune cell subpopulations, downregulation of E2F gene sets was detected in endothelial, myeloid and T cells. Similar data were obtained for the proliferation-driven, G2M checkpoint control gene set, critical for cell cycle progression; however, there was no significant difference for tumor and myeloid cells when correcting for multiple testing (Fig. 4a, Supplementary Fig. 5, and Supplementary Table 8). These data reveal that the cell cycle machinery is inhibited across different cell types in the tumor microenvironment, with the most pronounced effect on T cells. Interestingly, decreased E2F target and G2M checkpoint gene set expression in tumor cells and T cells post-treatment was only found in responding patients with either CCCA (23 of 37 patients) or US response (18 of 37 patients; Supplementary Table 8). In addition, upregulation of the TNFa gene signaling through the NFkB pathway was observed in tumor, stroma and immune cells post-treatment. When looking at stromal and immune subpopulations, we saw an increase in hypoxia, TNFa, inflammatory and/or IL2 stat5 signaling gene sets post-treatment in endothelial cells, fibroblasts, myeloid cells and T cells. These data suggest there may be a link between hypoxia and inflammatory response gene programs in the post-treatment tumor microenvironment. The decrease in E2F gene and G2M checkpoint control gene sets in endothelial cells is potentially linked to the detected increase in hypoxia associated with stromal and immune cells.
a Gene Set Enrichment analysis, GSEA (hallmark gene sets) based on differential gene expression between baseline and surgery samples using single-nuclei RNA-seq (only data on E2F targets and G2M checkpoint gene sets are presented). b GSEA (hallmark gene sets) based on genes identified in differentially accessible peaks between baseline and surgery samples using single-nuclei ATAC-seq (only data on E2F targets and G2M checkpoint gene sets are presented). Color denotes the significance level (Q value). The X-axis denotes the normalized enrichment score (NES) of each of the gene sets shown on the Y-axis. Source data are provided as supplementary Table 8, 11.
Post-treatment changes of chromatin accessibility in the tumor, stromal and immune cell populations
Changes in chromatin accessibility in tumor, stromal and immune cells post-treatment were analyzed by snATAC-seq. After rigorous filtering to remove low-quality nuclei, snATAC-seq profiles for 289,850 cells were obtained (Supplementary Figs. 3, 6). A higher % of differentially accessible peaks were detected in the post-treatment tumor (3.04% downregulated and 8.87% upregulated) and stromal (8.3% downregulated and 5.13% upregulated) compared with the immune (0.37% downregulated and 0.39% upregulated) cells. (Supplementary Tables 9, 10). Moreover, in tumor cells, peaks were principally upregulated, whereas in stromal cells, they were generally downregulated. These results were primarily driven by patients with CCCA (n = 23).
GSEA based on genes residing in the differentially accessible peaks revealed that E2F and G2M gene set downregulation post-treatment, in populations of tumor and immune cells together with the subpopulations of endothelial, myeloid and T cells, was associated with changes in the chromatin accessibility of these genes (Fig. 4b, Supplementary Fig. 7 and Supplementary Table 11). Interestingly, the association between E2F or G2M gene set downregulation and altered chromatin accessibility (tumor cells and T cells) was driven by the CCCA patient subset (n = 23).
Finally, the post-treatment increases in hypoxia, TNFa signaling via NFkB, inflammatory gene and/or IL2 stat5 signaling gene sets detected in endothelial cells, fibroblasts, myeloid cells and T cells were associated with changes in chromatin accessibility (Supplementary Fig. 7 and Supplementary Table 11). These data suggest that post-treatment changes in transcriptional programs reflect altered chromatin accessibility of the genes controlling these programs.
Confirmation of decreased immune cell proliferation post-treatment
To validate the findings from snRNAseq, which suggested decreased proliferation of tumor and immune cells, multiplex immunohistochemistry (mIHC) was employed to stain formalin fixed paraffin embedded (FFPE) tissues for cytokeratin positive tumor cells (CK), CD45 (total leukocytes), Ki67 (proliferating cells), programmed death- ligand 1 and 2 (PD-L1 and PD- L2) and 4’,6-diamidino-2-phenylindole (DAPI) on 36 baseline biopsies and 33 surgical tumor samples from 37 patients. We observed a marked reduction in the % of Ki67 + tumor (median: 3.6, range: 0–25.5 at base- line, median: 0, range: 0-9.1 at surgery) and immune cells (median: 1.8, range: 0–9.2 at baseline, median: 0.2, range: 0–2 at surgery) post-treatment (Fig. 5). These results validate the snRNA-seq data on proliferation; however, no significant post-treatment difference was observed in the % of PD-L1 + and PD-L2 + tumor or immune cells.
a Immune (CK-/CD45 + ), stroma (CK-/CD45-/Ki67-) and tumor (CK + /CD45-) were defined. The percentage of the respective cells expressing Ki67, PDL1 and PDL2, respectively, is shown on the Y-axis. Each sample is represented by one point; patients with samples in both time points are connected via a line. CK: cytokeratin, DAPI: 4’,6-diamidino-2-phenylindole, PD-L1: Programmed death-ligand 1, PD-L2: Programmed death-ligand 2. Box plots show the median (horizontal line), interquartile range (box = 25th to 75th percentile), and whiskers extending to the smallest and largest values within 1.5 × IQR from the lower and upper quartiles, respectively; points beyond the whiskers represent outliers. Two-sided unpaired Wilcoxon was used, no adjustments were made for multiple comparisons. b Visual representation of stroma/tumor segmentation masks, classifications and cell-level segmentation for mIHC analysis.
Changes in immune and T-cell subpopulations post-treatment
The snRNA-seq data revealed decreased proliferation of immune cells, which was confirmed by the mIHC data showing a decrease in Ki67 + immune cells (%). To further explore these changes post-treatment, re-evaluation of the single-nuclei data confirmed no significant changes in the % of total immune cells (out of all cells analyzed) between baseline (10%) and surgery (8%). Moreover, no differences were detected from baseline to surgery in B cells (16% to 14%), myeloid cells (48% to 52%) and T cells (37% to 34%) relative to total immune cells. This suggested that the observed decrease in immune cell proliferation was not associated with a significant overall decrease in immune cells nor the major B cell, myeloid and T cell subpopulations. This parallels the observed absence of significant changes in TILs (%) post-treatment (Fig. 1d).
The snRNA-seq data identified a decrease in T cell proliferation post-treatment, suggesting it might be informative to investigate changes in the T cell subpopulations. Thus, we focused on the CD8 + , CD4 + , CD8 + CD103 + and CD4 + FOXP3 T cell subpopulations (Fig. 6a). We observed a post-treatment decrease in the expression of E2F target gene sets in CD8 + (q < 0.05) but not in CD4 + FOXP3 + T-cells (q = 0.85). There was a trend toward decreased expression of G2M checkpoint gene set in CD8 + (q = 0.07) but not in CD4 + FOXP3 + T-cells (q = 0.85).CD8 + CD103 + tissue-resident memory T cells had characteristically high expression of HAVCR2 (TIM-3) and low expression of genes such as SELL, KLRG1 and KLF2 as has been previously described (Savas et al., 2018) (Fig. 6b). No significant changes were detected in the % of T cells or CD8 + T cells post-treatment although there was a significant increase in the % of CD4 + cells (Fig. 6c). An examination of key CD8 + and CD4 + T cell subpopulations focused on the CD8 + CD103 + tissue-resident memory T cells (from now on referred to as TRM cells) and the CD4 + FOXP3 + regulatory T cells. TRM cells are important effectors of local immune surveillance and long-term protection against tumors, while regulatory T cells are well-known for their immunosuppressive activities. These data revealed a 37% decrease in the % of TRM cells post-treatment, which was predominantly driven by patients with CCCA (63% decrease) and US response (66% decrease) (Fig. 6c and Supplementary Fig. 8). Similar results were observed for CD4 + FOXP3 + cells (Fig. 6c and Supplementary Fig. 8).
a UMAP representation of T cells subtypes according to the expression of CD4, CD8, CD103 (ITGAE) and FOXP3. b Expression of marker genes related to CD4, CD8, regulatory (FOXP3) and tissue-resident memory (CD103) T-cells, checkpoint inhibitors (HAVCR2/TIM-3, PDCD1, CTLA4), Granzyme B (GZMB), perforin (PRF1) and genes such as SELL, KLRG1 and KLF2 that are downregulated in tissue-resident memory T-cells. c Barplots representing the percent change of cell type ratios between baseline and surgery samples. Each computed ratio is shown on the Y-axis, and the change percent is shown on the X-axis. Analyses were performed in all patients, patients with Complete Cell Cycle arrest (CCCA_Yes) and without CCCA (CCCA_No). d, e Box plots comparing the percentage of Ki67 and CD103 positive cells before and after treatment by means of immunofluorescence quantification in 7 patients. Cells are split in CD4 positive, CD8 positive and either CD4 or CD8 positive cells (Total T cells) and quantifications have been split in intratumoral and stroma tissue. Each dot represents a patient sample, box plots are arranged by time point, and dots are colored by the CCCA value of each patient. Box plots show the median (horizontal line), interquartile range (box = 25th to 75th percentile), and whiskers extending to the smallest and largest values within 1.5 × IQR from the lower and upper quartiles, respectively; points beyond the whiskers represent outliers. Two-sided unpaired Wilcoxon was used, no adjustments were made for multiple comparisons. f Representative images of tumor infiltrating lymphocytes (TIL) and tumor cells in FFPE breast cancer sections stained by multiplex immunohistochemistry (mIHC) to detect CD4 + (orange) and CD8 + (red) T cells positive or negative for CD103 (green), cytokeratin (CK, white) positive epithelial tumor cells and Ki67 (cyan) for proliferating tumor and immune cells. mIHC slides were scanned at 20x magnification on a Vectra PolarisTM (Akoya Biosciences®) and immune cells were quantified in the tumor and stromal compartments using InForm, phenoptR and PhenoptrReports (Akoya Biosciences®). All samples from 7 patients processed using mIHC was performed at the same time in one experiment. g Pseudo-colored pathology views showing the mIHC staining for CD103, CD8 and CD4. Red boxes identify CD103 + CD8 + whereas yellow boxes identify CD103 + CD4 + cells.
As a next step to validate our snRNA-seq data, we performed mIHC analysis of paired biopsy and surgical samples from a subset of patients (n = 7). We confirmed that the percentage of KI67 + T cells significantly decreased post-treatment (Fig. 6d, f) in both the intratumoral and stromal tissue compartments (p = 0.016 and 0.047, respectively). Further, focusing on the CD8 + T cells, we observed a significant decrease in CD103-positive cells after treatment again in both intratumoral and stromal tissue compartments (p = 0.016) (Fig. 6e–g). The loss of TRM cells post-treatment is readily visible in areas with TIL aggregates (Fig. 6f). The percentage of CD39-positive cells was not significantly increased or decreased post-treatment; however, as observed in the sequencing data, the mIHC images confirm a significant decrease in the percentage of Ki67 + CK + cells (Supplementary Figs. 9, 10). Overall, the mIHC data further support our observation on the dual effect of treatment on tumor cell proliferation and adaptive immunity (Supplementary Table 13).
FELINE trial
In order to understand post-treatment changes of transcriptional profiles in the tumor, stroma and immune cell populations induced by endocrine therapy alone (letrozole) versus endocrine therapy and CDK4/6 inhibitor (letrozole and ribociclib), publicly available snRNA-seq data from baseline and post-treatment tumor samples of patients in the FELINE trial11 were analyzed. In both the letrozole alone and the letrozole plus ribociclib arms, we detected a decrease in early or late estrogen response gene sets in tumor cells and an increase in the hypoxia gene set in tumor, immune and stromal cells. In the letrozole alone arm, significant downregulation was observed in E2F target and G2M checkpoint gene set expression in tumor and stromal cells, which included endothelial cells and fibroblasts, whereas in the letrozole plus ribociclib arm, this downregulation was additionally observed in immune cells, including T-cells, B-cells and macrophages/myeloid lineage cells (Supplementary Dataset / Table 12). This suggests that the decrease in T-cell proliferation is driven by adding ribociclib to endocrine therapy. Post-treatment increases in TNFa signaling via NFkB and IL2 stat5 signaling gene sets were observed in tumor and stromal cells in both the letrozole alone and letrozole plus ribociclib arms, whereas increases in these gene sets in immune cells were detected only in the letrozole plus ribociclib arm (Supplementary Tables 12). These data provide independent confirmation that adding CDK4/6 inhibitors, in this case ribociclib, to endocrine therapy has immunomodulatory effects. We were unable to analyze the effect of letrozole alone or letrozole plus ribociclib on TRMs due to the extremely low number of CD8 cells (n = 327) available from the FELINE trial data.
Discussion
The present study provides a detailed analysis of the effects of palbociclib and endocrine therapy on the transcriptional programs and chromatin accessibility of immune, stromal, and tumor cells in the breast cancer tumor microenvironment. Employing snRNA-seq & ATAC-seq, significant downregulation of proliferation markers and gene sets related to cell cycle regulation were observed, notably both in tumor cells and T cells. These findings are further supported by chromatin accessibility data, which detected a robust link between transcriptional downregulation and chromatin state alterations.
The chromatin accessibility analyses provide further insight into the molecular mechanisms underlying these observations. snATAC-seq data revealed reduced accessibility at key regulatory regions of proliferation-associated genes in both tumor and immune cells post-treatment. This decreased accessibility was most pronounced at E2F target genes and G2M checkpoint regulators, suggesting a mechanistic link between chromatin state and transcriptional regulation. This concordance between chromatin accessibility and altered gene expression highlights the integrated nature of epigenetic and transcriptional responses to CDK4/6 inhibition. Palbociclib blocking of CDK4/6 disrupts the activity of transcription factors such as E2F, leading to widespread chromatin remodeling and thereby contributes to the profound anti-proliferative effects observed in the present study.
Previous neoadjuvant studies, such as NeoPalAna12 and NeoMONARCH9 detected upregulation of interferon gamma and inflammatory response gene set expression post-treatment in the analysis of bulk tumor gene expression. We obtained similar results in the analysis of bulk RNA-seq and in addition, found increases in hypoxia-related gene expression. Moreover, using snRNA-seq, we demonstrated that increases in hypoxia and inflammatory response gene sets were principally driven by stromal (endothelial and fibroblast) and immune (myeloid and T) cells but not tumor cells. Previous studies have suggested that increases in inflammatory response and interferon gamma genes may be linked with resistance to CDK4/6 inhibitors13,14.
Preclinical studies have shown that CDK4/6 inhibition can promote anti-tumor immunity by enhancing tumor antigen presentation and inducing anti-proliferative effects, particularly on regulatory T cells, thereby helping to relieve immunosuppression in the tumor microenvironment8. We did not evaluate the effect of palbociclib and endocrine therapy on tumor antigen presentation, but did observe a decrease in CD4 + /FOXP3 + regulatory T cells similar to Goel et al. Furthermore, based on snRNA & ATAC-seq and mIHC data from human samples, we found a significant decrease in T cell proliferation, including both CD4+ and CD8 + T cell subpopulations. Interestingly, mIHC data suggest that this decrease mainly occurs in intratumoral cells compared to stromal CD4 + and CD8 + T cells. More importantly, we detected an important, significant decrease in TRM cells. Using murine models and human peripheral blood samples (breast cancer patients and healthy donors), Heckler et al. reported that palbociclib inhibition of CDK4/6 can facilitate global CD8 + T cell memory formation15. Recent single-cell sequencing and pseudo-time trajectory analyses revealed that CD8 + T cells from mice treated with CDK4/6 inhibitors exhibit a transcriptomic signature suggestive of stem-like differentiation15,16. These findings suggest a potential shift from fewer anergic/exhausted effectors to stem-like memory precursors. These stem-like T cells are particularly important biomarkers with the potential to identify responders to immune checkpoint inhibitors, as they are thought to play a pivotal role in mediating sustained anti-tumor immunity. The above-mentioned preclinical model studies did not examine alterations in the percentage of TRM cells.
A phase 1b study evaluating administration of the PD1 inhibitor pembrolizumab with the CDK4/6 inhibitor abemaciclib in patients with metastatic breast cancer showed high rates of lung and liver toxicity that precluded further development of this combination. Combining different CDK4/6 and immune checkpoint inhibitors, as well as altering the schedule/sequence of administration for each class of drug, could amplify the immune response without increasing toxicity. In addition, CDK4/6 inhibitors, including palbociclib, have been shown to augment T cell activation and increase T cell infiltration in murine lung tumors17. Our study did not detect an increase in sTILs post-treatment, with our snRNA-seq data supporting this observation, with no general increase seen in immune cells or T cells post-treatment.
The discrepancies between the data shown here and the preclinical studies (above) could be due to several factors, including differences between human versus mouse tumor biology or variation in treatment duration and effective palbociclib dose in human versus murine tumors. Moreover, mice are typically treated with palbociclib monotherapy, whereas in humans, including in our NeoRHEA trial, treatment includes a combination of endocrine therapy and palbociclib.
Our data indicate that treatment with palbociclib and endocrine therapy diminishes adaptive anti-tumor immunity by reducing T cell proliferation and the generation of TRM cells at the tumor site. This may also reflect a reduction of their numbers in the peripheral immune system. This observed decrease of TRM cells in responder patients raises concerns about the potential long-term effects of this treatment on antitumor immunity. TRM cells play an essential role in maintaining tissue homeostasis and are responsible for rapidly reactivating antigen-specific responses to recurrent or persistent tumor antigens18,19. TRM cells have been shown not only to express surface integrins but also various immune checkpoint molecules across many different organs20. Several studies found that CDK4/6 inhibition reduced immune checkpoint expression on both CD4+ and CD8 + T cells8,17. The observed reduction in TRM cell subpopulations in our study could be attributed to a similar effect. Another hypothesis is that in patients responding to treatment, a larger proportion of tumor cells are eradicated, leading to decreases in antigen presentation. As a result, the T cells might not be activated to proliferate and respond. Moreover, reduction of the TRM cell pool in responding patients could signal clearance of the tumor with a subsequent dampening of immune activation and thereby lead to the observed reduction in the TRM pool. While it has been shown that NFkB signaling is important for establishing and maintaining TRM during an infection, timing is also a critical factor. Enhancing NF-kB signaling during the contraction phase of the immune response has a negative effect on TRM differentiation21. TNF signaling, through inducible activation of NF-kb, interfered with TGF-b signaling that was critical for TRM imprinting and maintenance. This data highlights the importance in timing of NF-kB activation and potential dual effects on TRM differentiation and maintenance. The downregulation of proliferation markers in both tumor and immune cells, coupled with the decrease in TRM cells observed in this study, suggests that while endocrine therapy and palbociclib can effectively target tumor cell proliferation, its bystander effect may critically impair anti-tumor immunity. This dual effect could have significant implications for clinical outcome, particularly in terms of long-term effective anti-tumor immune surveillance and tumor recurrence.
Currently, there are no data on the effect on T-cells and T-cell subpopulations, including TRM cells, for the combination of endocrine therapy and other CDK 4/6 inhibitors (including ribociclib, abemaciclib and the newer CDK4 selective inhibitor PF-07220060). This study was not designed to examine differences between palbociclib, ribociclib and abemaciclib, nor differences in the basic pharmacological characteristics of these drugs. Palbociclib exerts greater inhibition on CDK6 compared to ribociclib, abemaciclib and PF-0722006022, and it is known that CDK6 is crucial for T cell proliferation17. Thus, one could speculate that palbociclib exerts a more pronounced adverse effect on T cells compared to the other CDK4/6 inhibitors, which in turn could at least partially explains the failure of palbociclib to reduce breast cancer recurrences when it was added to endocrine therapy as adjuvant treatment for women with high-risk early breast cancer in the PALLAS5 and PENELOPE-B trials6. Consideration of CDK4/6 inhibitor effects on the immune response are an important component of the long-term benefit that this patient group may derive from treatment.
One limitation of our study is that all patients were treated with endocrine therapy and palbociclib, with no patients receiving endocrine monotherapy. Thus, it is not possible to determine based on these data how endocrine therapy versus palbociclib contributes to the observed changes in transcriptional programs and chromatin accessibility. To partly address this limitation, we analyzed publicly available snRNA-seq data from the letrozole alone versus letrozole plus ribociclib arms of the neoadjuvant FELINE trial. A decrease in T-cell proliferation was observed in the letrozole plus ribociclib but not the letrozole alone arm. The low number of CD8+ cells in this trial precluded any analysis on the effect of these drugs on TRM cells. Another limitation of our study is that the pre-treatment samples were biopsies, whereas the post-treatment samples were surgical specimens. It is known that transcriptional analysis of different sample types (biopsy vs. surgical sample) can lead to false-positive results, due to the different handling of samples and selectivity of tissue biopsies23,24. Previous reports found that FOS and FOSB genes were upregulated in paired breast cancer samples following two weeks of aromatase inhibitor presurgical treatment, as well as in control samples receiving no presurgical systemic therapy23. However, it has been shown that CDK4/6 inhibition increases the expression of Activator Protein-1 (AP-1) transcription factors, including FOS, FOSB, and JUNB, in breast cancer cell lines25. Therefore, although it is not possible to rule out that the observed changes in FOS and FOSB gene expression are unrelated to endocrine or palbociclib treatment, our findings in human samples support these preclinical observations, suggesting that these expression changes may indeed be treatment-related. Finally, in the study by Gao et al., changes in proliferation and ESR1 signaling were only observed in the treatment and not the control arm. On the other hand, there are studies showing concordance between breast tumor biopsies and surgical samples for gene expression, protein analysis and TILs evaluation26,27,28,29,30. We cannot exclude, however, that some of our findings may be influenced by sampling bias; therefore, independent validation in another cohort with an appropriate control group is warranted. Another limitation is that the study of immune cells in our cohort is hampered by the fact that we used OCT instead of fresh samples and that luminal tumors have less immune infiltration compared to triple-negative tumors31. Nevertheless, our study used single-cell technologies to reveal interesting and potentially clinically important changes in immune cells post CDK4/6 inhibitor treatment. Future research needs to focus on understanding differential effects on the immune response and more specifically, on T cells by the various CDK4/6 inhibitors, known to exert different degrees of CDK4 versus CDK6 inhibition. Moreover, it is important that we more thoroughly understand the conditions where CDK4/6 inhibitors enhance or diminish the immune response. Investigating the mechanistic basis of these divergent effects will be critical for optimizing therapeutic strategies with the potential to maximize anti-tumor efficacy while preserving or enhancing immune functionality. Integrating comprehensive immunophenotyping and functional assays in future studies should provide deeper insights into the immunomodulatory effects of CDK4/6 inhibitors and inform the development of combination therapies that leverage their antiproliferative effects while mitigating potential immunosuppression.
Methods
NeoRHEA clinical study
NeoRHEA (clinical trials gov identifier NCT03065621) is an open-label, single-arm, phase 2 study, including patients with ER + /HER2- early breast cancer candidates for neoadjuvant therapy. The study protocol is provided as supplementary material. All patients have been treated with 4 cycles of endocrine treatment and palbociclib, plus three to seven days of the same treatment (bridge period), so that they were on treatment at the time of surgery. A total of 100 patients were recruited between July 2017 and March 2019 in 5 hospitals in Belgium. Out of 100 patients, two have withdrawn consent, and one had bone metastases at the time of diagnosis and thus were excluded from the analyses. All remaining 97 patients have provided informed consent to participate in the study and related translational research. The study was approved by the Institut Bordet Ethical Committee. The full study protocol is provided as supplementary material. A CONSORT flow diagram for the NeoRHEA study, including the bulk RNA, and single-nuclei RNA and ATAC-seq analyses, can be found in Supplementary Fig. 11.
Criteria for ultrasound response
The main method for evaluation of ultrasound (US) response was bi-dimensional (longest diameter x greatest perpendicular diameter of primary tumor) ultrasound, performed before and after study treatment. Response criteria were those of the World Health Organization as evaluated through US32. Patients were evaluated at the end of the 4-month neoadjuvant treatment and were classified as having tumors with complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD). Patients with CR or PR were classified as responders, whereas patients with SD or PD as non-responders.
Ki67 and sTILs central pathology review
Tumor biopsies have been collected at baseline (before treatment) and after 4 months of treatment (at surgery). Histopathological assessment of all formalin-fixed parrafin embedded (FFPE) tumor samples collected before and after neoadjuvant treatment were centrally performed, including: % tumor cellularity, Ki67 status by immunohistochemistry (IHC) and stromal Tumor Infiltrating Lymphocytes (sTILs). Ki67 and PanCytokeratine (PanCK) staining were performed using the monoclonal mouse anti- Human Ki67 antigen Clone MIB-1 (Dako Omnis) and monoclonal mouse anti-human cytokeratin clone AE1/AE3 Ready-To-Use (Dako Autostainer/Autostainer Plus), respectively. Briefly, PanCytokeratine staining was used to select only the Ki67 staining within the tumor cells, excluding Ki67-expressing lympho- cytes. In order to do so, adjacent sections stained with each antibody were scanned using a digital slide scanner Nanozoomer (Hamamatsu) that converts glass slides into high-resolution digital data by high-speed scanning, and subsequently aligned using Visiopharm (Visopharm®), an histopathology image analysis software for can- cer diagnostics and research with Artificial Intelligence (AI) and deep learning technologies in Augmented Pathology®.
Of note, this procedure allows us to have a precise percentage (%) of Ki67 expression on a scale of 1–100%. Therefore, it allowed us to evaluate the presence or absence of Complete Cell Cycle Arrest (CCCA) in our patients’ tumors, defined as centrally reviewed Ki67 ≤ 2.7% at surgery. The percentage of sTILs was centrally evaluated using Hematoxylin and Eosin (H&E) slides according to the guidelines of the TILs working group33.
Bulk RNA sequencing
RNA extracted from frozen tumor and FFPE samples with at least 25% centrally- assessed tumor cellularity was processed for RNA-sequencing. Strand-specific cDNA libraries were constructed using the NEBNext Ultra directional RNA library Preparation Kit for Illumina paired-end sequencing on a NovaSeq 6000 instrument (Illumina). Briefly, starting from 200 ng of total RNA, rRNA was depleted using the Ribo-Zero Magnetic Kit (Epicenter, Wisconsin) according to the manufacturer’s instructions. Library quality control and quantification was performed using the Fragment Analyzer (Advanced Analytical) and the QUBIT® (Invitrogen). Clusters were generated in a cBot Cluster Generation System (Illumina) using the Kits TruSeq PE Cluster Kit v3-cBot-HS and HiSeq PE Cluster Kit v4 cBot and were sequenced on the Illumina NovaSeq 6000 in 2 x 100bp paired-end mode. In total matched (baseline + surgery) FFPE from 18 patients, matched frozen from 31 patients and unmatched frozen baseline from 29 patients were sequenced. Salmon quantifying the expression of transcripts was used in mapping-based mode using a hg38 reference genome.
Single-nuclei sequencing
Multiome Wet Lab
We used the 10x Chromium Single Cell Multiome ATAC + Gene Expression kit. Frozen optimal cutting temperature (OCT) embedded baseline (pre-treatment) biopsy and surgery tumor samples with at least 25% tumor cellularity were selected for single-nuclei (sn)RNA-seq and ATAC-seq analysis. Briefly, samples were processed into the single-nuclei dissociation with protocol C “Multiome” - adapted version of “Nuclei Isolation from Complex Tissues for Single Cell Multiome ATAC + Gene Expression Sequencing”. Nuclei were first isolated by multiple steps of centrifuging and lysis, then they were sorted, and lastly, they were permeabilized again by means of centrifuging and lysis. Then, extracted nuclei underwent snRNA-seq & ATAC-seq using Chromium Next GEM Single Cell. In order to retain optimal nuclei membrane quality, we modified the aforementioned protocol by using a 10X lysis buffer instead of a 1X lysis buffer for the isolation step. In total, samples from 37 patients were successfully processed (matched samples from 26 patients, unmatched baseline samples from 4 patients and unmatched surgery samples from 7 patients).
Sequencing
The sequencing of the samples was done at the BRIGHTcore Genomics core facility of Uni- versite Libre de Bruxelles (ULB). 10x Genomics-generated cDNA libraries were sequenced on the Illumina NovaSeq 6000 instrument. The sequencing metrics for the GEX was: Read 1-28 cycles, I7-10 cycles, I5-10 cycles, Read 2-90 cycles. The sequencing metrics for the ATAC was: Read 1-50 cycles, I7-8 cycles, I5-24 cycles, Read 2-49 cycles.
Bioinformatics methods
To arrive at count matrices for both RNA-seq and ATAC-seq data, various components of Cell Ranger ARC v2.0, provided by 10X Genomics, were used. More specifically, to demultiplex raw base call files generated by Illumina sequencers “cellranger-arc mkfastq” was used for both ATAC and RNA flow cells. Doing that resulted in a handful of fastq files for each sample, which were then used as an input to “cellranger-arc count” using the hg38 reference for both the RNA and ATAC assays. The latter trimmed and aligned all fastq files to the reference genome in use and then performed filtering, peak calling and counting for both the ATAC and RNA molecules. Doing these steps, we produced count matrices for both the RNA-seq and ATAC-seq samples. Having arrived at count matrices for each cell, RNA counts were processed with the standard Seurat (version 4.3) workflow (normalize data, find variable genes, scale data, PCA, find cell clusters using sNN with resolution set to 0.5 and lastly run dimensionality reduction projections such as t-SNE and UMAP) while ATAC counts were processed again with Signac by finding the most frequently observed peaks, calculating singular value decompositions and dimensionality reduction projections. Before normalizing any of the produced count matrices, we first applied some quality control filters to discard bad-quality nuclei. Having predicted cell type annotation for the RNA partition of our data, the snATAC-seq had predicted annotations transferred from the snRNA-seq dataset. Cell types were identified by expression patterns of known in the literature biomarkers and tumor cells were identified by profiling the copy number aberrations of epithelial cells via infercnv. Concerning the snATACseq, peaks were called for all cells after quality control filtering via MACS2. After quantifying reads in the snATAC-seq data, we were able to assess the activity of transcription factor motifs by cell, utilizing chromVAR using motifs from the JASPAR 2022 transcription factor database. After this, differential tests comparing genes and peaks between baseline and surgery conditions were performed. These included differential gene expression (DGE) analyses, differential peak accessibility analyses, differential motif activation analyses, as well as subsequent over gene set enrichment analyses (GSEA). Lastly, we performed an analysis linking peaks with genes that were concurrently expressed and accessible.
Quality control for snRNA-seq & ATACseq
Samples with less than 400 or more than 16,000 nuclei were immediately excluded, as this indicates suboptimal wet lab processing of the respective samples. Samples passing that filter were included in the rest of the droplet quality control process. Droplets not identified as nuclei by cellranger were excluded. For the snRNAseq, regarding the number of unique molecular identifiers (UMIs), droplets with less than 400 or more than 25,000 were excluded and regarding the number of genes, droplets with less than 200 or more than 10,000 were also excluded. We computed mitochondrial and ribosomal genes percent for each of the droplets, and those exceeding 2% of mitochondrial genes or 1% of ribosomal genes were excluded. For the snATAC-seq droplets with less than 1000 or more than 100,000 numbers of UMIs were excluded. Peaks were then called inputting fragments of nuclei from all samples together via MACS2 so that the peak set is common for all samples, and peaks with width less than 20 base pairs or more than 10,000 base pairs were excluded.
Cell type annotation via markers and copy numbers
High level labels
Initially, we spit our cell population into three categories - epithelial, stromal, and immune – based on respective markers. For epithelial cells we used Keratin 19 (KRT19), Estrogen Receptor 1 (ESR1) and Ankyrin Repeat Domain 30 A (ANKRD30A); for stromal cells we used Platelet And Endothelial Cell Adhesion Molecule 1 (PECAM1), Fibroblast Activation Protein Alpha (FAP) and Actin Alpha 2, Smooth Muscle (ACTA2) and lastly for immune cells we used CD68 Molecule (CD68), Membrane Spanning 4-Domains A1 (MS4A1) and Protein Tyrosine Phosphatase Receptor Type C (PTPRC). Cells were labeled based on their cluster associations with each of the markers, meaning that for each cluster, we carefully inspected which of the selected markers were mostly expressed and then manually labeled all cells of the respective cluster to the cell type whose markers were most expressed.
Tumor vs normal cells
Having identified the three major cell type populations, we then profiled copy number aberrations of the epithelial cells specifically to distinguish the malignant cells from the normal epithelial ones. To do that we created a reference set of 500 immune and stromal cells that were thought to be copy number neutral. For each patient, we selected proportionally in relation to the rest of the patients the least variable in terms of expression of immune and stromal cells, resulting in 500 cells in total. Then, for each sample, we separated the epithelial cells from the rest and profiled their copy number status using the previously generated set of reference cells via infercnv in subclusters analysis mode, without clustering by groups and by using an HMM (hidden Markov model) model and denoising technique. Having profiled the copy number status of the epithelial cells by employing a KNN (k nearest neighbor) classification strategy for each sample, we compared the copy number profile of each cell against the profile of the reference set of cells and the profile of the top 5% most aberrant cells of the same sample. Cells whose copy number profile resembled most the one of the reference set of cells were labeled as normal epithelial cells, while the rest were labeled as tumor cells.
Stromal and Immune cells subclassification
Stromal cells were further labeled as either fibroblasts, myofibroblasts or endothelial cells again by using specific markers and leveraging the cluster information. More specifically, FAP was used to label fibroblasts, ACTA2 to label myofibroblasts, and lastly, PECAM1 to label endothelial cells. Immune cells were classified into T, B and myeloid cells by using CD3 Epsilon Subunit Of T-Cell Receptor Complex (CD3E), MS4A1 and CD68 respectively.
Immune T cells subclassification
All immune T cells were labeled as either CD4 or CD8 based on their expression of CD4 Molecule (CD4) and CD8 Subunit Alpha (CD8A). CD8 cells were further labeled as tissue resident memory (TRM) cells based on their expression of Integrin Subunit Alpha E (ITGAE) or CD103, and CD4 cells further labeled as FOXP3-positive regulatory cells.
Differential gene expression and gene set enrichment analyses
For bulk RNA sequencing data, DESEq2 was used for DGE analysis with fold change shrinkage as implemented in the lfcShrink function, of note for patients with both OCT and FFPE samples available in any of the time points, the sample with paired counterpart in the other time point was in use. To take into account the effect of samples originating from tissues with different conservation methods (OCT vs FFPE) the conservation method was added in the generalized model comparing expression of genes. In the single-nuclei cohort, after robustly set cell type labels for each of the cells included in our cohort, we then performed DGE analyses between baseline and surgery cells for each of the cell type subpopulations. For these analyses, we used the FindMarkers Seurat command with default settings the log-fold threshold, as well as the minimum percentage of cells expressing each gene to zero. To infer biological conclusions, using the DGE analyses, we subsequently performed GSEA using the Hallmark gene sets. For all the GSEA analyses, the clusterProfiler Bioconductor with the fgsea method was in use.
Differential peak accessibility analyses
To gain insights into changes in chromatin accessibility in our set of cells post-treatment, we performed differential peak accessibility analyses again for all identified cell types. The Seurat FindMarkers command was in use however, this time the test utilized logistic regression with the total number of fragments in each of the peaks added as a latent variable to mitigate the effect of differential sequencing depth on the results. Log-fold threshold was set to zero and the minimum percentage of cells with each of the peaks being accessible was set to 5%.
Gene set enrichment analysis of peaks
To perform a conventional GSEA analysis using the differentiated peak, there is no need to split each set of peaks into upregulated and downregulated peaks; however, a fold change of genes and not peaks is needed. Having both a set of differentiated genes and peaks, we select the genes residing in the differentiated peaks and use the fold change as calculated in the DGE analysis of these specific genes as an input to the GSEA analysis using the Hallmark gene set.
Multiplex immunohistochemistry PD-L1, PD-L2
Multiplex immunohistochemistry (mIHC) was performed in the context of an ongoing collaboration between Institut Bordet and Rejuveron. mIHC to stain for CK, CD45, Ki67, Programmed death-ligand 1 and 2 (PD-L1 and PD-L2) and 4’,6-diamidino-2-phenylindole (DAPI) was performed on 36 baseline and 33 surgery FFPE tumor samples from 37 patients. The markers included in this panel aimed to validate our snRNA-seq findings on the decrease in the proliferation of tumor and immune cells. Moreover, we wanted to study changes in the expression of the PDL1 and PDL2 ligands of the immune-checkpoint receptor programmed cell death protein 1 (PD1), which is the most clinically relevant checkpoint in cancer immunotherapy (Pardoll et al.34). Immunofluorescence images were first unmixed using InForm, and then subsequent cell quantifications were done using QuPath (version 0.5.1). Cell segmentation was performed by employing the stardist algorithm, filtering out cells with a detection probability lower than 50%. For each of the markers, a boolean classifier was trained, classifying each cell as positive or negative. All classifiers were combined in one composite classifier, with which the final classification was performed. Cells were labeled as tumor if they were classified as panCK positive and CD45 negative, immune if they were classified as panCK negative and CD45 positive and stromal if they were classified as panCK negative, CD45 negative and Ki67 negative.
Multiplex immunohistochemistry CD103, CD39
Multiplex immunohistochemistry (mIHC) was performed on FFPE tumor tissues collected at baseline and post-surgery from seven patients, selected based on their high tumor infiltrating lymphocyte (TIL) scores. The mIHC panel included: CD4, CD8, CD39, CD103, CK and Ki67 plus DAPI for nuclear staining. The markers selected in this panel aimed to validate our snRNA-seq findings on the decrease in T-cell proliferation and TRMs. CD39 was included in the panel based on published data showing that co-expression of CD103 and CD39 identifies a subset of tumor-reactive T cells in solid tumors35. Of note, the mIHC panel optimization was done according to the procedure we have previously detailed36 before processing the samples from the 7 NeoRHEA patients. Image acquisition was achieved using a Vectra PolarisTM (Akoya Biosciences®), and subsequent analysis and quantification was performed using InForm, phenoptR and PhenoptrReports (Akoya Biosciences®). Images were preprocessed for spectral unmixing to identify marker signals, followed by tissue segmentation to differentiate tumor and stroma regions using machine learning algorithms trained on manual annotations. Nuclear segmentation was performed using the DAPI staining. Phenotyping categorized cellular subpopulations of tumor cells (pan-cytokeratin or CK + ), CD4 + T cells, CD8 + T cells, and total T cells (CD4 + plus CD8 + ). Tissue-resident memory T cells (TRM) were identified as CD8 + T cells co-expressing CD103. Proliferating cells (tumor and immune) were identified by Ki67 positivity. CD39 expression was assessed across all subpopulations, with CD4 + and CD8 + T cells the principal positive subpopulations; however, a large proportion of vessels also stained positive, increasing the background for this marker.
FELINE trial
FELINE trial (clinicaltrials.gov identifier NCT02712723) randomized in a 1:1:1 ratio 120 post-menopausal women with node-positive or ›2 cm ER + /HER2- breast cancer to a 6-month neoadjuvant treatment with either letrozole alone or letrozole and intermittent administration of ribociclib or letrozole and continuous administration of ribociclib. snRNA-seq using 10X genomics was performed on tumor biopsies collected at baseline (pre-treatment), after 14 days of treatment and at the end of treatment (surgery). We analyzed snRNA-seq data from 23 patients with letrozole alone and 39 patients with letrozole and ribocilib combining the intermittent and continuous ribociclib schedule that we have retrieved from Griffiths et al., 2025. We only used data from tumor biopsies at baseline and surgery that represented similar timepoints with the NeoRHEA study. We used the count matrices and cell type annotations provided in ref. 11. We have performed DGE analyses between baseline and surgery cells for each of the cell type subpopulations. To infer biological conclusions, using the DGE analyses, we subsequently performed GSEA using the Hallmark gene sets as we did in the NeoRHEA study.
Statistics and reproducibility
The study primary objective was to develop biomarkers to predict resistance to neoadjuvant treatment of endocrine therapy and palbociclib using RNA sequencing of the baseline tumor biopsy. The following few assumptions were made: The overall response rate would be around 75%-80%. The resistance rate would be therefore around 20%-25%. According to simulations carried out by Dobbin et al.37, a sample size of at least 20 subjects in each class would be required to develop a binary predictor using high-dimensional data. Therefore, to get 20 subjects in the resistant group with an expected 22.5% resistance rate, around 90 evaluable subjects would be needed. To take into account a rate of 10% of non-evaluable subjects, 100 eligible subjects would be required for inclusion in the study. For all the analyses between baseline and surgery samples using either bulk RNA-seq or snRNA-seq and ATAC-seq data, as well as for all mIHC analyses, no statistical method was used to predetermine sample size.
NeoRHEA data availability
The bulk and single-nucleus RNA-seq data generated and analyzed in this study have been deposited in the European Genome-phenome Archive (EGA) under the accession number ID – EGAD50000002038. Access to the data is controlled and subject to approval by the Data Access Committee, composed of Michail Ignatiadis, Christos Sotiriou, and David Venet. There are no restrictions on eligibility for data access, provided that the submitted proposal is approved by the committee. For data access or related queries, please contact David Venet and Michail Ignatiadis. Data access requests are typically processed within 3–6 months, depending on the nature of the request. The data will remain available for the duration of the approved proposal.
Raw images from mIHC experiments and count matrices from bulk RNA-seq, and snRNA-seq, and ATAC seq data have been uploaded in Zenodo DOI 10.5281/zenodo.16760008. Source data are provided as Source Data files.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
References
Ignatiadis, M. & Sotiriou, C. Luminal breast cancer: from biology to treatment. Nat. Rev. Clin. Oncol. 10, 494–506 (2013).
Morrison, L., Loibl, S. & Turner, N. C. The CDK4/6 inhibitor revolution — a game-changing era for breast cancer treatment. Nat. Rev. Clin. Oncol. 21, 89–105 (2023).
Johnston, S. R. D. et al. Abemaciclib combined with endocrine therapy for the adjuvant treatment of HR+, HER2−, node-positive, high-risk, early breast cancer (monarchE). J. Clin. Oncol. 38, 3987–3998 (2020).
Slamon, D. et al. Ribociclib plus endocrine therapy in early breast cancer. N. Engl. J. Med. 390, 1080–1091 (2024).
Mayer, E. L. et al. Palbociclib with adjuvant endocrine therapy in early breast cancer (PALLAS): interim analysis of a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 22, 212–222 (2021).
Loibl, S. et al. Palbociclib for residual high-risk invasive HR-positive and HER2-negative early breast cancer — the PENELOPE-B trial. J. Clin. Oncol. 39, 1518–1530 (2021).
Brandão, M. & Ignatiadis, M. CDK4/6 inhibitors as neoadjuvant treatment in breast cancer — what can we learn? Ann. Oncol. 29, 2274–2278 (2018).
Goel, S. et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017).
Hurvitz, S. A. et al. Potent cell-cycle inhibition and upregulation of immune response with abemaciclib and anastrozole in neoMONARCH, phase II neoadjuvant study in HR+/HER2− breast cancer. Clin. Cancer Res. 26, 566–580 (2020).
Griffiths, J. I. et al. Serial single-cell genomics reveals convergent subclonal evolution of resistance as patients with early-stage breast cancer progress on endocrine plus CDK4/6 therapy. Nat. Cancer 2, 658–671 (2021).
Griffiths, J. I. et al. Cellular interactions within the immune microenvironment underpin resistance to cell cycle inhibition in breast cancers. Nat. Commun. 16, 2132 (2025).
Ma, C. X. et al. NeoPalAna: Neoadjuvant palbociclib, a cyclin-dependent kinase 4/6 inhibitor, and anastrozole for clinical stage 2 or 3 estrogen receptor–positive breast cancer. Clin. Cancer Res. 23, 4055–4065 (2017).
De Angelis, C. et al. Activation of the IFN signaling pathway is associated with resistance to CDK4/6 inhibitors and immune checkpoint activation in ER-positive breast cancer. Clin. Cancer Res. 27, 4870–4882 (2021).
Pascual, T. et al. Cell-cycle inhibition and immune microenvironment in breast cancer treated with ribociclib and letrozole or chemotherapy. NPJ Breast Cancer 10, 25 (2024).
Heckler, M. et al. Inhibition of CDK4/6 promotes CD8 T-cell memory formation. Cancer Discov. 11, 2564–2581 (2021).
Lelliott, E. J. et al. Combined BRAF, MEK, and CDK4/6 inhibition depletes intratumoral immune-potentiating myeloid populations in melanoma. Cancer Immunol. Res. 9, 136–146 (2021).
Deng, J. et al. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 8, 216–233 (2018).
Savas, P. et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 24, 986–993 (2018).
Virassamy, B. et al. Intratumoral CD8+ T cells with a tissue-resident memory phenotype mediate local immunity and immune checkpoint responses in breast cancer. Cancer Cell 41, 585–601 (2023).
Christo, S. N., Park, S. L., Mueller, S. N. & Mackay, L. K. The multifaceted role of tissue-resident memory T cells. Annu. Rev. Immunol. 42, 317–345 (2024).
Teixeiro, E. et al. Different T cell receptor signals determine CD8+ memory versus effector development. Science 323, 502–505 (2009).
Fassl, A., Geng, Y. & Sicinski, P. CDK4 and CDK6 kinases: from basic science to cancer therapy. Science 375, eabc1495 (2022).
Gao, Q. et al. Major impact of sampling methodology on gene expression in estrogen receptor-positive breast cancer. JNCI Cancer Spectr. 2, pky005 (2018).
López-Knowles, E. et al. Heterogeneity in global gene expression profiles between biopsy specimens taken peri-surgically from primary ER-positive breast carcinomas. Breast Cancer Res. 18, 39 (2016).
Watt, A. C. et al. CDK4/6 inhibition reprograms the breast cancer enhancer landscape by stimulating AP-1 transcriptional activity. Nat. Cancer 2, 34–48 (2021).
Saghir, H. et al. How reliable are gene expression-based and immunohistochemical biomarkers assessed on a core-needle biopsy? A study of paired core-needle biopsies and surgical specimens in early breast cancer. Cancers 14, 4000 (2022).
Crozier, J. A. et al. High concordance of 70-gene recurrence risk signature and 80-gene molecular subtyping signature between core needle biopsy and surgical resection specimens in early-stage breast cancer. J. Surg. Oncol. 125, 596–602 (2022).
Buisseret, L. et al. Reliability of tumor-infiltrating lymphocyte and tertiary lymphoid structure assessment in human breast cancer. Mod. Pathol. 30, 1204–1212 (2017).
You, K. et al. Comparison of core needle biopsy and surgical specimens in determining intrinsic biological subtypes of breast cancer with immunohistochemistry. J. Breast Cancer 20, 297–303 (2017).
Pölcher, M. et al. Concordance of the molecular subtype classification between core needle biopsy and surgical specimen in primary breast cancer. Arch. Gynecol. Obstet. 304, 783–790 (2021).
Denkert, C. et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 19, 40–50 (2018).
Miller, A. B., Hoogstraten, B., Staquet, M. & Winkler, A. Reporting results of cancer treatment. Cancer 47, 207–214 (1981).
Salgado, R. et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an international TILs working group 2014. Ann. Oncol. 26, 259–271 (2015).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Duhen, T. et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 9, 2724 (2018).
Boisson, A. et al. Fluorescent multiplex immunohistochemistry coupled with other state-of-the-art techniques to systematically characterize the tumor immune microenvironment. Front. Mol. Biosci. 8, 673042 (2021).
Dobbin, K. K. & Simon, R. M. Sample size planning for developing classifiers using high-dimensional DNA microarray data. Biostatistics 8, 101–117 (2007).
Acknowledgements
This study was supported by an educational grant from Pfizer and Rejuversen a grant from Fondation Contre le Cancer No 2018-118 and a grant from L’association Jules Bordet No 2020-02. MI was the recipient of all the above grants. Pfizer had no role in study design, data collection and analysis or manuscript writing.
Author information
Authors and Affiliations
Contributions
M.I. designed and supervised the study. A.P. performed all bioinformatic and most statistical analyses. S.M. performed the wet lab experiments for the bulk and single cell analyses. F.P.D., E.A., L.B., I.V., T.B.H., A.A., C.S., P.V., and P.N. contributed with patient data. D.L. and R.S. performed the pathology analyses, including TILs. M.P., A.S., and L.A. performed statistical analyses. T.V., F.M., T.C., F.R., and K.W.G. performed mIHC analyses. A.P., E.A., T.V., K.W.G., and M.I. wrote the manuscript. All authors contributed to data interpretation and provided comments, read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
MI reports consultancy roles for Seattle Genetics, Daichi, AstraZeneca, Menarini/Stemline, Gilead Sciences, and Novartis (all unrelated to the present work), Rejuveron Senescence Therapeutics, grant/research support (to institution) from Roche, Inivata Inc, and Natera Inc (all unrelated to the present work), Pfizer, and uncompensated roles for EORTC (EORTC Board of Directors 2018–2021, Chair EORTC Breast Cancer Group 2021–2025). FPD reports grants from Fondation Belge Contre le Cancer, institutional payments from Roche, Pfizer, AstraZeneca, Eli Lilly, Novartis, Amgen, Daiichi Sankyo, Pierre-Fabre, Gilead, Seagen, and MSD, and support for attending meetings from Amgen, Roche, Teva, Pfizer, Daiichi Sankyo, AstraZeneca, Gilead, and MSD (all unrelated to the present work). ORCID ID: 0000-0002-5429-7888.
L.B. reports Research funding to my Institution from Astra Zeneca/MedImmune Advisory role for Domain Therapeutics, iTeos Therapeutics, AstraZeneca Salary partly covered by the Fondation contre le Cancer (Belgium) Member of the IMMUcan Consortium (EORTC) Travel grants from GILEAD, AstraZeneca, Roche (all unrelated to the present work).
E.A. reports Research grant to my institution: Gilead Advisory Board for AstraZeneca Receipt of honoraria or consultation fees: Eli Lilly, AstraZeneca, Abscint, Bayer Travel/meeting grants: Eli Lilly, AstraZeneca, Daiichi Sankyo, Abscint, Menarini, Gilead (all unrelated to the present work).ORCID ID: 0000-0003-0295-7511. A.A. reports Advisory role: Amgen, AstraZeneca, Bayer, Daiichi, EISAI, Genomic Health, Hengrui, Innate, Ipsen, Leo Pharma, Lilly, Merck, MSD, Novartis, Pfizer, Seattle Genetics, Menarini Speaker fees: Amgen, AstraZeneca, Bayer, Daiichi, EISAI, Genomic Health, Ipsen, Leo Pharma, Lilly, Merck, MSD, Novartis, Pfizer, Seattle Genetics, Hikma, Sothema. Research grants to the Institute: BMS, Roche (all unrelated to the present work). R.S. reports Advisory Board role for BMS, Roche, Exact Sciences, Daicchii Sankyo, Astra Zeneca, Case45. Research funding by Roche, Puma, Merck, BMS and Travel and congress-registration support by Roche, Merck, Astra Zeneca (all unrelated to the present work).F.M. and T.P.C. are employes of Rejuversen. The remaining authors declare no other competing interests.
Peer review
Peer review information
Nature Communications thanks Masakazu Toi and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Papagiannis, A., Majjaj, S., Duhoux, F.P. et al. Palbociclib and endocrine therapy diminish adaptive anti-tumor immunity in early breast cancer: The NeoRHEA phase 2 study. Nat Commun 16, 11659 (2025). https://doi.org/10.1038/s41467-025-66590-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66590-2