Abstract
The dimethylation of histone H3 lysine 4 (H3K4me2) plays an important role in developmental phase transitions in plants, such as regeneration from the callus and the initiation of flowering. H3K4me2 in plants is correlated with transcriptionally repressed states, which can be accounted for by transcription-coupled active H3K4me2 demethylation, but the converse molecular pathway by which H3K4me2 represses transcription remains largely unexplored. Here, we show that H3K4me2 colocalizes with the H2A variant H2A.Z and H2A ubiquitination (H2Aub). Our genetic analyses reveal that H3K4me2 functions upstream but not downstream of these marks. H3K4me2 also partially colocalizes with the facultative heterochromatin mark H3K27me3, and loss of H3K4me2 causes a decrease in the colocalized H3K27me3. Interestingly, in genes with diel H3K4me2 oscillation, H3K4me2 oscillates in antiphase with transcription but in phase with H2A.Z. In addition, the genetic manipulation of H3K4me2 affects the oscillating profiles of H2A.Z, suggesting the efficient relay from H3K4me2 to H2A.Z. Notably, the diel oscillation of H2Aub is much weaker than that of H2A.Z despite the overall similarity in their distributions. These results suggest that H3K4me2 orchestrates H2A.Z and H2Aub with distinct dynamics. We propose that H3K4me2 promotes stepwise progression of chromatin toward repressive states in plants.
Similar content being viewed by others
Introduction
Methylation of histone H3 lysine 4 (H3K4) is a widely conserved modification among eukaryotes. H3K4 methylation has three states depending on the number of methyl groups: H3K4 monomethylation (H3K4me1), dimethylation (H3K4me2), and trimethylation (H3K4me3). H3K4 methylation generally marks actively transcribed genes; therefore, H3K4 methylation has been thought to play a role in activating transcription1. Indeed, H3K4me3 is strongly positively correlated with the transcription output of the genes in diverse species1. However, H3K4me2 is negatively correlated with transcription in plants, including the monocot rice and the eudicot Arabidopsis thaliana2,3. The removal of H3K4me2 by LYSINE-SPECIFIC DEMETHYLASE 1-LIKE 3 (LDL3) is necessary for the activation of genes involved in shoot regeneration from callus4. LDL3 binds to the phosphorylated C-terminal domain of RNA polymerase II and demethylates H3K4me2 in a transcription-coupled manner5. These findings suggest a possibility that transcription and H3K4me2 repress each other in plants2,3,4,5,6,7. While the effect of transcription on H3K4me2 is beginning to be understood mechanistically5, the general function of H3K4me2 on transcription remains ambiguous.
Multiple histone methyltransferases in plants catalyze H3K4me1, H3K4me2, and H3K4me3 with distinct specificities8,9. In A. thaliana, ARABIDOPSIS TRITHORAX 3 (ATX3), ATX4, and ATX5 are mainly responsible for H3K4me28,9. Although the loss of function of ATX3/4/5 or LDL3 alters H3K4me2 localization at thousands of genes, that does not result in widespread changes in gene expression5,8, suggesting that H3K4me2, as H3K4me1 and H3K4me3, functions depending on chromatin contexts9,10,11.
Chromatin states are defined by the combinatorial presence of histone modifications and variants. H3K27me3, the trimethylation of histone H3 lysine 27, is a facultative heterochromatin mark, which is catalyzed by Polycomb repressive complex 2 (PRC2)12. Mono-ubiquitination of histone H2A (H2Aub) and H2A.Z are also localized in facultative heterochromatin13, and involved in the deposition of H3K27me314,15,16, although they are also enriched across the genome in contrast to H3K27me3. Previous studies showed that H2Aub contributes to transcriptional repression via PRC2-mediated H3K27me3 deposition, and it is proposed that H2Aub represents stand-by states for gene activation or repression15,17. H2A.Z is enriched near the transcription start sites of highly expressed genes, whereas it accumulates throughout the gene bodies of lowly expressed genes18,19. H2A.Z levels show negative correlations with gene expression, similar to H3K4me22,20. Interestingly, recent studies in A. thaliana demonstrated that the deposition of H2A.Z and H2Aub is promoted by H3K4me3, which is an active mark, and these contradictory trends are discussed in relation to bivalent chromatin modifications10,11. On the other hand, H3K4me2 is known to be negatively correlated with transcription2,5, but the possible connections between H3K4me2 and H2A.Z or H2Aub have not been tested.
To explore the molecular function of H3K4me2 in plants, we focused on the colocalization of H3K4me2 with H2A.Z, H2Aub, and H3K27me3. Using a genetic approach with mutants of genes encoding H3K4me2 methyltransferases and demethylases, we showed that an increase in H3K4me2 induces the accumulation of H2A.Z and H2Aub and that a decrease in H3K4me2 reduces H2A.Z, H2Aub, and H3K27me3. This pathway was one-way; H2A.Z, H2Aub, and H3K27me3 act downstream of H3K4me2. We also found that H3K4me2 daily oscillates in phase with H2A.Z but in antiphase with transcription, highlighting tight cooperation between H3K4me2 and H2A.Z in diel transcriptional regulation. In contrast, H2Aub does not oscillate during the day, despite its similarity to H2A.Z in the static profiles and responses to H3K4me2. We propose that H3K4me2 coordinates H2A.Z and H2Aub on distinct timescales, contributing to the stepwise establishment of repressive chromatin states in plants.
Results
H3K4me2 colocalizes with H2A.Z and H2Aub, and partially with H3K27me3
To identify chromatin marks that are functionally linked to H3K4me2, we first explored chromatin modifications that colocalize with H3K4me2. Chromatin immunoprecipitation sequencing (ChIP-seq) in the wild type (WT) plants revealed that H2A.Z and H2Aub colocalized with H3K4me2 (Fig. 1a–e). These three chromatin marks were mainly accumulated in protein-coding genes and partially in non-protein-coding genes and pseudogenes, but did not accumulate in transposable element genes (Fig. 1a). Hereafter, we focused on chromatin marks in protein-coding genes. The distribution patterns of H3K4me2 within genes were similar to those of H2A.Z and H2Aub; these three marks mainly accumulated around the transcription start site (TSS), whereas some genes had broader accumulation of these marks throughout the gene bodies (Fig. 1a, d, e), which is consistent with previous reports on the localization patterns of H3K4me221, H2A.Z22, and H2Aub15,23 in A. thaliana. H2A.Z and H2Aub levels were positively correlated with H3K4me2 levels across genes (Fig. 1b). When comparing accumulation levels among subregions within gene bodies, H2A.Z and H2Aub levels presented high correlations with H3K4me2 between the same subregion from the TSS to the transcription end site (TES) (Fig. 1c). These observations indicate that H2A.Z and H2Aub accumulate in the same genes and the same regions as H3K4me2. Consistent with previous reports21, H3K4me2 and H3K27me3 were colocalized only in a subset of genes (Fig. 1a, b, e). H3K4me2 levels in the genic region from the TSS to approximately 30% downstream of the TSS were negatively correlated with local H3K27me3 levels, whereas H3K4me2 levels further downstream were positively correlated with global H3K27me3 levels (Fig. 1c).
a Metaplots and heatmaps of H3K4me2, H2A.Z, H2Aub, and H3K27me3 in WT. All genes (n = 37,898) are divided into 4 types: protein-coding genes (PCGs; n = 27,443), non-protein-coding genes (non-PCGs; n = 5613), transposable element genes (TEs; n = 3901), and pseudogenes (PSGs; n = 941). Genes are sorted in descending order by H3K4me2 levels. b Scatter density plots showing the relationship between H3K4me2 levels and other chromatin mark levels (square root of RPKM). Each dot represents a protein-coding gene. Colors represent density. c Heatmaps showing Spearman’s rank correlation coefficient between H3K4me2 and other chromatin marks (H2A.Z, H2Aub, and H3K27me3) calculated for each segmented gene region. Each gene region (TSS to TES) is divided into 100 equal parts. Higher coefficient values indicate greater similarity across all protein-coding genes between each pair of the 100 parts. d Histograms showing the similarity of distribution pattern between H3K4me2 and H2A.Z (red), H3K4me2 and H2Aub (green), and H3K4me2 and H3K27me3 (blue) per gene. The similarity is calculated by Spearman’s rank correlation coefficient between read numbers mapped in 100 equally sized subregions of each gene. Higher coefficient values indicate similar intragenic distribution. e A representative genome browser view of ChIP-seq signals of H3, H3K4me2, H2A.Z, H2Aub, and H3K27me3 in WT.
Compared with H3K4me1 and H3K4me3, H3K4me2 showed greater positive correlations with H2A.Z and H2Aub at both the whole-gene level and within subgenic regions (Supplementary Fig. 1a, b). H3K4me2 colocalized with H3K27me3 in a subset of genes, whereas H3K4me3 was strongly exclusive to H3K27me3 (Supplementary Fig. 1a). To compare H3K4me2 and H3K4me3 regarding their relationship to H2A.Z, H2Aub, and H3K27me3, we classified genes into four groups based on their H3K4me2 and H3K4me3 levels. Group 1, which showed high levels of both H3K4me2 and H3K4me3, exhibited higher accumulation of H2A.Z and H2Aub (Supplementary Fig. 1c–e). Notably, even in group 2, characterized by low H3K4me3 and high H3K4me2 levels, H2A.Z and H2Aub were accumulated, and H3K27me3 was also highly enriched (Supplementary Fig. 1c–e). Consistent with previous reports15,20, H2A.Z and H2Aub show strong colocalization, while their colocalization is modest for H3K27me3 (Supplementary Fig. 1f). Overall, we found strong colocalization of H3K4me2 with H2A.Z, H2Aub, and, to a lesser extent, with H3K27me3.
H2A.Z and H2Aub levels increase along with H3K4me2 in ldl3
The colocalization of H3K4me2 with H2A.Z, H2Aub, and H3K27me3 prompted us to investigate potential crosstalk among these chromatin marks. To determine whether H3K4me2 affects the distribution of H2A.Z, H2Aub, and H3K27me3, we perturbed the H3K4me2 pattern in plants using ldl3 mutants. Previous studies have shown that a defect in the histone demethylase LDL3 causes an increase in H3K4me2 levels in thousands of actively transcribed genes4,5.
We performed ChIP-seq for H3K4me2, H2A.Z, H2Aub, and H3K27me3 in WT and ldl3. We defined genes with increased H3K4me2 in ldl3 in both independent experiments as LDL3-target genes (n = 7115; Fig. 2a and Supplementary Data 1). Focusing on LDL3-target genes, H2A.Z and H2Aub levels increased in ldl3, especially in the gene body, as did H3K4me2 (Fig. 2b–d and Supplementary Fig. 2a, b). Approximately 87% and 80% of LDL3-target genes showed increased H2A.Z and H2Aub, respectively (Supplementary Fig. 2c). The increase in H2A.Z and H2Aub levels in ldl3 were proportional to that in H3K4me2 levels (Fig. 2e). In contrast, H3K27me3 levels were generally low in LDL3-target genes and hardly changed in ldl3 (Fig. 2b–e and Supplementary Fig. 2a–c). To examine whether the change in H2A.Z and H2Aub levels in ldl3 is due to changes in the transcription of their regulators, the expression of H2A.Z-related genes and H2Aub-related genes in ldl3 were compared with those in WT. The expression of H2A.Z-related genes (e.g., HTA8) and H2Aub-related genes (e.g., AtBMI1A) did not significantly change in ldl3 (Supplementary Fig. 2d). These results support the idea that the increase in H3K4me2 itself, rather than secondary effects of the ldl3 mutation, induces the accumulation of H2A.Z and H2Aub.
a–e ChIP-seq for H3K4me2, H2A.Z, H2Aub, and H3K27me3 in WT and ldl3. a Overlap of genes with increased H3K4me2 in ldl3 compared to WT (H3K4me2 change (ldl3–WT) > 2) between two biological replicates. The overlapping genes are defined as “LDL3-target genes (n = 7115)”. b Scatter plots showing the levels of H3K4me2, H2A.Z, H2Aub, and H3K27me3 comparing WT and ldl3. Each dot represents a protein-coding gene. Red dots indicate LDL3-target genes (n = 7115). c Box plots showing the changes in H3K4me2, H2A.Z, H2Aub, and H3K27me3 (ldl3–WT) in LDL3-target genes (n = 7115) and other genes (7115 genes were randomly selected from genes other than LDL3-target genes). The p-values from two-sided Mann-Whitney U tests are shown. d Metaplots showing the averaged distribution of H3K4me2, H2A.Z, H2Aub, and H3K27me3 in WT (black) and ldl3 (red) in LDL3-target genes (n = 7115) and all protein-coding genes (n = 27,443). e Box plots showing the relationship between the change in H3K4me2 and the change in H2A.Z, H2Aub, and H3K27me3 in ldl3 compared to WT. Histograms above the boxplots indicate the number of genes in each category of H3K4me2 change. The values of scatter plots, metaplots, and box plots represent RPKM. An additional series of the ChIP-seq analyses for (b and d) showed reproducible results (Supplementary Fig. 2a, b). Box plots in (c and e) display the median (center line), the interquartile range (IQR; box bounds), and the maximum and minimum values within 1.5 IQR (whiskers).
H2A.Z, H2Aub, and H3K27me3 levels decrease along with H3K4me2 in atx3/4/5
As a complementary approach, we next examined whether a decrease in H3K4me2 levels affects the localization of H2A.Z, H2Aub, and H3K27me3 using triple mutants of the H3K4 methyltransferases ATX3, ATX4, and ATX5. H3K4me2 drastically decreased in atx3/4/5, which is consistent with our previous study9. The 5,842 genes with decreased H3K4me2 levels in atx3/4/5 in both independent experiments were defined as ATX3/4/5-target genes (Fig. 3a and Supplementary Data 1). Notably, in ATX3/4/5-target genes, H2A.Z, H2Aub, and H3K27me3 levels also decreased in atx3/4/5 (Fig. 3b–d and Supplementary Fig. 3a, b), with 69% to 84% of the ATX/3/4/5-target genes showing these changes (Supplementary Fig. 3c). The decrease in H2A.Z, H2Aub, and H3K27me3 levels in atx3/4/5 were proportional to that in H3K4me2 levels (Fig. 3e). We confirmed that the expression of H3K27me3-related genes (e.g., CURLY LEAF (CLF)), H2A.Z-related genes, and H2Aub-related genes showed no significant or only minor changes in atx3/4/5 (Supplementary Fig. 3d).
a–e ChIP-seq for H3K4me2, H2A.Z, H2Aub, and H3K27me3 in WT and atx3/4/5. a Overlap of genes with decreased H3K4me2 in atx3/4/5 compared to WT (H3K4me2 change (atx3/4/5–WT) < –5) between two biological replicates. The overlapping genes are defined as “ATX3/4/5-target genes (n = 5842)”. b Scatter plots showing the levels of H3K4me2, H2A.Z, H2Aub, and H3K27me3 comparing WT and atx3/4/5. Each dot represents a protein-coding gene. Blue dots indicate ATX3/4/5-target genes (n = 5842). c Box plots showing the changes in H3K4me2, H2A.Z, H2Aub, and H3K27me3 (atx3/4/5–WT) in ATX3/4/5-target genes (n = 5842) and other genes (5842 genes were randomly selected from genes other than ATX3/4/5-target genes). The p-values from two-sided Mann-Whitney U tests are shown. d Metaplots show the averaged distribution of H3K4me2, H2A.Z, H2Aub, and H3K27me3 in WT (black) and atx3/4/5 (blue) in ATX3/4/5-target genes (n = 5842) and all protein-coding genes (n = 27,443). e Box plots showing the relationship between the change in H3K4me2 and the change in H2A.Z, H2Aub, and H3K27me3 in atx3/4/5 compared to WT. Histograms above the boxplots indicate the number of genes in each category of H3K4me2 change. The values of scatter plots, metaplots, and box plots represent RPKM. An additional series of the ChIP-seq analyses for (b and d) showed reproducible results (Supplementary Fig. 3a, b). Box plots in (c and e) display the median (center line), the IQR (box bounds), and the maximum and minimum values within 1.5 IQR (whiskers).
Although we previously showed that ATX3/4/5 are mainly responsible for H3K4me2, they also contribute to H3K4me39. To distinguish the effect of decreased H3K4me2 and H3K4me3 in atx3/4/5, we classified all protein-coding genes into four groups based on H3K4me2 changes and H3K4me3 changes in atx3/4/5: genes with a reduction in both H3K4me2 and H3K4me3 (group A; n = 1342), genes with a reduction in H3K4me3 only (group B; n = 1258), genes with a reduction in H3K4me2 only (group C; n = 6079), and genes with no reduction in either mark (group D; n = 18,764) (Supplementary Fig. 3e). The decrease in H2A.Z and H3K27me3 levels in atx3/4/5 was observed in group A and group C, but not in group B, indicating that the reduction in H2A.Z and H3K27me3 levels are primarily associated with the reduction in H3K4me2 rather than H3K4me3 (Supplementary Fig. 3f). On the other hand, the decrease in H2Aub levels was more prominent in group A than group C (Supplementary Fig. 3f), leaving open the possibility that H3K4me3 also plays a role in depositing H2Aub in ATX3/4/5-target genes. It should be noted, however, that group A genes also showed more reduction of H3K4me2 than group C genes (Supplementary Fig. 3f). Furthermore, the ldl3 mutant exhibits increased H3K4me2 levels, accompanied by increased H2Aub (Fig. 2b–e and Supplementary Fig. 2a–c), despite almost no change in H3K4me35, which strongly supports the conclusion that H3K4me2, rather than H3K4me3, promotes H2Aub deposition. Taken together, the results from the ldl3 mutant and the atx3/4/5 mutant suggest that H3K4me2 promotes the accumulation of H3K27me3 in a restricted manner at ATX3/4/5-target genes, while more consistently facilitating the accumulation of H2A.Z and H2Aub.
Alterations in H3K27me3, H2A.Z, or H2Aub levels do not affect H3K4me2
We then tested the converse effect. First, a quintuple mutant of genes encoding Jumonji-C-domain-containing proteins (JMJs) with H3K27me3 demethylase activity, i.e., JMJ30, JMJ32, JMJ11/EARLY FLOWERING 6 (ELF6), JMJ12/RELATIVE OF EARLY FLOWERING (REF6), and JMJ1324,25, and a single mutant of the gene encoding the H3K27me3 methyltransferase CLF26, were used to investigate whether H3K27me3 regulates H3K4me2. The jmj30 jmj32 elf6 ref6 jmj13 quintuple mutant (jmj quintuple) exhibited ectopic accumulation of H3K27me3, but H3K4me2 did not change (Fig. 4a, b, Supplementary Fig. 4a and Supplementary Data 1). In addition, the decrease in H3K27me3 levels in thousands of genes in clf did not cause any change in H3K4me2 patterns (Fig. 4c, d, Supplementary Fig. 4b and Supplementary Data 1). These results demonstrate that altering H3K27me3 levels does not affect the localization patterns of H3K4me2.
a Scatter plots showing H3K27me3 and H3K4me2 levels comparing jmj30 jmj32 elf6 ref6 jmj13 quintuple mutants (jmj quintuple) and WT. Each dot represents a protein-coding gene. Red dots indicate JMJ-target genes (n = 3048, as shown in Supplementary Fig. 4a). b Metaplots showing the averaged distribution of H3K27me3 and H3K4me2 in JMJ-target genes (n = 3048) in WT and jmj quintuple. c Scatter plots showing H3K27me3 and H3K4me2 levels comparing clf and WT. Each dot represents a protein-coding gene. Blue dots indicate CLF-target genes (n = 2160, as shown in Supplementary Fig. 4b). d Metaplots showing the averaged distribution of H3K27me3 and H3K4me2 in CLF-target genes (n = 2160) in WT and clf. e Scatter plots showing H2A.Z and H3K4me2 levels comparing pie1 and WT. Each dot represents a protein-coding gene. Blue dots indicate SWR1-C-target genes (n = 6096, as shown in Supplementary Fig. 5c). The regions from TSS to TSS + 500 bp were analyzed. f Metaplots showing the averaged distribution of H2A.Z and H3K4me2 in SWR1-C-target genes (n = 6096) in WT, arp6, and pie1. g Scatter plots showing H2Aub and H3K4me2 levels comparing WT and atbmi1a/b/c. Each dot represents a protein-coding gene. Blue dots indicate AtBMI1-target genes (n = 4982, as shown in Supplementary Fig. 6a). h Metaplots showing the averaged distribution of H2Aub and H3K4me2 in AtBMI1-target genes (n = 4982) in WT and atbmi1a/b/c. The values of scatter plots and metaplots represent RPKM.
To test the effect of H2A.Z on H3K4me2, we used the mutants of PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 (PIE1) and ACTIN-RELATED PROTEIN 6 (ARP6), which encode the components of SWI2/SNF2-Related 1 chromatin remodeling complex (SWR1-C) responsible for H2A.Z deposition27. As most H2A.Z peaks were in the TSS to TSS + 500 bp region of genes (Supplementary Fig. 5a), H2A.Z and H3K4me2 in this region were investigated. H2A.Z changes in arp6 and pie1 were highly correlated (Spearman’s rank correlation coefficient ρ = 0.888; Supplementary Fig. 5b); therefore, we defined SWR1-C-target genes (n = 6096) as the overlap of genes with decreased H2A.Z levels in both arp6 and pie1 (Supplementary Fig. 5c and Supplementary Data 1). Although H3K4me2 levels were slightly disturbed in SWR1-C-target genes in pie1 and arp6, the overall pattern of H3K4me2 was unchanged, and there was no clear correlation between H2A.Z changes and H3K4me2 changes in both arp6 and pie1 (Fig. 4e, f and Supplementary Fig. 5d, e).
To address whether a decrease in H2Aub levels affects H3K4me2 levels, we analyzed the effects of the loss of AtBMI1A, AtBMI1B, and AtBMI1C, which are components of the PRC1 complex responsible for the monoubiquitination of H2A14,28,29. We performed ChIP-seq for H3K4me2 and H2Aub in the atbmi1a atbmi1b atbmi1c triple mutant (atbmi1a/b/c). Despite the changes in H2Aub levels in AtBMI1-target genes, H3K4me2 levels did not change in atbmi1a/b/c (Fig. 4g, h, Supplementary Fig. 6a and Supplementary Data 1).
We also investigated the interplay between H2A.Z and Polycomb repressive marks. A previous study revealed that H3K27me3 decreases in pie116. We found that H2A.Z did not change in atbmi1a/b/c, jmj quintuple, or clf, indicating that Polycomb repressive marks do not affect the deposition of H2A.Z (Supplementary Fig. 6b–g). Furthermore, to clarify the crosstalk between H2Aub and H3K27me3, we performed H2Aub ChIP-seq in jmj quintuple and clf. H2Aub levels were unchanged in both mutants, suggesting that H2Aub deposition is independent of H3K27me3 (Supplementary Fig. 6h–k).
Overall, we found that H3K4me2 consistently colocalized with H2A.Z and H2Aub. These chromatin marks responded proportionally to the changes in H3K4me2 (Figs. 2, 3) but not vice versa (Fig. 4), demonstrating that they act downstream of H3K4me2.
H2A.Z oscillates in phase with H3K4me2 in the context of the diel oscillation
We showed that H3K4me2 regulates the accumulation of H2A.Z and H2Aub on chromatin. However, changes in H2A.Z and H2Aub levels in ldl3 and atx3/4/5 were lower than those in H3K4me2 (Figs. 2, 3), whereas H2A.Z and H2Aub strongly colocalized with H3K4me2 in WT (Fig. 1). This implies that there are unknown common factors upstream of H3K4me2, H2A.Z, and H2Aub, in addition to the regulation of H2A.Z and H2Aub by H3K4me2. Furthermore, the timescale over which H2A.Z and H2Aub change after changes in H3K4me2 is still unknown. To address these issues, we analyzed the diel oscillation of these chromatin marks in the WT plants. In A. thaliana, H3K4me2 levels were reported to show diel oscillation in core clock genes such as LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), and TIMING OF CAB EXPRESSION 1 (TOC1)6. At least in the core clock genes, H3K4me2 exhibits diel oscillation in antiphase with transcription6. This let us consider the possibility that H2A.Z and H2Aub also oscillate following H3K4me2 oscillation and that these chromatin marks synergistically regulate transcriptional oscillation. Because it has already been reported that H3K27me3 does not show diel oscillation in Arabidopsis species6,30, we focused on H3K4me2, H2A.Z, and H2Aub and investigated the genome-wide diel oscillation of these chromatin marks.
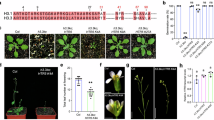
ChIP-seq for H3, H3K4me2, H3K4me3, H2A.Z, and H2Aub and mRNA-seq were performed using WT seedlings sampled at Zeitgeber Time 0 (ZT0), ZT6, ZT12, and ZT18 after growth for 14 days under a 12-h light/12-h dark cycle (Fig. 5a). We identified 781 genes with H3K4me2 oscillation (called “H3K4me2 diel oscillating genes (DOGs)”), 1180 genes with H3K4me3 oscillation (“H3K4me3 DOGs”), 345 genes with H2A.Z oscillation (“H2A.Z DOGs”) and 29 genes with H2Aub oscillation (“H2Aub DOGs”) (Fig. 5b and Supplementary Data 2). H3K4me2 DOGs significantly overlapped with H2A.Z DOGs, as well as H3K4me3 DOGs and mRNA DOGs (Fig. 5c). To dissect the temporal relationship between H3K4me2 and other chromatin marks, H3K4me2 DOGs were classified into four gene groups on the basis of the time when H3K4me2 had reached its maximum value: genes with ZT0-peak (n = 337), ZT6-peak (n = 149), ZT12-peak (n = 209), and ZT18-peak (n = 86) (Supplementary Data 3). For all ZT-peak groups, H2A.Z oscillated in phase with H3K4me2, whereas H3K4me3 and mRNA oscillated in antiphase with H3K4me2 (Fig. 5d). Consistent with this observation, the temporal change in H3K4me2 was positively correlated with that in H2A.Z and negatively correlated with that in H3K4me3 and mRNA for most of the H3K4me2 DOGs, including core clock genes (Fig. 5e and Supplementary Fig. 7). On the other hand, H2Aub oscillation was much weaker and not synchronous with H3K4me2 (Fig. 5d, e and Supplementary Fig. 7). These results uncovered the difference between H2A.Z and H2Aub in the diel oscillation, in addition to the tight temporal association of H2A.Z with H3K4me2.
a Sampling scheme for time-course ChIP-seq and mRNA-seq. b–e Analyses of time-course ChIP-seq and mRNA-seq in WT. b Overlap of genes with H3K4me2, H3K4me3, H2A.Z, and H2Aub oscillation between two biological replicates. These overlapping genes are defined as “H3K4me2 diel oscillating genes (DOGs)”, “H3K4me3 DOGs”, “H2A.Z DOGs”, and “H2Aub DOGs”, respectively. c Overlap between H3K4me2, H3K4me3, H2A.Z, H2Aub, and mRNA DOGs. Numbers at grid intersections indicate the actual number of overlapping genes; numbers in parentheses indicate the expected number of overlapping genes if the same number of genes is randomly chosen from all protein-coding genes (n = 27,443). Circle size indicates the similarity (Jaccard index) of the two gene sets. Asterisks indicate significant enrichment (Bonferroni-corrected p-value from one-sided Fisher’s exact test < 0.05). d Time-course changes in H3, H3K4me2, H3K4me3, H2A.Z, H2Aub, and mRNA for H3K4me2 DOGs (n = 781). H3K4me2 DOGs are classified into 4 groups based on the time when H3K4me2 reached its maximum. Chromatin mark and mRNA levels are standardized to a mean of 0 and a standard deviation of 1 across four time points for each gene. e Histograms showing correlations of temporal changes between H3K4me2 and others (H3, H3K4me3, H2A.Z, H2Aub, and mRNA) for H3K4me2 DOGs (n = 781). For each gene, the Pearson correlation coefficient is calculated between the levels of H3K4me2 and those of other chromatin marks or mRNA across four time points. f Violin plots showing the amplitude changes (atxr3 – WT) of H3, H3K4me2, H3K4me3, H2A.Z, and mRNA in H3K4me2 DOGs (n = 781) and H3K4me2 non-DOGs (n = 781; randomly selected from genes other than H3K4me2 DOGs). Amplitudes were calculated as the difference between the maximum and minimum values across four time points for each gene: for chromatin marks, amplitudes were defined as (maximum RPKM – minimum RPKM), whereas for mRNA, they were defined as (square root of maximum RPKM – square root of minimum RPKM). Box plots display the median (center line), the IQR (box bounds), and the maximum and minimum values within 1.5 IQR (whiskers). The p-values from two-sided Mann-Whitney U tests are shown. n.s.: not significant (p > 0.05).
The attenuation of H3K4me2 oscillation induces the mirrored attenuation of H2A.Z oscillation
H3K4me2 affects the localization of H2A.Z but not vice versa (Figs. 2, 3, 4e, f). Therefore, we speculated that the diel oscillation of H3K4me2 also regulates that of H2A.Z. An H3K4 methyltransferase, ARABIDOPSIS TRITHORAX-RELATED3 (ATXR3)/SET DOMAIN GROUP 2 (SDG2), has been reported to regulate the oscillation of H3K4me3 in core clock genes (e.g., CCA1, LHY, and TOC1)6,31. We noticed that H3K4me2 DOGs were enriched in genes with decreased H3K4me3 in atxr3 (Supplementary Fig. 8a, b). These findings imply that ATXR3 is involved in the diel oscillation of H3K4me2/3. Therefore, we performed time-course ChIP-seq and mRNA-seq using atxr3 mutants to change the diel oscillation pattern of H3K4me2 and investigate the effect on H2A.Z oscillation. Consistent with previous reports6,31, the oscillation of H3K4me3 in most core clock genes was attenuated in atxr3 (Supplementary Fig. 9). In addition to H3K4me3, H3K4me2 oscillation was weakened (Supplementary Fig. 9), likely reflecting the contribution of ATXR3 on H3K4me2 oscillation9,32. Importantly, while H3K4me3 and H3K4me2 oscillate in antiphase in WT, loss of H3K4me3 in atxr3 did not induce accumulation of H3K4me2 (Supplementary Fig. 9). On the contrary, H3K4me2 oscillation was attenuated in atxr3 (Fig. 5f and Supplementary Fig. 10a, b). Furthermore, the oscillation of H2A.Z and mRNA was significantly attenuated in atxr3 (Fig. 5f and Supplementary Fig. 10c, d). The loss of H2A.Z in atxr3 showed a higher genome-wide correlation with that of H3K4me2 at the same time than that of H3K4me3 in H3K4me2 DOGs (Supplementary Fig. 10e), indicating that H2A.Z oscillation is more strongly linked to H3K4me2 oscillation than H3K4me3 oscillation.
In contrast to atxr3, approximately 25% of H3K4me2 DOGs overlapped with LDL3-target genes, which did not represent significant enrichment (Supplementary Fig. 11a). We also performed time-course ChIP-seq using the ldl3 mutants. Focusing on H3K4me2 DOGs regulated by LDL3, the attenuation of H3K4me2 diel oscillation was not observed in ldl3 partially because both the maximum and the minimum H3K4me2 levels across the four time points significantly increased (Supplementary Fig. 11b). However, the ldl3 mutation led to changes in the spatiotemporal localization pattern of H3K4me2 in some core clock genes (e.g., LHY, CCA1, and TOC1), which in turn induced similar alterations in H2A.Z (Supplementary Fig. 11c). These results further strengthened the conclusion that H3K4me2 promotes the accumulation of H2A.Z not only in the static states but also in the dynamic states, such as diel oscillation.
Discussion
Through genetic analyses, here we demonstrated that H3K4me2 generally regulates the localization of H2A.Z and H2Aub, while influencing H3K27me3 accumulation at a subset of genes (Figs. 2, 3). We also showed that H3K4me2 exhibits diel oscillation even in genes other than core clock genes. Interestingly, H3K4me2 oscillated in phase with H2A.Z but in antiphase with H3K4me3 and mRNA (Fig. 5a–e). This result aligns with our observations in the static states and suggests that H3K4me2, rather than H3K4me3, influences H2A.Z dynamics. Furthermore, the attenuation of H3K4me2 oscillation in the atxr3 mutant also reduced H2A.Z oscillation (Fig. 5f). In addition, in the ldl3 mutant, H2A.Z oscillated while exhibiting an altered intragenic pattern similar to that of H3K4me2 in core clock genes (Supplementary Fig. 11). These results suggest that H3K4me2 has a strong impact on regulating H2A.Z.
Furthermore, despite the modest changes in H2A.Z levels in the mutants with altered H3K4me2 levels, H2A.Z exhibited strong co-oscillation with H3K4me2 (Fig. 5d, e). This result suggests not only that H3K4me2 regulates H2A.Z but also that shared regulatory factors may coordinate the regulation of both H3K4me2 and H2A.Z. The SWR1-C has been reported to be recruited to core clock genes by the Evening Complex, a key component of the Arabidopsis circadian clock33. These components may also recruit factors responsible for H3K4me2 methylation or demethylation.
Our findings indicate that H2A.Z, H2Aub, and H3K27me3 are controlled by H3K4me2; however, their regulation does not appear to be parallel because (i) H3K4me2 colocalizes much more strongly with H2A.Z and H2Aub than with H3K27me3 (Fig. 1), (ii) both H2A.Z and H2Aub increase in ldl3, but H3K27me3 does not (Fig. 2), (iii) H2A.Z and H2Aub positively regulate H3K27me3 levels15,16,17, but not vice versa (Supplementary Fig. 4a, b and Supplementary Fig. 6d–k), and (iv) H2A.Z exhibits diel oscillation synchronously with H3K4me2 whereas H2Aub and H3K27me3 do not6 (Fig. 5). The findings (i)-(iii) suggest that H3K4me2 primarily regulates H2A.Z and H2Aub, and H3K27me3 acts downstream of this module. H2A.Z and H2Aub within gene bodies are involved in the repression of inducible genes11,17,19,20,23, suggesting that the chromatin enriched with H2A.Z or H2Aub represents pre-repressive states. Furthermore, the finding (iv) implies that the dynamics of H2Aub and H3K27me3 may lag behind those of H2A.Z. Based on these observations, we propose a regulatory model regulating a stepwise establishment of repressive chromatin states. H3K4me2 first recruits H2A.Z, creating intermediate states (Fig. 6, middle). This is followed by the deposition of H2Aub, which primes repression, and finally by the accumulation of H3K27me3, establishing stable repression (Fig. 6, bottom). Conversely, H3K4me2 is removed by LDL3 on actively transcribed genes, leading to active chromatin states with H3K4me15 (Fig. 6, top). Therefore, H3K4me2 represents the bifurcation point between active and repressive states, the latter of which is further established by H2A.Z and Polycomb repressive marks.
In active states, LDL3 associates with Ser2/Ser5-phosphorylated RNA polymerase II (RNAPII S2P/S5P) and cotranscriptionally demethylates H3K4me2 to H3K4me1 (top). Under low transcriptional activity, H3K4me2 accumulates, promoting H2A.Z deposition and leading to intermediate states (middle). Subsequent accumulation of H2Aub, followed by H3K27me3 deposition, establishes stably repressed states (bottom).
The molecular mechanism by which H3K4me2 recruits downstream chromatin marks remains unknown, but ALFIN1-LIKE (AL) family proteins can be candidates, as they have been shown to bind not only to H3K4me3 but also to H3K4me2 in vitro34,35. Some AL members bind to ARP6, a component of SWR1-C, which incorporates H2A.Z into nucleosomes10,11,36,37. In addition, some AL proteins also bind to PRC1 components10,38. AL proteins are conserved in plants but are absent in both animals and fungi, which aligns with the idea that H3K4me2 negatively regulates transcription by promoting the deposition of H2A.Z and H2Aub in a plant-specific manner34. Further studies are needed to clarify the molecular mechanism by which H3K4me2 promotes the deposition of H2A.Z, H2Aub, and H3K27me3.
Although H3K4 methylation is highly conserved across eukaryotes, our findings suggest that its downstream transcriptional regulatory mechanisms have diverged between plants and animals. These results highlight the importance of H3K4me2 in regulating chromatin states and raise intriguing questions about how H3K4me2 influences its downstream long-term chromatin marks, which is conserved from animals but in plants used to memorize environmental stimuli.
Methods
Plant materials and growth conditions
Arabidopsis thaliana strain Columbia-0 (Col-0) was used as the wild type (WT). atx3-1 (GK-128H01), atx4 (SALK_060156), atx5 (SAIL_705_H05) and ldl3-1 (GABI_092C03) were previously described5,9. The jmj32-1 (SALK_003313), elf6-3 (SALK_074694), ref6-1 (SALK_001018), jmj30-2 (GK-454C10), and jmj13G (GK-113B06) were previously described24. Other mutants used in this study were described in the following previous reports: hta9-1 (SALK_054814), hta11-2 (SALK_031471)39, clf-28 (NASC: N639371)40, atbmi1a-1 (SALK_145041), atbmi1b (WiscDsLox437G06)28, atbmi1c-1 (SALK_148143)41, arp6-3 (WiscDS_Los289_29), and pie1-3 (SAIL_78_C11)42.
For ChIP-seq and mRNA-seq, seeds were sown on Murashige and Skoog (MS) plates and kept in the dark at 4 °C for a few days, then plants were grown for 14 days under long-day conditions (8 h dark/16 h light cycle) at 22 °C. For ChIP-seq in atbmi1a/b/c or time-course ChIP-seq and mRNA-seq, seeds were sown on MS plates and kept in the dark at 4 °C for 4 days, then plants were grown for 14 days under a 12-h light/12-h dark cycle at 22 °C.
Library preparation and sequencing for ChIP-seq
The enhanced chromatin immunoprecipitation (eChIP) approach43 was adopted in this study. eChIP-seq was performed with slight modifications to our previously described method5,44. ~ 0.6 g of seedlings were sampled, frozen with liquid nitrogen, ground into a powder using a mortar and pestle, and cross-linked with 15 mL of Fixation Buffer (phosphate-buffered saline (PBS) containing 1% formaldehyde, 1 mM Pefabloc SC (Roche), cOmplete proteinase inhibitor (Roche) and 0.3% Triton X-100) for 10 min at room temperature (RT). To stop the cross-linking reaction, glycine (final concentration: 200 mM) was added and incubated for 5 min at RT. The cell lysate was centrifuged at 5000 × g for 5 min at 4 °C, and the supernatant was discarded. The pellet was washed with 4 mL of ice-cold PBS, resuspended with 0.8 mL of Buffer S (50 mM HEPES-KOH (pH7.5), 150 mM NaCl, 1 mM Ethylene Diamine Tetraacetic Acid (EDTA), 1% Triton X-100, 0.1% sodium deoxycholate and 1% Sodium Dodecyl Sulfate (SDS)) containing cOmplete proteinase inhibitor, and incubated for 10 min at 4 °C. The chromatin in the cell lysate was fragmented by sonication using S220 Focused-ultrasonicator (Covaris) and milliTUBE 1 ml AFA Fiber (Covaris) with following settings (time: 20-25 min, peak power: 140, duty factor: 5.0, cycles/burst: 200 and avg. power: 7.0). After centrifuging at 20,000 × g for 3 min at 4 °C, the supernatant was discarded and the pellet was resuspended with Buffer F (50 mM HEPES-KOH (pH7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.1% sodium deoxycholate) containing cOmplete proteinase inhibitor. For the antibody reaction, 1-2 µL of antibodies were added to the chromatin solution, and this mixture was incubated overnight at 4 °C. The following antibodies were used; anti-H3 (ab1791, Abcam), anti-H3K4me1 (ab8895, Abcam), anti-H3K4me2 (ab32356, Abcam), anti-H3K4me3 (ab8580, Abcam), anti-H3K27me3 (MABI0323, Cosmo Bio), anti-H2A.Z (ref. 45) and anti-H2AK119ub (#8240, Cell Signaling) antibodies. The anti-H2AK119ub antibody was reported to recognize Arabidopsis H2AK121ub (K120, excluding the N-terminal methionine)28. The chromatin-antibody mixture was incubated with Dynabeads M-280 Sheep anti-Mouse IgG (Thermo Fisher Scientific) for anti-H3K27me3 or Dynabeads Protein G (Thermo Fisher Scientific) for other antibodies for 2 h at 4 °C with rotation. The beads were washed once with low-salt RIPA buffer (50 mM HEPES-KOH (pH7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate and 0.1% SDS) containing cOmplete proteinase inhibitor, twice with medium-salt RIPA buffer (low-salt RIPA buffer with NaCl concentration changed from 150 mM to 300 mM) or high-salt RIPA buffer (from 150 mM to 500 mM) containing cOmplete proteinase inhibitor, once with ChIP wash buffer (10 mM Tris-HCl (pH7.8), 250 mM LiCl, 1% Igepal CA-630, 1 mM EDTA, 1% sodium deoxycholate) and once with TE buffer. The chromatin was detached from the beads by adding ChIP elution buffer (10 mM Tris-HCl (pH 7.8), 300 mM NaCl, 5 mM EDTA, and 0.5% SDS) with 2 µL of RNase (10 mg/ml; NIPPON GENE) and incubating for 30 min at 37 °C. Then, 2 µL of Proteinase K (20 mg/ml; Thermo Fisher Scientific) was added and incubated for over 3 h at 65 °C. The immunoprecipitated DNA was purified using the Monarch PCR & DNA Cleanup Kit (New England Biolabs). The libraries for ChIP-seq were prepared using ThruPLEX DNA-Seq Kit (Takara Bio) and DNA Unique Dual Index Kit (Takara Bio) following the manufacturer’s protocol and purified using SPRIselect beads (Beckman Colter). The libraries were sequenced by the HiSeq X or NovaSeq X Plus sequencer (Illumina). Two biological replicates were analyzed using independently grown plants.
For time-course ChIP-seq, 0.6 g of seedlings were sampled at ZT0, ZT6, ZT12, and ZT18 (Fig. 5a). The procedure of ChIP-seq was the same as above.
Library preparation and sequencing for mRNA-seq
For mRNA-seq in WT, ldl3, and atx3/4/5, three seedlings were sampled for three biological replicates. A seedling was frozen with liquid nitrogen and ground into a powder using Automill (Token) with two stainless steel beads at 1600 r/min for 60 sec. Total RNA was purified using the RNeasy Plant Mini Kit (Qiagen). The libraries for mRNA-seq were prepared using KAPA RNA HyperPrep Kit (Roche) following the manufacturer’s protocol and sequenced by NovaSeq X Plus sequencer (Illumina).
For time-course mRNA-seq, three seedlings were sampled at ZT0, ZT6, ZT12, and ZT18 for three biological replicates. The procedure of mRNA-seq was the same as above.
Sequence data processing
The sequenced data of ChIP-seq was trimmed of adapters using Trimmomatic (v.0.39)46, then mapped to the TAIR10 reference genome using Bowtie2 (v.2.4.5)47. Multi-mapped reads were removed from mapped reads, and the number of reads on each gene was counted using the “coverage” command of BEDtools (v.2.30.0)48.
The sequenced data of mRNA-seq was trimmed of adapters using Trimmomatic (v.0.39)46, then mapped to the TAIR10 reference genome using STAR (v.2.7.9a)49. The number of reads on each gene was counted using the “coverage” command of BEDtools (v.2.30.0)48.
Heatmaps and metaplots were generated using deepTools (v.3.5.1)50. Genome browser views of ChIP-seq signals were displayed with Integrative Genomics Viewer (v.2.14.1)51. Other figures were generated using Python libraries: pandas (v.2.2.3), matplotlib (v.3.3.4), and seaborn (v.0.13.0). For the detection of H2A.Z peaks against H3, MACS2 (v2.2.7.1) with default parameters was used (Supplementary Fig. 5a)52.
Statistical analysis
The changes in gene expression levels between WT and ldl3 (Supplementary Fig. 2d) and WT and atx3/4/5 (Supplementary Fig. 3d) were tested with the Wald test using an R package DESeq2 (v.1.34.0)53. The enrichment between H3K4me2 DOGs, H3K4me3 DOGs, H2A.Z DOGs, H2Aub DOGs, and mRNA DOGs was evaluated with one-sided Fisher’s exact test with Bonferroni-correction using a Python library SciPy (v.1.12.0) (Fig. 5c). The enrichment between H3K4me2 DOGs and LDL3-target genes was evaluated with one-sided Fisher’s exact test using SciPy (v.1.12.0) (Supplementary Fig. 11a). Two-sided Mann-Whitney U tests were performed using SciPy (v.1.12.0) for evaluating the differences between two gene groups (Figs. 2c,3c, 5f and Supplementary Figs. 3f, 8b, 11b).
Definition of diel oscillating genes (DOGs) of chromatin marks and mRNA
H3K4me2 DOGs were selected with the following 2 criteria; When comparing any two time points (e.g., ZT0 and ZT6), (1) There was a genomic region within that gene where H3K4me2 was significantly enriched at one time point compared to another time point (likelihood ratio ≥ 1000), and (2) the H3K4me2 level (RPKM) in that gene at one time was 1.2 times higher than that at another time point. As for criterion 1, the “bdgdiff” command of MACS3 (v.3.0.1) was used to detect H3K4me2-enriched regions52. H3K4me3 DOGs, H2A.Z DOGs, and H2Aub DOGs were also selected similarly.
mRNA DOGs were selected with the following three criteria: (1) the expression level significantly changed across four time points (ZT0, ZT6, ZT12, and ZT18), with the adjusted p-value of the likelihood ratio test across four time points being < 10–5, (2) the mean expression (RPKM) across four time points was > 2, and (3) the fold change (max/min) was > 1.5. The likelihood ratio test was performed using DESeq2 (v.1.34.0)53. Two independent biological replicates were analyzed in parallel, and the selected genes in both replicates were defined as mRNA DOGs.
Sources of reanalyzed data
The following data was reanalyzed in this study: ChIP-seq for H3K4me1, H3K4me2, and H3K4me3 in WT, atx3/4/5, and atxr3 (NCBI: PRJNA732996)9.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
ChIP-seq and mRNA-seq data generated in this study have been deposited in the NIH BioProject database under accession number PRJNA1256242.
Code availability
Codes are available on GitHub (https://github.com/TakumiNoyori/Arabidopsis_H3K4me2).
References
Ruthenburg, A. J., Allis, C. D. & Wysocka, J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30 (2007).
Liu, Y. et al. H3K4me2 functions as a repressive epigenetic mark in plants. Epigenetics Chromatin 12, 40 (2019).
Liu, B. et al. The H3K4 demethylase JMJ1 is required for proper timing of flowering in Brachypodium distachyon. Plant Cell 36, 2729–2745 (2024).
Ishihara, H. et al. Primed histone demethylation regulates shoot regenerative competency. Nat. Commun. 10, 1786 (2019).
Mori, S. et al. Cotranscriptional demethylation induces global loss of H3K4me2 from active genes in Arabidopsis. EMBO J. 42, e113798 (2023).
Malapeira, J., Khaitova, L. C. & Mas, P. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 109, 21540–21545 (2012).
Wang, Y. et al. The Arabidopsis DREAM complex antagonizes WDR5A to modulate histone H3K4me2/3 deposition for a subset of genome repression. Proc. Natl. Acad. Sci. USA 119, e2206075119 (2022).
Chen, L.-Q. et al. ATX3, ATX4, and ATX5 Encode Putative H3K4 Methyltransferases and Are Critical for Plant Development. Plant Physiol. 174, 1795–1806 (2017).
Oya, S., Takahashi, M., Takashima, K., Kakutani, T. & Inagaki, S. Transcription-coupled and epigenome-encoded mechanisms direct H3K4 methylation. Nat. Commun. 13, 4521 (2022).
Su, X.-M. et al. ALFIN-like proteins link histone H3K4me3 to H2A ubiquitination and coordinate diverse chromatin modifications in Arabidopsis. Mol. Plant 18, 130–150 (2025).
Xu, L. et al. H3K4me3 binding ALFIN-LIKE proteins recruit SWR1 for gene-body deposition of H2A. Z. Genome Biol. 26, 1–18 (2025).
Kim, D.-H. & Sung, S. Polycomb-mediated gene silencing in Arabidopsis thaliana. Mol. Cells 37, 841–850 (2014).
Jamge, B. et al. Histone variants shape chromatin states in Arabidopsis. Elife https://doi.org/10.7554/elife.87714 (2023).
Yang, C. et al. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23, 1324–1329 (2013).
Zhou, Y., Romero-Campero, F. J., Gómez-Zambrano, Á, Turck, F. & Calonje, M. H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 18, 69 (2017).
Carter, B. et al. The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis. Plant Cell 30, 1337–1352 (2018).
Kralemann, L. E. M. et al. Removal of H2Aub1 by ubiquitin-specific proteases 12 and 13 is required for stable Polycomb-mediated gene repression in Arabidopsis. Genome Biol. 21, 144 (2020).
Yelagandula, R. et al. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158, 98–109 (2014).
Coleman-Derr, D. & Zilberman, D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8, e1002988 (2012).
Gómez-Zambrano, Á, Merini, W. & Calonje, M. The repressive role of Arabidopsis H2A.Z in transcriptional regulation depends on AtBMI1 activity. Nat. Commun. 10, 2828 (2019).
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M. & Jacobsen, S. E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10, R62 (2009).
Zilberman, D., Coleman-Derr, D., Ballinger, T. & Henikoff, S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature 456, 125–129 (2008).
Yin, X. et al. H2AK121ub in Arabidopsis associates with a less accessible chromatin state at transcriptional regulation hotspots. Nat. Commun. 12, 315 (2021).
Yamaguchi, N. et al. H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat. Commun. 12, 3480 (2021).
Yan, W. et al. Dynamic and spatial restriction of Polycomb activity by plant histone demethylases. Nat. Plants 4, 681–689 (2018).
Goodrich, J. et al. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51 (1997).
Choi, K. et al. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134, 1931–1941 (2007).
Bratzel, F., López-Torrejón, G., Koch, M., Del Pozo, J. C. & Calonje, M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859 (2010).
Li, W. et al. Overexpression of AtBMI1C, a polycomb group protein gene, accelerates flowering in Arabidopsis. PLoS ONE 6, e21364 (2011).
Nishio, H., Nagano, A. J., Ito, T., Suzuki, Y. & Kudoh, H. Seasonal plasticity and diel stability of H3K27me3 in natural fluctuating environments. Nat. Plants 6, 1091–1097 (2020).
Song, Q. et al. Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol. 20, 170 (2019).
Guo, L., Yu, Y., Law, J. A. & Zhang, X. SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 107, 18557–18562 (2010).
Tong, M. et al. The evening complex establishes repressive chromatin domains Via H2A.Z deposition. Plant Physiol. 182, 612–625 (2020).
Lee, W. Y., Lee, D., Chung, W.-I. & Kwon, C. S. Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 58, 511–524 (2009).
Liang, X. et al. Family-wide characterization of histone binding abilities of PHD domains of AL proteins in arabidopsis thaliana. Protein J. 37, 531–538 (2018).
Sijacic, P., Holder, D. H., Bajic, M. & Deal, R. B. Methyl-CpG-binding domain 9 (MBD9) is required for H2A.Z incorporation into chromatin at a subset of H2A.Z-enriched regions in the Arabidopsis genome. PLoS Genet. 15, e1008326 (2019).
Potok, M. E. et al. Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition. Nat. Commun. 10, 3352 (2019).
Molitor, A. M., Bu, Z., Yu, Y. & Shen, W.-H. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 10, e1004091 (2014).
Xu, M., Leichty, A. R., Hu, T. & Poethig, R. S. H2A.Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of H3K4me3. Development 145, https://doi.org/10.1242/dev.152868 (2018).
Doyle, M. R. & Amasino, R. M. A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol. 151, 1688–1697 (2009).
Bratzel, F. et al. Regulation of the new Arabidopsis imprinted gene AtBMI1C requires the interplay of different epigenetic mechanisms. Mol. Plant 5, 260–269 (2012).
Rosa, M., Von Harder, M., Cigliano, R. A., Schlögelhofer, P. & Mittelsten Scheid, O. The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 25, 1990–2001 (2013).
Zhao, L. et al. Integrative analysis of reference epigenomes in 20 rice varieties. Nat. Commun. 11, 2658 (2020).
Yabe, K. et al. H3K9 methylation regulates heterochromatin silencing through incoherent feedforward loops. Sci. Adv. 10, eadn4149 (2024).
To, T. K. et al. RNA interference-independent reprogramming of DNA methylation in Arabidopsis. Nat. Plants 6, 1455–1467 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Ramírez, F., Dündar, F., Diehl, S., Grüning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
The computations were partially performed on the NIG supercomputer at NIG, Japan. We thank Taiko To for kindly providing H2A.Z antibodies. We thank Nobutoshi Yamaguchi, Taku Sasaki, Taiko To, and Akihisa Osakabe for kindly providing mutant seeds. This work was supported by grants from Human Frontier Science Program (HFSP) (RGP0025/2021) to T.K., Japan Science and Technology Agency (JST) CREST (no. JPMJCR15O1) to H.K. and T.K., Japan Society for the Promotion of Science (JSPS) (nos. 21H04977 to H.K. and T.K., 23H00365 and 24K21268 to T.K., and 20H05913, 23K23565, and 25K02252 to S.I.).
Author information
Authors and Affiliations
Contributions
T.N., S.M., S.O., H.N., H.K., S.I. and T.K. conceived the study. T.N., S.M. and S.O. performed the experiments. T.N. conducted the analysis and made figures. T.N. drafted, and T.N., S.M., H.N., H.K., S.I. and T.K. edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Noyori, T., Mori, S., Oya, S. et al. H3K4me2 orchestrates H2A.Z and Polycomb repressive marks in Arabidopsis. Nat Commun 16, 11657 (2025). https://doi.org/10.1038/s41467-025-66645-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66645-4