Abstract
Prostate cancer is a highly heterogeneous disease, driven by genomic and transcriptional changes that impact disease progression and metastatic potential. The interplay between clonal evolution, transcriptional plasticity, and tumour microenvironment is, however, poorly understood. Here, we leverage and integrate single-nuclei RNA sequencing and whole-genome sequencing from 43 spatially distinct tumour samples from five patients with locally advanced prostate cancer to reconstruct clonal evolution trajectories and transcriptional changes driving metastasis at single-cell resolution. We find extensive clonal heterogeneity, including both monophyletic and polyphyletic metastatic dissemination, and ongoing clonal evolution in the primary tumour after metastatic spread. Metastatic seeding converges on disease trajectories involving both genomic and transcriptional changes, including androgen receptor independence and activation of estrogen-, WNT- and JAK-STAT- pathway activity, in spatially distinct areas. Our findings suggest an intricate interplay between clonal evolution and cellular plasticity driving metastatic seeding and point toward more integrative prognostic markers for improved patient management.
Similar content being viewed by others
Introduction
Prostate cancer is characterised by a prolonged disease course and substantial molecular and clinical heterogeneity1,2. While most patients do not experience fatal outcomes, approximately one in eight patients progresses to aggressive metastatic disease. High intra- and inter-tumoral genetic diversity complicates efforts to unravel the evolutionary trajectories driving metastasis. Effective management of high-risk primary prostate cancer requires a deep understanding of the intricate genetic, phenotypic, and tumour microenvironment (TME) factors that contribute to heterogeneity and metastatic potential, particularly in the context of multimodal therapy concepts, including neoadjuvant and adjuvant treatments as well as targeted systemic therapies.
Our understanding of prostate cancer evolution has primarily come from bulk genome sequencing studies. While most of these studies have focused on single-biopsy tumour material, multi-region bulk sequencing studies have revealed significant intratumoural heterogeneity linked to distinct tumour microenvironments3,4. Different metastatic seeding patterns have been identified from locally advanced prostate cancer, mainly involving monoclonal dissemination (a single primary subclone seeds all metastatic lesions), but multi-region sampling approaches have shown that polyclonal dissemination is not uncommon1,5,6,7.
While it is believed that high clonal diversity can serve as a strong indicator of future tumour progression2, the metastatic dissemination in prostate cancer is, nevertheless, often driven by a single dominant lineage3,8. The characteristic properties of seeding clones in the primary tumour are poorly understood, but both intrinsic and extrinsic factors play an important role, including tumour microenvironment (TME)9 and signalling pathway activities10. This highlights the need for more comprehensive approaches to unravel the intricate clonal architecture and population evolutionary dynamics of both primary and metastatic lesions.
The cancer cells in prostate cancer are thought to arise from a luminal epithelial cell of origin, situated with different cell types, including luminal, basal, hillock, and club cells, as well as stromal and immune cells11,12,13. Recent advancements in single-cell and spatial transcriptomic sequencing technologies have advanced our ability to study this cellular and molecular heterogeneity, providing insights into the spatial relationship between diverse genetic and phenotypic profiles of both tumour cells and TME in primary prostate cancer14,15,16,17. Despite these advancements, there has been a relative paucity of studies focusing on both the genomic and transcriptional evolutionary aspects of prostate cancer at single-cell resolution. Understanding the evolutionary dynamics of prostate cancer is crucial, as it can reveal the mechanisms driving tumour progression, metastasis, and resistance to therapy.
Here, we integrate single-nuclei transcriptomics with whole-genome DNA sequencing (WGS) from multiregional sampling to explore the evolutionary pathways of prostate cancer and elucidate convergent clonal and phenotypic routes and processes involved in metastatic dissemination.
Results
Pathology-guided whole genome sequencing and single-cell transcriptomics of advanced prostate adenocarcinoma
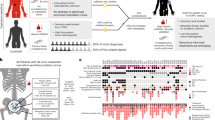
We examined 43 spatially distinct specimens from five patients with locally advanced high-grade prostate adenocarcinoma with a pathology-guided single-cell transcriptomics workflow to study tumour heterogeneity and metastatic potential (Fig. 1a). Four patients were treatment-naive, while patient PCAL25 received neoadjuvant androgen deprivation hormone therapy six weeks before surgery. Additionally, patient PCAL03 had synchronous bone metastasis (Supplementary Fig. 1A). Following radical prostatectomy and pelvic lymph node dissection, the whole prostate specimens were processed following a whole prostate cryopreservation approach18. Ten regions with high cancer cell content (>80%) were selected and marked in a topographic pathology report (Fig. 1b). Tissue punches were obtained from these areas and, along with corresponding regional lymph node metastasis samples (LNM), subjected to nuclei isolation, followed by single-nuclei RNA sequencing (snRNA) and low-pass bulk WGS (Fig. 1c). The spatial distances from the centre of all prostate samples per patient (centroid) to the analysed area were calculated based on the topographic pathology report.
a Five locally advanced prostate cancer patients with radical prostatectomy: PCAL37, PCAL34, PCAL25, PCAL10 and PCAL03. Created in BioRender. Weischenfeldt, J. (2025) https://BioRender.com/jglk9y9. Prostate organs were sliced according to the schematic. Prostate zones are coloured. PZ., Peripheral Zone (light orange); TZ., Transition Zone (purple); CZ., Central Zone (red); AFS, Anterior Fibromuscular Stroma (green); Tumour (grey); LNM, lymph node metastasis (brown). b Representative example of pathologist 3D prostate slices and areas selected for analysis (red dots). Prostate zones are marked in dashed lines, coloured as in (a), and tumour-containing areas in grey. c Single nuclei were extracted from fresh-frozen tissue punches and used for both snRNA-seq and bulk low-pass WGS. Right, spatial 3D representation of selected areas relative to the centroid of the prostate. Created in BioRender. Weischenfeldt, J. (2025) https://BioRender.com/5tar688. d UMAP showing the integrated data from all five patients. Semi-supervised integration was used to integrate datasets based on cell-type identity. Four major cell type clusters were present with the following cell type groupings: immune compartment (mast cells (light pink), MNPs (dark purple), T cells (dark pink) and B cells (coral)); stromal compartment (fibroblasts (light blue), endothelial cells (dark blue)); normal epithelial cells (luminal epithelial (LE, light green), club epithelial (CE, olive green), hillock epithelial (HE, yellow), basal (amber) and glial cells (mint green)); malignant epithelial cells (malignant (teal green), proliferating (light purple) and ciliated (orange)). e Barplot showing cell-type composition across areas for each patient, coloured by cell type as in (d). Tiles above show the number of cells captured per area, tumour content, TME entropy, distance from centroid, prostate anatomical zones, and Gleason grade. f Dot plot of cell-type specific marker genes. Dot size represents the percentage of cells within a group expressing the marker, and colour indicates scaled average expression within the cell group. Avg.exp., average expression. g Cell type variation. Left - bar plot of the Gini coefficient by cell type across all samples with cell types and number of samples for LE (43), Fibroblasts (35), Basal (10), Endothelial (34), MNPs (37), T cells (24), CE (6), Mast (6), B cells (13), HE (2), Glial (1), Malignant (43), Proliferating (22), Ciliated (10). Middle - box plot of cell type proportions, aggregated across all samples. Coloured by cell type as in (d). Right - tile plot of the percentage of samples in which each cell type is present. Panels (d–g) represent the integrated data of 356,860 single nuclei from 43 samples from all five patients. Box plots show the median (line), interquartile range (box), whiskers extending to the most extreme data points within 1.5× the interquartile range, and individual points beyond the whiskers representing outliers.
After quality-based filtering, including the removal of doublets, nuclei with high mitochondrial content, and ambient RNA (“Methods”), we retained more than 356,000 single nuclei from 43 samples (Supplementary Data 1). Each patient contributed between 5 and 9 areas and at least one LNM sample (Fig. 1d, e and Supplementary Fig. 1B). Malignant cells were predicted using Numbat19, and cell type assignment was determined by consensus among several reference-based cell-type prediction approaches, including scPred20 and SELINA21. As expected, the majority of cells captured (64.9%) were cancer cells (Fig. 1d). Two distinct cancer cell populations (Ciliated and Proliferating cells) comprising 0.46% and 1.0% of cancer cells, were defined by motile cilia markers (FOXJ1, DNAI1, DNAH11) and proliferation markers (MKi67), respectively (Fig. 1f). In addition, the proliferating cells exclusively expressed EZH2, a component of the polycomb repressive complex 2 (PRC2), which is implicated in lineage plasticity and cell cycle progression in prostate cancer22. We also detected normal luminal- (LE, 8%), hillock- (HE, 1.29%), club- (CE, 0.6%), and basal epithelial cells (0.1%). The immune compartment consisted of mononuclear phagocytic cells (MNPs, 3.04%), T cells (3.95%), B cells (2.2%) and mast cells (0.08%), while the stromal compartment consisted of fibroblasts (11%) and endothelial cells (3%) (Supplementary Data 2). The PCAL25 tumour exhibited a high degree of fibrosis, as indicated by a substantial stromal compartment contribution of 33.9%, compared to the other patients (median 9.27%, range 4.71–20.4%, Fig. 1e and Supplementary Fig. 1D). In contrast, PCAL37 contained the highest proportion of tumour cells (81%), except for the lymph node sample (55 tumour cells). This patient also exhibited minimal immune compartment contribution compared with the other patients (averaging between 5.9 and 13.5%). We found a high degree of heterogeneity at the tumour microenvironment (TME) level, both between and within patient samples, with TME entropy ranging from 0.3 to 2.7 (indicating low and high TME heterogeneity, respectively) (Fig. 1e). To estimate the spatial distribution of cell types, we computed their Gini coefficient (Fig. 1g), a metric for inequality and cellular heterogeneity. The Gini coefficients for most cell types were normally distributed across patients (Supplementary Fig. 1C), indicating similar variability in their spatial distribution among patients. Malignant and ciliated cells were evenly distributed across areas, as indicated by their low Gini coefficient (Gini coefficient 0=perfect equality, 1=perfect inequality), whereas LE and B cells were among the cell types exhibiting higher Gini coefficients, reflecting greater cellular heterogeneity.
Reconstructing tumour phylogenies at spatially distinct single-cell resolution reveals extensive multi-lineage branching
We examined intra-tumour heterogeneity at the genetic level by integrating our multi-region, high-resolution snRNA-seq and WGS data. To reconstruct the phylogenetic trees for each tumour, we first inferred copy number alteration (CNA) profiles for all areas. We integrated genomic segments from both area-specific snRNA-seq and bulk low-pass WGS data (Supplementary Figs. 2–4), to build phylogenetic trees representing local tumour evolution (Fig. 2a–c, and Supplementary Fig. 5). For comparison, we built phylogenetic trees from single-biopsy bulk-WGS data (Supplementary Fig. 5, 6, and Supplementary Data 3)23.
a Heatmap showing copy number gains and losses across the genome of PCAL10 (n = 46,404 cells). Gains are depicted in red and losses in blue, colour intensity corresponds to the level of copies lost or gained. Baseline, 2 N or 4 N, is neutral (Neu, white). The left side bars show clustering of cells, coloured by clone ID, area ID, and ploidy. 2 N, diploid; 4 N, tetraploid. b UMAP of all cancer cells for patient PCAL10 (n = 46,388 cells). Cells are coloured by clone ID as in A. Cells clustering together from the same area are circled, coloured by area ID as in (a). Tumour areas are LNM1 (dark red); T02 (red); T03 (orange); T04 (light orange); T05 (yellow); T06 (light green); T07 (teal); T09 (blue); T10 (purple). c Reconstructed phylogenetic trees of PCAL10 tumour. Top left, WGS-based tree from a bulk DNA single biopsy (dashed grey rectangle). Colour indicated the closest clone found in a single-cell tree. Right, multi-region scRNA-seq based tree. Branch length and the number on the branch indicate the number of events. Major events are marked, such as WGD (yellow star) and MSH2 locus deletions: focal deletion (chr2:42.8-47.9 Mbp, red arrow), large locus deletion (chr2:42.5-69.3 Mbp, orange arrow) and whole 2p arm (blue arrow). Seeding clones are represented with a dashed red rectangle. d Pathologist report for PCAL10 showing the clonal composition of cancer cells in each captured area. A lymph node is also shown, with an arrow pointing from a seeding clone. Subclones deriving from the same lineage are shown within the clone. Prostate zones are marked in dashed lines and tumour spread in grey. PZ., Peripheral Zone (light orange); TZ., Transition Zone (purple); CZ., Central Zone (red); AFS, Anterior Fibromuscular Stroma (green). e Mean pathway activity for each clone in each area (n = 46,388 cells) in colour scale from −1 (blue) to 1 (red). Clone colour marked as in c, the seeding clone is highlighted. f Box plots showing pathway activities in seeding (PCAL10-C2 clone, n = 24,412 cells, green) versus non-seeding cells (n = 19,073 cells, grey) for Androgen (P = 9 × 10–24) JAK-STAT (P = < 1 × 10–120), Oestrogen (P = 2.4 × 10–114), and WNT (P < 1 × 10–120). Statistical comparisons between seeding and not seeding cells were performed using a two-sided Wilcoxon rank-sum test. Effect size was estimated using rank-sum correlation. Asterisks show p values; δ, effect size. g Mean activities of selected transcription factors (TFs) per clone in colour range from −2 (blue) to 2 (red) in for clones separated by proliferation marker (pink), AR positive (yellow) or none (white), based on values in (e). Violin plot shows AR activity for each clone (n = 46,388 cells). The central line indicates the median; the box limits represent the interquartile range (IQR; 25th–75th percentiles); whiskers extend to the minimum and maximum values. Box plots show the median (line), interquartile range (box), and whiskers extending to the most extreme data points within 1.5×interquartile range. Clone colour marked as in (c).
All patient tumours exhibited extensive genetic heterogeneity at the single-cell level, with numerous intra-prostatic lineages (Supplementary Fig. 5). Our single-biopsy bulk-WGS tree structures were overall in agreement with the single-cell multi-region phylogenetic trees, but only captured a minor fraction of the clonal diversity. We note that the branch length of our high-resolution phylogenetic trees is based on the number of CNAs, and is, therefore, only a simplistic estimate of the actual evolutionary timing. In the case of PCAL37, clone PCAL37-C18 represented the most recent common ancestor (MRCA), characterised by copy number losses of chromosomes 8p (NKX3-1), 17p (TP53), 18, and smaller deletions on chromosomes 1q, 2p, and 4q, along with an 8q arm gain including MYC (Supplementary Fig. 5A, B). PCAL37 diverged into two main branches, one marked by a 19q13 amplification, which further evolved into multiple lineages. Additionally, we found support for a subclonal shattering of chromosome 19 in this clade (Supplementary Fig. 4). We identified several lines of evidence for convergent evolution, for example, with clones PCAL37-C11 and PCAL37-C6 both acquiring independent subclonal 12p (CDKN1B) losses. The PCAL37-C1 clone, responsible for metastatic seeding to the lymph node, constituted the majority of the PCAL37 tumour (cancer cell fraction (CCF) of 0.498). Areas T05 and T08, which were spatially close in the prostate (1 unit pairwise distance, with a median 2.6 units, and a standard deviation of 1.2 units for all areas from patient PCAL37), were also similar at the expression and clonal composition level (Fig. 1e, Supplementary Fig. 5A, B and 7A, B). Clones detected in more than one area were from areas in close spatial proximity, as expected. Interestingly, the MRCA clone (PCAL37-C18) was located at the basis of the prostate, far from the largest tumour lesion, suggesting extensive intra-tumour dissemination.
We found numerous subclonal and recurrent copy number gains in PCAL34 (Supplementary Fig. 5C, D). Truncal events included losses of chromosomes 8p and 13, 8q gain, and small deletions on chromosomes 2, 5, and 6. The earliest branching event was characterised by whole-genome doubling (WGD). The other clade remained diploid but further split into multiple lineages, all characterised by large genomic gains of chromosomes 1q21.2-44, 3, and 7. Tumour cells in area T09 contained both tetraploid and diploid clades while a distinct lineage, comprising clones PCAL34-C6 and PCAL34-C5, was restricted to area T06 (Supplementary Fig. 7C). This clone acquired additional copy number gains across the genome, including on chromosomes 3, 6, 12, 14, 17q, 19 and 21 (Supplementary Fig. 4). The observed convergent evolution of independent copy number gains of chromosomes 3 and 7 suggests a selective advantage in this prostate tumour. This is supported by the retention of four copies of these chromosomes after WGD, while most of the genome was reduced to a 3 N state (Supplementary Fig. 5D). The PCAL34-C14 clone from area T10 seeded the lymph node and acquired additional gains of chromosomes 5p and 14.
PCAL03 was the largest tumour (Supplementary Fig. 1A). Despite an early WGD event, PCAL03 surprisingly showed the lowest clonal diversity (Supplementary Fig. 5E, F). We found 8p deletion and gains of chromosomes 8q and 20q to be early events, with WGD followed by several whole chromosome losses, present in the majority of the clones, which was supported by our DNA-based WGS copy number profiles across the examined areas (Supplementary Fig. 3A). The largest clone PCAL03-C2 (CCF 0.579) spanning four areas (Supplementary Fig. 7E, F), acquired additional rearrangements on chromosome 11p, and gave rise to smaller subclones PCAL03-C3, PCAL03-C5 and PCAL03-C7, CCF of 0.043, 0.276 and 0.127, respectively (Supplementary Fig. 5E, F). A branching event gave rise to one leaf clone spanning areas T09 and T10 and another leaf giving rise to metastatic seeding, with an additional 4q focal gain and 7p loss following lymph node dissemination. We note that two different LNMs were subjected to single-biopsy deep WGS and to snRNA-seq from PCAL03. Interestingly, while the phylogenetic trees were comparable in the primary tumour, the single-biopsy WGS-based phylogenetic tree inferred a different seeding clone (Supplementary Fig. 7C), suggesting polyphyletic metastatic dissemination, where several distinct clones in the primary tumour independently seed metastases.
Acquired mismatch repair deficiency and JAK-STAT signalling in metastasising lineage
Genomic instability is associated with aggressive disease and metastatic competency24,25. We found a particularly striking example of copy number instability in PCAL10, with early truncal deletions of chromosomes 6q13-23.3, 8p, and 18, followed by WGD in one branch, which was not detected in the single-biopsy bulk WGS tree, while the other branch remained diploid (Fig. 2a, c). In the diploid lineage, clone PCAL10-C2, with the least percentage of genome altered (PGA), emerged as the largest clone (CCF = 0.459), spanning three areas with the highest proportion of proliferating tumour cells (Fig. 1, Supplementary Fig. 8). We also identified monophyletic seeding from PCAL10-C2 to the lymph node. Expression-based clustering identified T04 as the most likely metastatic seeding area (Fig. 2b–d). Interestingly, we found the lowest Gleason grade in this area (Fig. 1e), suggesting that pathology-based tumour grading should be accompanied by genomic-based analysis to identify clones with metastatic potential. A second branching event in the diploid lineage gave rise to clones with major genomic losses/gains, all confined to areas T09 and T10. Moreover, both areas contained clones from diploid and tetraploid lineages (Fig. 2d), pointing to local migration and intermixing of distinct clonal lineages within an individual prostate tumour. To further investigate somatic alterations in these regions, we performed deeper WGS on areas T02 and T09. Area T02, which contained the PCAL10-C2 clone, exhibited a biallelic deletion at the MSH2/6 locus (2p21-p16.3, expression log2 fold-change = −3.7, T02 versus T09) and concomitant increased levels of the DNA mismatch repair (MMR) mutational signatures SBS15 and SBS21, and a microsatellite instability (MSI) phenotype compared with area T09 (Supplementary Fig. 9).
WGD is a prevalent event observed across various cancer types and is strongly associated with genomic instability, metastatic disease and poor clinical outcomes26,27. Relying solely on bulk WGS makes it challenging to accurately determine the subclonal composition of a sample. For example, our multi-region WGS alone indicated area T05 to be 4 N, with subclonal single-copy losses of chromosomes 1, 3 and 20 (Supplementary Fig. 10), but our snRNA-seq-based clonality revealed two distinct cell clusters in almost equal proportions, with divergent loss of chromosomes 1 and 3 in clone PCAL10-C5 and chromosome 20 in clone PCAL10-C4.
Metastatic seeding is a crucial step in the progression of localised prostate cancer to lethal metastatic disease. Therefore, we aimed to identify pathway activity unique or shared across subclones within the entire tumour, as well as specific to the seeding clone, using decoupleR28. As expected, pathway activity was similar between clones derived from the same lineage within an area (Fig. 2e). For instance, clones PCAL10-C4 and PCAL10-C5 clustered together based on pathway activity, in contrast to PCAL10-C2 from areas T02, T04, and T06. We found enriched JAK-STAT activity, associated with lineage plasticity and resistance to Androgen Deprivation Therapy (ADT)29, exclusive to the PCAL10-C2 clone, regardless of the area (Fig. 2e, f). Notably, the seeding clone and metastatic clones (in the LNM) exhibited low androgen activity. Based on mean androgen activity, we grouped clones into androgen-positive and androgen-negative, which was further confirmed by inferred AR transcription factor (TF) activity (Fig. 2g and Supplementary Fig. 11A). AR-positive clones had higher NCOA3 activity, a steroid receptor coactivator30, while AR-negative clones were enriched in SHOX2 and MECP2, both of which promote cell growth and proliferation31,32. These findings suggest that AR-negative copy number unstable lineages can drive the clonal evolution towards metastatic seeding.
Early chromothripsis followed by polyphyletic metastatic seeding to regional lymph nodes
Chromothripsis is a one-off genomic shattering process, often encompassing simultaneous disruption of several cancer genes and associated with aggressive disease33. Although the clonality analysis included a limited number of samples (see Methods), our clonal reconstruction of PCAL25 primary tumour identified clone PCAL25-C3 in area T10 as the MRCA, with TP53 loss and chromothripsis of chromosome 6 (Fig. 3a and Supplementary Fig. 3C). We found the MRCA to branch off, leading to parallel multi-lineage dissemination to two regional lymph nodes, LNM1 and LNM2. Single-cell analysis revealed that metastatic tumour clades originated from polyphyletic clonal lineages within the primary tumour (Fig. 3b). In contrast, only a single monophyletic metastatic spread was detectable in the single biopsy bulk WGS tree (Fig. 3a). We observed two clades seeding LNM1, a direct descendant of PCAL25-C3 in T10, and another with acquired WGD in areas T02 and T05. This was followed by multiple subclonal amplifications in chromosomes 7, 10, 11, 13, and 20, as well as large deletions in chromosomes 2, 4, 9, 15, 16q, and 22q (Supplementary Fig. 5H). We traced a third metastatic seeding to LNM2 back to area T10 (Fig. 3a, b).
a Reconstructed phylogenetic trees of PCAL25 tumour. Top left, WGS-based tree from a bulk DNA single biopsy (dashed rectangle). Right, multi-region scRNA-seq based tree (see Fig. 2 for details). Dotted red line represents clones detected within the lymph node. WGD (yellow star) and chromothripsis (blue triangle) is shown. b Pathologist report for PCAL25 showing the clonal composition of cancer cells in each captured area. Both lymph nodes are shown; arrows point from an area most likely containing the seeding clone. Subclones deriving from the same lineage are shown within the clone. Prostate zones are marked in dashed lines and tumour spread in grey. PZ., Peripheral Zone (light orange); TZ., Transition Zone (purple); CZ., Central Zone (red); AFS, Anterior Fibromuscular Stroma (green). c UMAP of all cancer cells of patient PCAL25. Cells are coloured by clone ID as in (a). Cells clustering together from the same area are circled and marked. Tumour areas are LNM1 (dark red); LNM2 (red); T02 (orange); T05 (light yellow); T06 (light green); T08 (teal); T10 (purple). d Mean pathway activity for each clone in each area. Clone colour marked as in (a). e Mean activities of selected transcription factors (TFs) per clone in the area. Violin plot shows AR activity. The central line indicates the median; the box limits represent the interquartile range (IQR; 25th–75th percentiles); whiskers extend to the minimum and maximum values. Clone colour marked as in (a). f UMAP showing cancer cells in lymph node 1. Top - coloured by clone ID, bottom—by ploidy. 4 N, tetraploid; 2 N, diploid. g Genome view plots for cancer cells of LNM1 grouped by ploidy. Top - genome-wide copy number segmentation for 4 N and 2 N groups; bottom—genome-wide b-allele frequency (BAF) of each ploidy group. Window size 1 Mb; coloured by ploidy. h UMAP of LNM1 cancer cells coloured by cell clusters. Clone separated into three UMAP-based cell clusters #1, #2 and #3. Bottom violin plot shows gene expression counts (y-axis) of selected genes for each cell cluster and clone (x-axis). Panels c, d, e, n = 23,081 cells from PCAL25. Panels f, g, h, n = 803 cells from PCAL25.
Next, we examined common pathway activities for clones of different lineages within each lymph node and seeding clones in the primary tumour. Again, we found low androgen activity in seeding clones (Fig. 3c, d). The PCAL25-C3 metastatic lineage displayed transcriptional plasticity, with exclusive WNT activity in the metastatic LNM1, while AR and JAK-STAT pathway activity was increased in the other metastatic area, LNM2. In support, we found differential enrichment of TFs in each LNM (Fig. 3e, Supplementary Fig. 11B), with LNM1 and the seeding area having higher activity of SATB2 and TCF7, a TF negatively regulated by AR34. Both TFs are implicated in activating the WNT pathway and epithelial-mesenchymal transition (EMT)34,35.
Samples with low tumour purity are often excluded from WGS analysis, as low CCF can hinder the detection of somatic mutations and lead to inaccurate reconstruction of clonal structures36. Using our snRNA-seq approach, we estimated LNM1 CCF to be 0.172. Despite low tumour purity, we were able to detect multi-lineage seeding events. In total, we identified 14% of the tumour cells to be diploid (114 cells) and 86% of tetraploid lineages (689 cells) (Figs. 3f, g, 2N cells (PCAL25-C2; C3; C4) in orange. 4 N cells (PCAL25-C1) in blue), providing further support for WGD following the initiating chromothripsis event on chromosome 6. The single-cell resolution also allowed us to explore further clonal heterogeneity (Fig. 3h and Supplementary Fig. 12). We detected heterogeneous expression of long non-coding RNAs, implicated in prostate cancer tumorigenesis, such as ARLNC1 and SChLAP137. All 4 N clones exhibited higher expression of ONECUT2, a TF implicated in suppression of AR and progression to lethal disease38. Notably, PCAL25-C2 from the 2 N lineage was the most distinct from all LNM1 resident clones, while PCAL25-C3/C4 clones clustered together with a subset of C1 cells.
These findings suggest convergent transcriptional programmes across different lineages, including AR suppression, JAK-STAT and WNT activity, even with distinct genotypes.
The impact of spatial organisation on clonal evolution and transcriptomic profiles in primary prostate cancer
The clonal composition of adjacent areas was more similar than distant ones, as expected (Figs. 2, 3). We also found a positive correlation between the phylogenetic distance between tumour cells and the TME composition (P = 0.01, R = 0.25, Pearson correlation, Supplementary Fig. 13A). To complement our centroid-based distance analysis (Fig. 1), we also computed pairwise Euclidean distance between each tumour area, to investigate the spatial relationships between individual clones. Neither the phylogenetic distance nor TME were proportional to the pairwise distance between areas (Supplementary Fig. 13B, C), suggesting a connection between TME and tumour cell composition at shorter distances, but also that tumour and normal cell types follow non-linear distributions at larger distances within a prostate tumour. We observed tumour cells preferentially in the peripheral and central zones, with higher clonal diversity (entropy) in the central zone (Supplementary Fig. 13E, D). In contrast, the transition zone was occupied by fewer tumour cells with lower clonal diversity (Supplementary Fig. 13F), suggesting that clonal expansions may be more constrained in this region.
Our spatially resolved analyses implied a strong interplay between transcriptional programmes and the clonal evolution in prostate cancer. AR signalling is the most common driver for prostate cancer growth, but accumulating evidence suggests AR-independent transcriptional reprogramming can drive lineage plasticity in tumour cells29,39. Overall, pathway activities exhibited varying degrees of intra- and inter-tumour heterogeneity, with, for example, Oestrogen, androgen and JAK-STAT pathway activity having high and low intra-tumour heterogeneity, respectively (Gini index, Supplementary Fig. 14F), and with the majority of pathways exhibiting patient-specific activities (pairwise variance test, Supplementary Data 4 and Supplementary Fig. 15). For example, PCAL25 (ADT-treated) was among the tumours with the lowest AR activity, as expected. To investigate the contribution of transcriptional programmes to the clonal evolution and metastatic seeding areas across all patient samples, we integrated and compared TME components, tumour cell states, which represent gene modules derived from a pan-cancer single-cell RNA-sequencing study40 including prostate cancer, and key pathways within and between patient samples using principal component analysis (PCA) (Fig. 4a). We found PC1 (20.79%) to be explained primarily by (positively correlated with) tumour content and negatively correlated with MNPs, TME diversity, Grade group and T cell proportion. PC2 (12.66%) was explained primarily by lymphocyte content and normal luminal prostate cells. The presence of areas with low tumour content, high immune cell content and high Gleason grade (Fig. 4a and Supplementary Fig. 16) suggests immunologically ‘warm’ areas in the prostate with tumour-immune interactions driving more aggressive cell phenotypes. High immune cell content also correlated with MSH2 homozygous deletion, whereas TP53 loss (17p) and MYC gain (8q) were both linked with immune cell content depletion and low JAK-STAT signalling (Supplementary Fig. 16B), supporting the previously established immune suppressive roles of TP53 and MYC mutations in cancer41,42. We also found a strong contribution of proliferating tumour cell content and JAK-STAT activity in PC2 (Fig. 4a). JAK-STAT activity, which we also found elevated in the seeding area of PCAL10 (Supplementary Fig. 14C–F), can drive immune evasion and increased lineage plasticity in prostate cancer in an AR-independent manner29. Changes in EMT signalling and mast cells correlated with PC3, whereas PC4 was mainly explained by ciliated tumour cells, endothelial and fibroblasts, as well as metal response, which is linked with homoeostasis, cell differentiation, and proliferation40. PC5 was explained by stress signalling and tumour areas towards the periphery of the tumour (high centroid distance).
a Features of each primary tumour area (n = 37) across all patients (n = 5) used for PCA (grey box, left). Heatmap of Pearson correlations between the first seven principal components (PCs; each explaining >5% variance) and the features included in the PCA, along with pathways activity, genomic aberrations and seeding information per area. Features marked with an asterisk (*) denote those used for the PCA. Right, PCA plot of the first two principal components. Each dot represents an individual sample, colour-coded by patient, with grey shading indicating tumour content. Patients are PCAL03 (pink); PCAL10 (light pink); PCAL25 (yellow); PCAL34 (green); PCAL37 (light blue). b Heatmap displaying significant correlations marked by an asterisk (P < 0.05, Pearson correlation) for seeding areas (n = 43,834 cells), proliferating (n = 2391 cells) and ciliated cells (n = 1075 cells). Pearson correlation coefficients (including those for Androgen activity) are displayed between these tumour-area metrics (rows) and the selected features (columns). c Schematic representation of tumour cell seeding to the lymph node, highlighting key cell types and pathways: ciliated cells (green cell); Proliferating cells (blue cell); upregulated AR (pink cell); upregulated WNT and oestrogen and downregulated AR (red cell).
Interestingly, we found metastatic seeding areas correlating with PC6 and PC7 together with ciliated and proliferating tumour cells. To further characterise these cells, we investigated their spatial properties and key signalling pathways. We found ciliated cells to be located preferentially near the centre of the prostate gland (anti-correlation with centroid distance), whereas the metastatic seeding areas were located preferentially towards the periphery (Fig. 4b, Supplementary Fig. 16). Ciliated tumour cells were also enriched for TRAIL (Tumour necrosis factor-related apoptosis-inducing ligand) pathway and depleted for WNT signalling. In agreement, WNT signalling is known to repress TRAIL-induced apoptosis and immune response modulation43.
Hormone signalling plays a pivotal role in prostate cancer dissemination. Whereas androgen response genes were reduced at the transcriptional level in the metastatic seeding area, we found significant enrichment of oestrogen pathway response genes (P = 0.017, R = 0.39, Pearson correlation, Fig. 4b), which are important for progression in primary untreated prostate cancer44,45. We also found seeding areas to exhibit a trend towards increased WNT and TRAIL signalling, both markers of cell plasticity.
These findings imply spatially distinct areas with different clonal and TME compositions, and suggest that seeding clones, phylogenetically closely related to the metastasis, are localised in spatially confined regions in the primary tumour. These regions tend to localise towards the periphery of the prostate, populated by tumour cells with elevated levels of oestrogen and WNT signalling and reduced androgen pathway signalling (Fig. 4c), accompanied by extensive and recurrent rewiring of transcriptional programmes in primary tumour cells.
Discussion
Metastasis is the single most important clinical event in the progression of prostate cancer. However, the characteristics of primary tumour cells with metastatic potential have been difficult to study due to the inherent heterogeneity of the disease. Here, we have approached this key question through a detailed investigation of the tumour cell types, TME composition, transcriptional changes and their spatial relationship at the single cell level. Reconstructing phylogenetic trees revealed both monophyletic and polyphyletic dissemination, with instances of several metastatic tumour clades from polyphyletic clonal lineages within the primary tumour. We find tumour areas that are phylogenetically closely related to the metastasis to exhibit distinct transcriptional and genomic characteristics, including androgen independence. This suggests that seeding clones are located in spatially confined regions, often located toward the periphery of the prostate. Despite our comprehensive multi-region sampling approach, we acknowledge that the spatial resolution of our sampling is, nevertheless, limited, and that the presence of metastatic clones in a given region does not exclude their existence elsewhere in the tumour. In extension, spatial relationships within a prostate tumour can be convoluted, following non-Euclidean paths, and additional analyses are warranted to explore and verify this finding. We also found that single biopsy bulk-WGS, the current standard for clonal evolution analysis, only captures a fraction of the tumour heterogeneity, including metastatic seeding areas. Combined with our finding that Gleason grade does not always correlate with metastatic potential, as supported by prior studies3,46 this has implications for clinical management of prostate cancer. Our findings imply that single-area biopsies assessed by genome sequencing and/or pathology grading will be suboptimal in detecting locally advanced disease. An important goal in prostate cancer genomics is to identify molecular alterations that can distinguish high-risk patients who require definitive treatment. We found extensive clonal divergence in all five analysed patients, with occasional polyphyletic seeding, and minimal convergence in somatic driver aberrations in the seeding clones. Although methods involving single-cell and spatially resolved analyses of prostate cancer genomics are gaining momentum, they are currently limited to academic research due to cost and lack of clinical evidence. Our results suggest that the current standards of single-biopsy panel sequencing or shallow WGS are likely to miss important metastatic seeding events, and that spatially resolved analyses may have clinical benefits. Moreover, the androgen-independent seeding may have direct implications for ADT treatment efficacy, as these cells are likely to harbour pre-existing resistance mechanisms against systemic treatment.
We also find substantial evidence for lineage plasticity as a key step in metastatic potential, including suppression of AR signalling and up-regulation of JAK-STAT, oestrogen-like signalling and the WNT pathway. Androgen independence is known to be an important step in disease progression, but has been identified primarily in castration-resistant prostate cancer (CRPC)47. Interestingly, a recent study identified four major subtypes, including a WNT- subtype, present in 5–7% of CRPC, and involving the transcription factor TCF739, a key cancer driver of lineage plasticity48—also expressed in WNT-activated cells in PCAL25. We note that this patient received prior ADT, which could impact the cellular plasticity towards a WNT-subtype. The identification of key characteristics of primary tumour regions with metastatic potential holds promise for integrative biomarker discovery. Lineage plasticity can give rise to distinct tumour cell populations and increase the ability of the tumour to change and adapt to treatments. We also identified two rare and poorly characterised cancer cell states40; ciliated and proliferating. The latter were characterised by high EZH2 and low AR expression, and a near-uniform spatial distribution, suggesting that they may represent a rare but important subtype of cancer cells. TME, such as hypoxia and stress, correlated with the presence of ciliated cells40. However, their significance in prostate cancer is poorly described, although ciliated epithelial cells have been reported in cribriform prostate cancer49. It is conceivable that the ciliated cancer cell state is an alternative differentiation endpoint triggered by a harsh local TME50. Future studies, including functional assays, will be needed to elucidate the function and relevance of these cells in prostate cancer.
While our study provides a step-change in our understanding of the genetic heterogeneity and transcriptional plasticity leading to intra-tumour heterogeneity and metastatic dissemination, it is not without limitations. First, while our study investigated more than 350,000 cells from 43 tumour areas, larger and better-powered study cohorts will be needed for validation. We find evidence for transcriptional plasticities across the cohort, but note that our cohort is not powered to draw broader conclusions on the prevalences of e.g. JAK-STAT-, Oestrogen- and WNT signalling. Second, additional transcriptional programmes and clonal evolution patterns not present in our cohort are likely to contribute to driving metastatic dissemination, e.g. stem-like and neuroendocrine programmes39. While CNAs were sufficient to identify the majority of the clonal heterogeneity, using single-nucleotide variants would provide even greater resolution, in particular for copy-number-stable tumours. Third, we used locoregional lymph nodes as metastatic sites to investigate dissemination patterns. Distant metastatic organs such as bone and liver are considered optimal proxies for aggressive metastatic disease. However, a recent study on genetic heterogeneity from multi-region primary and metastatic sites found that locoregional lymph nodes represented a good marker for aggressive disease1.
Future therapeutic interventions will need to address the intra-tumour heterogeneity of the disease. Prostate cancer is considered an immunologically ‘cold’ tumour, and clinical trials with immunotherapies have been disappointing51 in the metastatic setting. We found areas with low tumour content and high Gleason grade to be infiltrated with lymphocytes, suggesting the presence of immunologically warm areas inside the prostate tumour. We also observed tumour areas containing clones with homozygous MSH2 loss with higher lymphocyte infiltration. This finding aligns with previous studies52, which highlight the potential for immunotherapy for prostate cancer patients with MSH2-mutated tumours. Moreover, the extensive heterogeneity also implicates a high likelihood of resistant clades of tumour cells. Our study emphasises both the need to sample multiple regions of prostate tumours and motivates integration of both genetic and transcriptional biomarkers to identify and target clones with metastatic competency.
Methods
Patient material
Sample collection, consent and clinical data
Five prostate cancer patients with high-risk, locally advanced primary prostate adenocarcinomas were selected from the German ICGC cohort23. Written informed consent and ethical approval from the patients were obtained in accordance with ICGC guidelines. The study was approved by the Ethics Committee of the Ärztekammer Hamburg (Hamburg Medical Association) under protocols PV3552 and PV4679. The study followed the Declaration of Helsinki guidelines and did not use statistical methods to determine sample size due to its exploratory nature. All patients underwent radical prostatectomy with lymph node dissection as the primary treatment for prostate cancer, except for patient PCAL25, who received neoadjuvant androgen deprivation therapy (Gonadotropin-Releasing-Hormone (GnRH) analogue) 6 six weeks before surgery. Patient PCAL03 presented with oligometastatic disease with bone metastasis at the time of surgery. Two different synchronous LNMs were obtained from this patient, one for the initial single biopsy WGS and another LNM for multi-region snRNA-seq.
After surgery, following a predefined scheme as described in Gerhauser et al.23, specialised pathologists dissected each prostate. A single biopsy was isolated and subjected to deep bulk WGS23. The precise location of this sample in the 3D coordinate system of the prostate is unknown. For multi-region sample-isolation, tissue blocks were embedded in a tissue-freezing medium, cryopreserved and stored at −80 °C. 10 punches (1.5 mm in diameter) were taken from different areas of the primary tumour and matching lymph node metastases from each patient. In total, we obtained 66 samples: 50 primary tumour biopsies, 16 lymph node biopsies. The selected tumour areas had more than 80% of tumour cell content, according to the pathologist. Gleason grading was reported for each biopsy.
Single-nuclei transcriptomics
Tissue dissociation
Fresh-frozen prostate cancer tissue punches were processed using an adapted version of Pfisterer et al. protocol53. Shortly, frozen tissue punctures were mechanically dissociated using dounce-homogenizer: 5–10 strokes with pestle A, following 20–25 strokes with pestle B, in an ice-cold homogenization buffer (HB): NIM with 1 mM DTT (Invitrogen), 1X Protease inhibitors (Roche), 0.4 U/µl RNAse inhibitors (Takara), 0.2 U/µl Superasin (Invitrogen) and 0.1% (v/v) Triton X-100. To make HB we first made a nuclei isolation medium (NIM): 250 mM sucrose, 25 mM KCl, 5 mM MgCl2, and 10 mM tris buffer, pH 8. Homogenised tissue suspensions were filtered through a 40 µm mesh filter and centrifuged at 1000 × g for 8 min, 4 °C. To remove debris, samples were subjected to equilibrium density centrifugation: sample pellets were first resuspended in 250 µl HB and then mixed 1:1 with 50% iodixanol solution (50% v/v OptiPrepTM (Sigma), 25 mM KCl, 5 mM MgCl2, 10 mM tris buffer, pH 8.0, 1 mM DTT, 1X Protease inhibitors, 0.4 U/µl RNAse inhibitors, 0.2 U/µl Superasin). 500 µl of the resulting suspension was layered over 500 µl 29% iodixanol solution (29% v/v of 50% iodixanol, diluted with NIM, supplemented with DTT (1 mM), Protease inhibitors (1X), RNAse inhibitors (0.4 U/µl), Superasin (0.2 U/µl)) in a 2.2 ml Ultra-Clear Centrifugation Tube (Beckman Coulter). Samples were centrifuged at 14,000 × g for 22 min, 4 °C. Then, the remaining nuclei pellet was resuspended in 1X PBS, supplemented with 2.5 mM MgCl2, 1 mM DTT, 0.50% BSA and 0.2 U/µl RNAse inhibitors. Sample quality was assessed under a microscope and nuclei were counted with a hemocytometer.
snRNA-seq library preparation and sequencing
Single-nuclei suspensions containing 16,000 nuclei were loaded to the Chromium NextGEM chip (10X genomics), aiming to capture 10,000 nuclei per area. snRNA-seq libraries were constructed either with Chromium Next GEM Single Cell 3′ v3 or v.3.1 Reagent Kit, following manufacturer’s protocol. 10X single-nuclei libraries were sequenced on Illumina NovaSeq-6000 platform, using NovaSeq S4 flow cell with paired-end 150 bp mode (28 × 10 x 10 × 281 bp configuration), at the Department of Genomic Medicine at Rigshospitalet, Denmark. We sequenced on average 46,000 reads (range 22.5K-92K) per cell (Supplementary Data 1).
snRNA-seq processing
We used 10X Cell Ranger software (version 5.0.0) with default parameters for alignment to human genome reference hg38 (including intronic sequences), demultiplexing, barcode processing and gene x cell count quantification. To remove technical artefacts from ambient RNA contamination, CellBender54 (version 0.2.0) was used with the following parameters: 10,000 expected cells, 25,000 total droplets included, a low count threshold of 15, and all remaining parameters set to default. In samples with higher background RNA contamination, the low count threshold parameter was increased accordingly.
The resulting corrected and raw gene x cell count matrices were analysed with scCustomize package (R 4.2.0) to create Seurat objects (version 4.3.0). Cells with fewer than 500 genes and genes present in fewer than five cells were excluded from further analysis. The SCP package (version 0.5.1, available from: https://github.com/zhanghao-njmu/SCP) was used to compute quality control metrics and perform initial filtering. Doublets were predicted using scDblFinder (available from: https://github.com/plger/scDblFinder) with default parameters, and cells with mitochondrial transcript content exceeding 1% were excluded.
An additional filtering step was implemented to remove low-quality or dying cells, based on the assumption that mitochondrial reads should be minimal in single-nuclei data. Apoptosis scores for each cell were calculated as the ratio of mitochondrial percentage to total RNA counts. The mean apoptosis score was then calculated for each cluster, and clusters with a mean apoptosis score exceeding one standard deviation above the mean were excluded from further analysis.
The filtered and corrected snRNA-seq data were normalised using the “SCTransfrom” function from Seurat (version 4.3.0) with vst.flavour = “v2”, while regressing out mitochondrial percentages, keeping all other parameters at their default settings. We used the “FindVariableFeatures” function to identify 3000 variable genes, the “RunPCA” function for dimensionality reduction with npcs = 20, and the “FindNeighbors” function to identify nearest neighbours, exploring a range of resolutions between 1 and 2. Dimensionality reduction was further performed using the “RunUMAP” function based on the first 20 principal components. All the above functions are from the Seurat package (version 4.3.0), with default settings for the remaining parameters.
Cell annotation and Integration
scPred, version 1.9.220 was used for single-nuclei preliminary data annotation with publicly available prostate cancer datasets49,55. SELINA.py tool, version 0.121, with “--disease” mode and default parameters, was used for cell-type prediction. We labelled cell types using each dataset as a reference, which included a literature-based reference set13,16,56. In addition, “normal prostate” and “NSCLC” references, provided by SELINA, were used for prediction. For cell cluster identity assignment, we took the consensus of all predictions, except for the assignment of proliferating and ciliated cells. We calculated the G2M score and a custom literature-based ciliated gene-set using JASMINE (v1, available from: https://github.com/NNoureen/JASMINE). Cells that had a high ciliated or proliferating score (>threshold=median+one standard deviation) were assigned as ciliated or proliferating, respectively. To integrate all patient data, we utilised STACAS, version 2.2.1, a semi-supervised method developed by Andreatta et al.57. We employed cell type annotation as input, using the default parameters. Cancer cell fraction was subset, normalised with “SCTransform” normalisation (Seurat v5) on raw counts, and clustered for visualisation.
Pathway, transcription factor activity and cell state scores analysis
We computed pathway activity inference on single-cell count data with decoupler-py28, version 1.8.0, together with a curated collection of pathways from PROGENy58 and transcription factor (TF) database from CollecTRI59. For each pathway set, we used the top 500 genes ranked by p-value and Multivariate Linear Model (mlm) method for pathway activity analysis. TF enrichment scores were inferred by running the Univariate Linear Model (ulm) method. The resulting activity scores represent the predicted regulatory activity of each pathway or TF in individual cells, based on the expression of their downstream targets. These scores are relative within each dataset, making them comparable between patient samples. Scores reflect the direction and magnitude of pathway activity compared to other cells in the same patient’s sample, rather than providing an absolute measure across patients. Mean pathway/TF activity per cell group was calculated and visualised with pheatmap using R version 4.3.3., python version 3.9.16.
Cell state scores were calculated on normalised single-cell expression data using the AddModuleScore function from Seurat (version 4.3.0.1). Gene sets defining cell states were obtained from Barkley et al.40. We calculated the proportions of cells exhibiting cell state activity per sample, using two different cutoff strategies based on the overall distribution of activity values. For each area and each cell state, if the mean percentage of active cells for a given signature was <70%, a binary cutoff of 0 was applied. Otherwise, the signature’s mean activity was used as the threshold.
Gini index, Shannon entropy, weighted phylogenetic dissimilarity, cell population dissimilarity and physical distance
We assigned each biopsy sample a 3D coordinate on a Cartesian map and calculated pairwise Euclidean distances. The pathology-provided tumour diameter was used to estimate the unit conversion, with 1 unit corresponding to 1 cm. The resulting pairwise distances were as follows: PCAL37 (median: 2.6 units, sd: 1.2), PCAL10 (median: 2.8 units, sd: 1.4), PCAL03 (median: 3.6 units, sd: 1.7), PCAL25 (median: 3.0 units, sd: 1.4), PCAL34 (median: 2.9 units, sd: 1.3). For each patient, we calculated the geometric centroid of the prostate gland based on the 3D coordinates of its biopsies and we then calculated the Euclidean distances from each biopsy to the geometric centroid.
To quantify the distribution of cell types across samples, we calculated the Gini coefficient for the proportions of each cell type across all samples using the ineq R package (version 0.2-13) with the parameter type = “Gini”. Gini coefficients for each cell type were calculated on a per-patient basis using the same package and parameters.
We developed a pairwise clonal phylogenetic distance metric, termed the weighted phylogenetic distance, to assess clonal heterogeneity between two samples from the same patient. This metric ranges from 0 to 1, where a score of 0 indicates identical clonal composition and a score of 1 reflects no similarity in clonal composition between samples. The dissimilarity score between sample pairs was computed as follows: first, we calculated the normalised absolute difference in their clone proportions. These differences were then weighted by their phylogenetic distances, considering only clones present in both samples. The weighted differences were summed, and the resulting dissimilarity matrix was normalised by the product of the number of clones present in each sample.
To assess the pairwise cell type dissimilarity, we calculated the inverted Jaccard index between pairs of samples. The Jaccard index was computed for each pair of cell type composition vectors as follows: the intersection of the cell types present in both samples was divided by the union of cell types present in either sample, yielding a value between 0 and 1. A value of 0 indicates complete similarity, while a value of 1 indicates no shared cell types. To obtain a dissimilarity measure, the Jaccard index was inverted such that higher values represented greater dissimilarity. The pairwise Jaccard indices were calculated for all combinations of samples.
Shannon entropy was calculated to assess the diversity of TME cell types and clonal proportions per sample. The entropy was computed on the frequency distribution of all TME cell types and clone proportions for each sample, using the entropy function (version 1.3.1) (base-2 logarithm) and rounding the values to two decimal places.
CNA analysis and phylogenetic tree building
We used Numbat, version 1.3.2-119 with default parameters to classify tumour cells. Heterozygous germline SNPs, previously obtained from deep-WGS23, were used for genome phasing. Numbat was then applied again to identify CNAs from the tumour cells. To remove artefact CNAs, we utilised a “segs_consensus_fix” mode, for which we obtained high-confidence genome segments, either losses or gains, from low-pass WGS, with coordinates adjusted from deep-WGS, or area-specific high-confidence CNAs from the second Numbat run. To resolve areas with mixtures of two or more distinct clones, we further subset the data and ran Numbat “segs_consensus_fix” for each clone individually. To construct the phylogenetic trees, we first cleaned the CNA profiles by assigning deletions or amplifications to all cells in the area if the event was clonal based on WGS data. For distinct cell clusters, we assigned CNA events where the majority had supporting BAF and/or expression evidence. For PCAL37, most of the chromosome 19 CNAs were excluded from the analysis due to difficulties in accurately detecting clusters of small deletions in this region.
Next, we binned the genome for each clone and merged all data together per patient. For PCAL25, we excluded areas T06 and T08 due to inconclusive CNA profiles obtained from low-pass WGS, as well as the whole chromosome 1 from phylogenetic tree building due to inconclusive Numbat results. The resulting CNA matrix was then used as input for MEDICC2, version 1.0.2, to build the phylogenetic tree60. The cancer cell fractions’ clonal composition in each area was visualised using cloneMap, v1.0.0 (available from: https://github.com/amf71/cloneMap).
Single-cell pseudobulk BAF calculation
We utilised SNP pileup from Numbat to calculate BAF for each cell with a coverage minimum cutoff of 100. We next computed a mean BAF for each clone. For clone-specific BAF plotting, we used the Sequenza “windowBf” function, with a bin size of 1 Mb. The function calculates the allele frequency for each bin using a peak-finding algorithm to identify the highest values in the allele frequency distribution, ranging from 0 to 1. It reports for each bin the highest peak for the B-allele frequency, which corresponds to values equal to or lower than 0.5.
Bulk WGS
DNA extraction
After 10X genomics snRNAseq library prep, high molecular weight DNA was extracted from the same population of nuclei using Gentra Puregene Buccal Cell kit (Qiagen, 158845) following the manufacturer’s instructions. Briefly, nuclei were lysed using 300 µl of Cell Lysis Solution, followed by proteinase K treatment incubation at 55 °C, for 1 h. Next, samples were treated with RNase by adding 1.5 μl RNase A solution and incubation for 15 min at 37 °C. After 5 min on ice, 100 µl Protein Precipitation Solution was added to the lysate to remove proteins. After centrifuging for 1 min at 15,000 × g, precipitated proteins form a tight pellet. The supernatant was transferred into a clean 1.5 ml microcentrifuge tube with 300 µl isopropanol. A final wash with 300 µl of 70% ethanol was done to remove the remaining impurities. DNA was resuspended in 100 µl DNA Hydration Solution and incubated at 65 °C for 1 h to dissolve the DNA. DNA concentration was measured by Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Q32850), purity by Denovix spectrophotometer (DS-11Fx) and DNA integrity by electrophoresis in 0.8% agarose gel.
WGS Library preparation and sequencing
Whole genome sequencing libraries were prepared using 200 ng of genomic DNA following the NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England Biolabs, E7805S). Briefly, DNA was enzymatically fragmented for 12 min at 37 °C to get 450 bp fragments on average. After size selection, end repair, dA-tailing and UDI Adaptor Ligation, final libraries were amplified by 5 PCR cycles, quantified by Qubit ssDNA Assay Kit (Thermo Fisher Scientific, Q10212). Size distribution was assessed by Bioanalyzer High Sensitivity DNA Kit (Agilent, 5067-4626). Finally, libraries were combined at 2 nM concentration pool and paired-end sequenced using 300 cycles on NovaSeq 6000 S2 Reagent Kit v1.5, 300 cycles (Illumina, 20028314).
WGS pre-processing and somatic copy-number calling
FastQ files containing sequencing reads were quality-checked using FastQC version 0.11.8 (https://github.com/s-andrews/FastQC) and aligned to the human reference genome (GRCh38) with BWA MEM61 version 0.7.15. Data preprocessing followed the GATK best practices (Van der Auwera G, O’Connor B. Genomics in the Cloud. 2020), using GATK version 4.1.9.0. PCR and optical duplicates were identified using the “MarkDuplicates” tool, and base quality score recalibration was performed with “BaseRecalibrator” followed by “ApplyBQSR.” Coverage statistics were obtained using “CollectWgsMetrics” and mosdepth62 version 0.3.1. Final BAM files QC was conducted with “CollectAlignmentSummaryMetrics,”, “CollectBaseDistributionByCycle,” “CollectGcBiasMetrics,” “CollectInsertSizeMetrics,” and “QualityScoreDistribution.” QC results were manually reviewed using MultiQC62,63 version 1.9. The average sequencing depth was 16X for WGS and 0.3X for low-pass WGS.
To estimate tumour purity and ploidy and to obtain somatic copy-number alterations and genomic segments for low-pass WGS, ACE64 version 1.9.3 was used, with bin sizes of 100, 500, 1000 and ploidy of 2 and 4; for deeper sequenced WGS samples, Sequenza65 version 3.0.0 was used, with default parameters. Sex chromosomes were omitted from the analysis.
Variant calling and annotation
High-confidence somatic single-nucleotide variants (SNVs) were called for each deep WGS tumour-normal pair using MuTect2 (following GATK best practices, using GATK version 4.1.9.0) and Strelka66 version 2.9.10. MuTect2 was run using the “1000g_pon.hg38.vcf.gz” panel of normals and the “gnomAD” germline resource as additional controls. For each pair, a consensus callset was obtained using SomaticSeq67. Functional annotation of the variants was performed using the Ensembl Variant Effect Predictor 99.067,68.
Microsatellite instability
Microsatellite instability (MSI) status was calculated with MSIsensor2 with default parameters, tumour only mode (Niu, 2024, available from: https://github.com/niu-lab/msisensor2). Next, mutations were classified based on mutational signatures of Single Base Substitutions (SBS). SBS signatures were assigned to individual samples using SigProfilerAssignment (version 0.1.7)69. Using variant calling format (VCF) files, the assignment was conducted with COSMIC reference signatures (version 3.4) (available from https://cancer.sanger.ac.uk/signatures/).
Copy number analysis and clonal reconstruction from deep WGS
Subclonal copy number segments were identified by estimating cancer cell fractions (CCF) using B-allele frequency and depth ratio data. Allele-specific copy number profiles were generated using Sequenza65, which models the most likely sample cellularity (ρ) and ploidy (ψ).
Segmentation was performed jointly across all samples from each donor. This strategy ensures that copy number aberrations present in any sample are consistently represented across all related samples. As a result, segments not detected in a particular sample are assigned a CCF of zero, while segments observed in one or more samples reflect their respective CCFs.
We assumed that all subclones share the same overall ploidy but differ in cellularity. Subclonal segments were defined as those differing by ±1 copy from the clonal copy number state. To identify these, we applied a grid search for each segment65, to find the best-fitting subclonal cellularity values. This approach accounts for the presence of both clonal cancer cells and normal diploid cells23. Segment-specific CCFs were then approximated by dividing the local cellularity of each segment by the sample’s overall cellularity. We note that B-allele frequency data were only attainable for samples with deep whole-genome sequencing (coverage >30X). For samples with lower coverage, the depth ratio was used to estimate copy number states.
For each of the five patients, we inferred the number of subclones and their cellular fractions using DPclust70. To reconstruct phylogenetic relationships, we applied the “pigeonhole principle” (PHP) to mutational clusters. This principle states that if the combined CCFs of two mutations exceed 100%, at least one cell must carry both mutations—indicating they lie on the same branch of the phylogenetic tree.
Statistics & reproducibility
All quantification and statistical analysis were performed as described in the figure legends. No statistical method was used to predetermine sample size. Only data failing our objective QC criteria were excluded. No other data were excluded from the analyses, and the experiments were not randomised.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw transcriptomic and genomic data generated in this study are available on request on the European Genome-phenome Archive under accession number EGAS50000000927. All researchers can obtain access by submitting a project proposal to the Data Access Committee (DAC). Source data are provided with this paper. The sequencing data analysed in this study contain sensitive patient information and are therefore only available under restricted access to protect confidentiality. Access may be granted to qualified researchers upon request to the corresponding author (J.W.), with responses provided within 10 business days. Approved access will be valid for 12 months under a data use agreement. Publicly available prostate cancer datasets used in this study for cell type prediction include: human data from SCP86471, GSE18129416, GSE17603113; EGAD0000100834056. Databases used in this study include the curated pathway database PROGENy (https://saezlab.github.io/progeny/)58 transcription factor database CollecTRI (https://github.com/saezlab/CollecTRI)72. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Code availability
Source code for the analysis is available at https://bitbucket.org/weischenfeldt/prostate_multiregion_sc_clonality.
References
Warner, E. W. et al. Multiregion sampling of de novo metastatic prostate cancer reveals complex polyclonality and augments clinical genotyping. Nat. Cancer 5, 114–130 (2024).
Fernandez-Mateos, J. et al. Tumor evolution metrics predict recurrence beyond 10 years in locally advanced prostate cancer. Nat. Cancer 5, 1334–1351 (2024).
Singhal, U. et al. Integrative multi-region molecular profiling of primary prostate cancer in men with synchronous lymph node metastasis. Nat. Commun. 15, 4341 (2024).
Hong, M. K. et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat. Commun. 6, 6605 (2015).
Boutros, P. C. et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat. Genet. 47, 736–745 (2015).
Nurminen, A. et al. Cancer origin tracing and timing in two high-risk prostate cancers using multisample whole genome analysis: prospects for personalized medicine. Genome Med. 15, 82 (2023).
Rao, S. et al. Intra-prostatic tumour evolution, steps in metastatic spread and histogenomic associations revealed by integration of multi-region whole-genome sequencing with histopathological features. Genome Med. 16, 35 (2024).
Woodcock, D. J. et al. Prostate cancer evolution from multilineage primary to single lineage metastases with implications for liquid biopsy. Nat. Commun. 11, 5070 (2020).
Ge, R., Wang, Z. & Cheng, L. Tumor microenvironment heterogeneity an important mediator of prostate cancer progression and therapeutic resistance. NPJ Precis. Oncol. 6, 31 (2022).
He, Y. et al. Targeting signaling pathways in prostate cancer: mechanisms and clinical trials. Signal Transduct. Target. Ther. 7, 198 (2022).
Wang, X. et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 (2009).
Henry, G. H. et al. A cellular anatomy of the normal adult human prostate and prostatic urethra. Cell Rep. 25, 3530–3542.e5 (2018).
Song, H. et al. Single-cell analysis of human primary prostate cancer reveals the heterogeneity of tumor-associated epithelial cell states. Nat. Commun. 13, 141 (2022).
Erickson, A. et al. Spatially resolved clonal copy number alterations in benign and malignant tissue. Nature 608, 360–367 (2022).
Kiviaho, A. et al. Single cell and spatial transcriptomics highlight the interaction of club-like cells with immunosuppressive myeloid cells in prostate cancer. Nat. Commun. 15, 9949 (2024).
Hirz, T. et al. Dissecting the immune suppressive human prostate tumor microenvironment via integrated single-cell and spatial transcriptomic analyses. Nat. Commun. 14, 663 (2023).
Zhang, N. et al. High clonal diversity and spatial genetic admixture in early prostate cancer and surrounding normal tissue. Nat. Commun. 15, 3475 (2024).
Weischenfeldt, J. et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 23, 159–170 (2013).
Gao, T. et al. Haplotype-aware analysis of somatic copy number variations from single-cell transcriptomes. Nat. Biotechnol. 41, 417–426 (2023).
Alquicira-Hernandez, J., Sathe, A., Ji, H. P., Nguyen, Q. & Powell, J. E. scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 20, 264 (2019).
Ren, P. et al. Single-cell assignment using multiple-adversarial domain adaptation network with large-scale references. Cell Rep. Methods 3, 100577 (2023).
Park, S. H. et al. Going beyond Polycomb: EZH2 functions in prostate cancer. Oncogene 40, 5788–5798 (2021).
Gerhauser, C. et al. Molecular evolution of early-onset prostate cancer identifies molecular risk markers and clinical trajectories. Cancer Cell 34, 996–1011.e8 (2018).
Drews, R. M. et al. A pan-cancer compendium of chromosomal instability. Nature 606, 976–983 (2022).
Nguyen, B. et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 185, 563–575.e11 (2022).
Bielski, C. M. et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 50, 1189–1195 (2018).
Brown, L. M. et al. An elevated rate of whole-genome duplications in cancers from Black patients. Nat. Commun. 15, 1–18 (2024).
Badia-I-Mompel, P. et al. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2, vbac016 (2022).
Chan, J. et al. Lineage plasticity in prostate cancer depends on FGFR and JAK/STAT inflammatory signaling. Eur. J. Cancer 174, S4–S5 (2022).
Wang, Y. et al. Bufalin is a potent small-molecule inhibitor of the steroid receptor coactivators SRC-3 and SRC-1. Cancer Res. 74, 1506–1517 (2014).
Yang, W. et al. SHOX2 promotes prostate cancer proliferation and metastasis through disruption of the Hippo-YAP pathway. iScience 26, 107617 (2023).
Bernard, D. et al. The methyl-CpG-binding protein MECP2 is required for prostate cancer cell growth. Oncogene 25, 1358–1366 (2006).
Cortés-Ciriano, I. et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet 52, 331–341 (2020).
Siu, M. K. et al. TCF7 is suppressed by the androgen receptor via microRNA-1-mediated downregulation and is involved in the development of resistance to androgen deprivation in prostate cancer. Prostate Cancer Prostatic Dis. 20, 172–178 (2017).
Yu, W. et al. Oncogenic role of SATB2 in vitro: Regulator of pluripotency, self-renewal, and epithelial-mesenchymal transition in prostate cancer. Cells 13, 962 (2024).
Hong, W. S., Shpak, M. & Townsend, J. P. Inferring the Origin of Metastases from Cancer Phylogenies. Cancer Res. 75, 4021–4025 (2015).
Ramnarine, V. R. et al. The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur. Urol. 76, 546–559 (2019).
Rotinen, M. et al. ONECUT2 is a targetable master regulator of lethal prostate cancer that suppresses the androgen axis. Nat. Med. 24, 1887–1898 (2018).
Tang, F. et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 376, eabe1505 (2022).
Barkley, D. et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat. Genet. 54, 1192–1201 (2022).
Efe, G., Rustgi, A. K. & Prives, C. P53 at the crossroads of tumor immunity. Nat. Cancer 5, 983–995 (2024).
Dhanasekaran, R. et al. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 19, 23–36 (2022).
Farooqi, A. A. et al. Wnt and SHH in prostate cancer: trouble mongers occupy the TRAIL towards apoptosis. Cell Prolif. 44, 508–515 (2011).
Bonkhoff, H. Estrogen receptor signaling in prostate cancer: Implications for carcinogenesis and tumor progression. Prostate 78, 2–10 (2018).
Fixemer, T., Remberger, K. & Bonkhoff, H. Differential expression of the estrogen receptor beta (ERbeta) in human prostate tissue, premalignant changes, and in primary, metastatic, and recurrent prostatic adenocarcinoma. Prostate 54, 79–87 (2003).
Haffner, M. C. et al. Tracking the clonal origin of lethal prostate cancer. J. Clin. Invest. 123, 4918–4922 (2013).
Formaggio, N., Rubin, M. A. & Theurillat, J.-P. Loss and revival of androgen receptor signaling in advanced prostate cancer. Oncogene 40, 1205–1216 (2021).
Weber, B. N. et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011).
Wong, H. Y. et al. Single cell analysis of cribriform prostate cancer reveals cell intrinsic and tumor microenvironmental pathways of aggressive disease. Nat. Commun. 13, 6036 (2022).
Hunter, M. V., Moncada, R., Weiss, J. M., Yanai, I. & White, R. M. Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat. Commun. 12, 6278 (2021).
Wang, I. et al. Prostate cancer immunotherapy: a review of recent advancements with novel treatment methods and efficacy. Am. J. Clin. Exp. Urol. 10, 210–233 (2022).
Guedes, L. B. et al. MSH2 loss in primary prostate cancer. Clin. Cancer Res. 23, 6863–6874 (2017).
Pfisterer, U. et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat. Commun. 11, 5038 (2020).
Fleming, S. J. et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat. Methods 20, 1323–1335 (2023).
Karthaus, W. R. et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science 368, 497–505 (2020).
Tuong, Z. K. et al. Resolving the immune landscape of human prostate at a single-cell level in health and cancer. Cell Rep. 37, 110132 (2021).
Andreatta, M. et al. Semi-supervised integration of single-cell transcriptomics data. Nat. Commun. 15, 872 (2024).
Schubert, M. et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 9, 20 (2018).
Müller-Dott, S. et al. Expanding the coverage of regulons from high-confidence prior knowledge for accurate estimation of transcription factor activities. Nucleic Acids Res 51, 10934–10949 (2023).
Kaufmann, T. L. et al. MEDICC2: whole-genome doubling aware copy-number phylogenies for cancer evolution. Genome Biol. 23, 241 (2022).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Pedersen, B. S. & Quinlan, A. R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047 (2016).
Poell, J. B. et al. ACE: absolute copy number estimation from low-coverage whole-genome sequencing data. Bioinformatics 35, 2847–2849 (2019).
Favero, F. et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 26, 64–70 (2015).
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018).
Fang, L. T. et al. An ensemble approach to accurately detect somatic mutations using SomaticSeq. Genome Biol. 16, 1–13 (2015).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 1–14 (2016).
Díaz-Gay, M. et al. Assigning mutational signatures to individual samples and individual somatic mutations with SigProfilerAssignment. Bioinformatics 39, btad756 (2023).
Noorani, A. et al. Genomic evidence supports a clonal diaspora model for metastases of esophageal adenocarcinoma. Nat. Genet. 52, 74–83 (2020).
Regenerative potential of prostate luminal cells revealed by single-cell analysis (Human). Single Cell Portal: an interactive home for single-cell genomics data https://singlecell.broadinstitute.org/single_cell/study/SCP864.
CollecTRI: Gene regulatory network containing signed transcription factor-target gene interactions. https://github.com/saezlab/CollecTRI.
Acknowledgements
We thank the patients, staff and scientists who contributed to this study. M.M. was supported by a grant from Danish Cancer Society (R231-A14079), E.K. was supported by the Danish Cancer Society Young Talented Cancer Researchers grant (R339-A19512) and J.W. was supported by funding from Novo Nordisk Foundation (NNF200C0060141). This work is supported by the Canadian Institute of Health Research (CIHR) (FRN-153234, 158225, 168933 & 191847 to M.L.), the Ontario Institute for Cancer Research (OICR) Investigator Award through funding provided by the Government of Ontario (IA-031 to M.L.), the Princess Margaret Cancer Foundation, the Cancer Digital Intelligence (CDI) and the Ontario Graduate Student awards by the Government of Ontario to T.K. M.L. holds the Joey and Toby Tanenbaum/Brazilian Ball Chair. K.F. was funded by a clinical scientist fellowship awarded by the German Cancer Research Centre Clinician Scientist Programme, which is supported by the Dieter Morszeck Foundation. Initial sample collection was funded by the ICGC EOPC (01KU1001 A) and ICGC-Data Mining (01KU1505A) projects on early-onset prostate cancer (to TS, GS, CP) by the German Federal Ministry of Education and Research (BMBF).
Author information
Authors and Affiliations
Contributions
J.W. conceived the project, supervised and provided resources for the study. M.M. adapted single-nuclei protocol for fresh-frozen prostate cancer punches and processed all samples. E.K. & M.M. performed data preprocessing, annotated cells and integrated data. M.M. built phylogenetic trees. E.K. calculated centroid and pairwise distances, entropies, Gini and Jaccard indexes, weighted phylogenetic metric, cell state scores and PCA. F.G.R.G. prepared and sequenced bulk low-pass WGS libraries. A.L. & F.F. performed bulk low-pass WGS processing. M.M. & D.K. performed signature analysis. M.M. & F.F. performed BAF calculations. J.W, E.K. and M.M. wrote the manuscript. C.G., C.P. and T.S. provided funds and supported the manuscript writing. T.K., K.F., M.G., R.G.B., G.S., R.S., M.L., and S.M. contributed to data provisioning. All authors contributed to the review and the editing of the manuscript. All authors approved the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Srinivasa Rao, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mikutenaite, M., Karadoulama, E., Favero, F. et al. Clonal evolution and transcriptional plasticity shape metastatic dissemination routes in prostate cancer. Nat Commun 16, 11338 (2025). https://doi.org/10.1038/s41467-025-66704-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66704-w