Abstract
Early activation and phenotypic transformation of monocytes and macrophages are essential for inflammatory activities and tissue repair following myocardial infarction (MI). However, the involvement of histone succinylation in monocyte phenotypic regulation during MI remains poorly understood. Here we show that succinylation, particularly histone H3K23succ, is significantly upregulated in monocytes from both MI patients and male mouse models, correlating with enhanced inflammatory responses. We further reveal that histone acetyltransferase 1 (Hat1) acts as a succinyltransferase essential for catalytic activity, and is upregulated together with histone succinylation in proinflammatory monocytes. Deficiency in Hat1 expression improves cardiac function, reduces infarct size, and suppresses inflammatory responses in infarcted hearts after MI. Mechanistically, Hat1 modulates chromatin accessibility and recruits H3K23 succinylation to regulate proinflammatory gene expression in monocytes and macrophages post-MI. Our study reveals a critical role for histone succinylation in early MI progression and establishes that Hat1 acts as an epigenetic regulator promoting proinflammatory monocyte transformation, highlighting its therapeutic potential for MI treatment.
Similar content being viewed by others
Introduction
Myocardial infarction (MI) is the most severe manifestation of coronary artery disease and remains associated with high morbidity and mortality rates worldwide1. The critical consequences of ischemic cardiac tissue are exacerbated by early-onset excessive inflammatory responses and adverse cardiac remodeling2. The rapid expansion of immune cells derived from bone marrow (BM), particularly monocytes and macrophages, triggers a robust inflammatory cascade in the early progression of MI, which is caused by the transformation of BM hematopoietic stem cells from a static state to proliferation and differentiation states after heart attack3. Suppressing the early excessive inflammatory activities has been an effective therapeutic intervention that facilitates a better post-infarct recovery4. Thus, modulating the functional phenotypic transformation of BM and circulating monocytes before they are recruited to ischemic cardiac tissues is critical for ameliorating exaggerated inflammation, adverse cardiac remodeling, and cardiac dysfunction. Much effort has focused on reducing the pro-inflammatory monocyte subpopulation or alleviating inflammatory pathways after heart attack; however, ideal anti-inflammatory or immunomodulatory therapeutic effects remain unachieved. Therefore, understanding the endogenous mechanisms and elucidating the crucial factors that determine early monocyte-macrophage phenotype transformation may help to explore better therapeutic strategies post-MI.
Histone modifications are dynamic, reversible, and crucial in regulating gene expression, transcription, and DNA damage repair5. Lysine succinylation of nonhistone proteins and histones is a newly discovered post-translational modification6. Lysine succinylation is reportedly related to multiple diseases, such as malignant tumors7, Parkinson’s disease8, and metabolic diseases9. Similarly, nonhistone succinylation has been raised as a hazardous factor for cardiovascular disease and has been reported in hypertrophic cardiomyopathy and atrial fibrillation10,11. Increasing evidence shows that histone succinylation influences the nucleosome structure and genome-wide transcription. However, the involvement of specific genes in histone succinylation in MI remains poorly understood.
Histone acetyltransferases (HATs) regulate gene transcription by transferring an acetyl group onto specific lysine residues12. Histone acetyltransferase 1 (Hat1) is the first discovered HAT, which is in the nucleus and is involved in various cellular processes, including proliferation, DNA replication, and cellular metabolism13,14. Recent studies showed that Hat1 activation correlates with macrophage activity in viral infection15. In addition, Hat1 has been reported as a succinyltransferase and drives cancer initiation and progression by mediating the succinylation16. How Hat1 drives the monocyte phenotypic transition of ischemic heart disease at the early inflammatory stage remains unknown. Therefore, our objective was to investigate whether histone succinylation collaborates with Hat1 to regulate the phenotypic transition of remote monocytes and cardiac macrophages during the early stage of MI.
Here, we demonstrate that proinflammatory monocytes with high succinylation levels accumulate in patients and mice with MI during the early inflammatory stage. Upregulated global and histone succinylation are closely associated with inflammatory responses. Moreover, Hat1 was spatially and temporally coordinated with elevated histone succinylation during the process from activation to differentiation in monocytes post-MI. Reduced Hat1 in monocytes attenuated ischemic injury and facilitated cardiac repair by limiting inflammatory responses and monocytes toward the proinflammatory phenotype, and the mechanism may be that Hat1 acted as succinyltransferase to regulate proinflammatory gene transcription. Hat1 inhibition also protects against myocardial I/R injury, presenting a potential strategy for MI treatment.
Results
Circulating monocyte succinylation levels in patients with acute myocardial infarction (AMI) correlate with inflammatory responses
We measured serum succinate levels in patients with AMI and unstable angina (UA) before percutaneous coronary intervention. Data were collected from 98 patients, including 50 with UA and 48 with AMI. The clinical characteristics and demographic data are presented in Supplementary Data 1. Succinate levels were increased in patients with AMI (57.11 (57.79, 71.56) μM) compared with those with UA (41.85 (37.95, 46.45) μM; P < 0.001; Fig. 1a, Supplementary Data 1). A nonlinear association with AMI was suggested for succinate concentration (P for nonlinearity <0.001, P overall < 0.001; Fig. 1b). Subsequently, we evaluated the diagnostic performance of plasma succinate in AMI. Receiver operating characteristic (ROC) curve analysis showed that the area under the curve (AUC) for plasma succinate was 0.868 (Fig. 1c).
a Plasma levels of succinate in unstable angina (UA; n = 50) and acute myocardial infarction (AMI; n = 48; two-tailed Mann-Whitney U test). b Restricted cubic spline analysis shows the relationship between succinate concentration and AMI. A spline function term was introduced into the multivariate Logistic regression model and assessed overall effects and nonlinear trends using Wald χ2 test. The solid line represents the adjusted odds ratio (OR), and the shaded area indicates the 95% confidence interval. c Receiver operating characteristic (ROC) curves reveal the diagnostic efficacy of succinate for AMI. d Immunoblot analysis and quantification of Pan Ksucc levels in monocytes from patients with UA and AMI (n = 5; two-tailed unpaired Student t test). e Correlation of Pan Ksucc levels in human monocytes with plasma succinate levels in patients with AMI (n = 48; Spearman’s correlation analysis). f The levels of NLR, TNF-α, IL-6, and IL-1β in the plasma of AMI patients with Pan Ksucc levels greater than the median compared with those in patients with Pan Ksucc levels less than or equal to the median (high: n = 24, low: n = 24; two-tailed Mann-Whitney U test). g ROC curves reveal the diagnostic efficacy of plasma inflammatory markers for Pan Ksucc levels in monocytes from patients with AMI and UA. Statistical significance between curves was determined using DeLong’s nonparametric test. a, f Data presented as box plots, with 75th and 25th percentiles limits; centre line represents the median; bars represent maximal and minimal values. Results are presented as the mean ± SD of independent replicates. Pan Ksucc global succinylation, OR odds ratio, AUC area under the curve, NLR neutrophil-to-lymphocyte ratio, TNF-α tumor necrosis factor-α, IL interleukin. Source data are provided as a Source Data file.
Succinate accumulation in the tricarboxylic acid (TCA) cycle implies the production of many succinyl-CoA, which, as a substrate, mediates protein succinylation6,17. Thus, we validated global succinylation levels in peripheral blood monocytes from patients with AMI and UA. Next, monocytes were roughly isolated from the peripheral blood mononuclear cells (PBMCs). Global succinylation levels (Pan Ksucc) were increased (Fig. 1d) in patients with AMI compared with those in UA controls. Furthermore, we verified the correlation between succinate and succinylation levels. As expected, global succinylation in monocytes was positively correlated with plasma succinate levels (P < 0.001; Fig. 1e). The inflammatory activity of monocytes is related to the prognosis and severity of MI18. Therefore, we assessed the relationship between the global succinylation levels of monocytes and plasma inflammatory cytokine levels. We categorized patients who had Pan Ksucc levels above the median with AMI (or UA) as the “high” group, and those with Pan Ksucc levels below or equal to the median as the “low” group. The neutrophil to lymphocyte ratio (NLR), tumor necrosis factor (TNF)-α levels, and interleukin (IL)-6 levels in the Pan Ksucc high group exhibited significant elevation compared to those in the Pan Ksucc low group, whereas there was no significant difference observed in plasma IL-1β levels between the two groups (Fig. 1f) in AMI patients. In patients with UA, however, no significant difference in NLR, TNF-α, IL-6, or IL-1β was observed between the Pan Ksucc low- and high-level groups (Supplementary Fig. 1). Next, we used ROC curves to determine whether the inflammatory factors can reliably distinguish between high and low Pan Ksucc groups in patients with AMI and UA. NLR (AUC value = 0.865) and TNF-α (AUC value = 0.911) allowed the differentiation between the AMI group with high Pan Ksucc levels and those with low Pan Ksucc levels. However, IL-6 (AUC value = 0.693) and IL-1β (AUC value = 0.663) could not distinguish the low-Pan Ksucc group from the high-Pan Ksucc group in patients with AMI. In the UA group, Pan Ksucc levels exhibited limited discriminative power for identifying patients with heightened inflammatory states, as reflected by a modest AUC value compared with AMI (Fig. 1g). Pan Ksucc levels possess significantly greater discriminatory capacity for detecting high-inflammatory states in AMI patients compared to UA cases. These findings suggest that Pan Ksucc may potentially contribute to guiding targeted anti-inflammatory management in AMI.
Histone succinylation is increased in monocytes post-MI
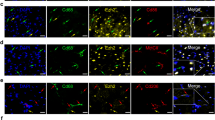
To elucidate the involvement of monocyte succinylation in the initial phase of AMI, we first investigated the succinylation levels in both BM and peripheral blood monocytes on Days 1 and 3 after MI. Global succinylation levels were increased significantly on Day 1 in BMs and Day 3 in peripheral blood monocytes, respectively. The band appearing near 17 kDa had a similar trend compared with global succinylation levels, which may represent histone H3 (Fig. 2a, b). Subsequently, immunoblots of bone marrow-derived macrophages (BMDMs) stimulated by lipopolysaccharide (LPS) and interferon-γ (IFN-γ) revealed that the succinylation levels of histone H3 exhibited a significant increase relative to acetylation levels (Supplementary Fig. 2a) and that H3K23succ was upregulated most significantly compared with other sites and H3K23ac (Supplementary Fig. 2b, c). Subsequently, we extracted histones from post-MI mouse monocytes and found that Pan Ksucc was elevated in BM monocytes on Day 1 and in circulating monocytes on Day 3 (Supplementary Fig. 3a, b). To examine changes in succinylation in infarcted hearts over 3 days, we performed multiplex immunofluorescence co-staining for Pan Ksucc, along with antibodies against macrophage markers (F4/80). Most macrophages exhibited Pan Ksucc expression in the infarct region (Fig. 2c). Next, we further found rising H3K23succ levels in monocytes of BM on Day 1 (Fig. 2d) and peripheral blood on Day 3 (Fig. 2e) post-MI. Immunofluorescence staining of infarcted hearts also showed H3K23succ levels were increased in macrophages (Fig. 2f). These results suggest that histone lysine succinylation is increased in remote monocytes and macrophages infiltrating the heart and is crucial in the dynamic activation of monocytes in the initial phase of MI.
a, b Immunoblot analysis and quantification of Pan Ksucc (global succinylation) level in monocytes sorted from bone marrow (a) and peripheral blood (b) of mice indicated days after MI (n = 3; one-way ANOVA followed by Bonferroni multiple comparisons test). c Immunofluorescence staining and quantification of the proportion of Pan Ksucc and F4/80 in cardiac tissues on Day 3 with sham operation or after MI (scale bar=50 μm; n = 6; two-tailed unpaired Student t test). d, e Immunoblots of the levels of H3K23succ (histone) in bone marrow monocytes on Day 1 (d) and circulating monocytes on Day 3 (e) sorted from MI mice (n = 3; two-tailed unpaired Student t test). The blots were derived from the same experiment and processed in parallel. Total histone H3 level was set as an internal reference. f Immunofluorescence staining and quantification of the proportion of H3K23succ and F4/80 in cardiac tissues on Day 3 with sham operation or after MI (scale bar=50 μm; n = 5; two-tailed unpaired Student t test). g Heatmaps of H3K23succ binding peaks in circulating monocytes from sham and MI mice. Color intensity reflects the relative read count. Genes exhibiting similar distribution patterns were subjected to clustering analysis using an algorithm, revealing the binding trends of histone succinylation modifications across all genes. h Bar graph showing the genomic distribution of differential H3K23succ peaks (MI vs sham) relative to the translation start site (TSS). i, j KEGG (i) and GO (j) analyses of upregulated genes with increased H3K23succ modification. Statistical significance was assessed using a two-sided hypergeometric test with Benjamini-Hochberg correction for multiple testing. k The GSEA pathways enriched in the MI mice. The indicative genes were enriched for Inflammatory response and Tnf-α signaling via nfκb, as defined by hallmarks. l The mRNA levels of H3K23succ-enriched inflammatory genes in sham and MI mice (n = 6; two-tailed unpaired Student t test). m ChIP–qPCR analysis of H3K23succ enrichment at the promoters of inflammatory genes in sham and MI mice (n = 4; two-tailed unpaired Student t test). Results are presented as the mean ± SD of independent replicates. DAPI 4ʹ,6-diamidino-2-phenylindole, GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, GSEA gene set enrichment analysis, NES normalized enrichment score, ChIP chromatin immunoprecipitation, PCR polymerase chain reaction. Source data are provided as a Source Data file.
H3K23succ targets the activation of the inflammatory signaling pathway
For the preliminary exploration of the potential functional significance of H3K23succ in monocytes post-MI, we performed cleavage under targets & tagmentation (CUT&Tag) analysis to identify the candidate pathways regulated by H3K23succ in peripheral blood monocytes from MI and sham surgery mice on Day 3. We used anti-H3K23succ antibodies and found significant enrichment of H3K23succ peaks in circulating monocytes from 3-day-MI mice compared with sham mice (Fig. 2g). Notably, monocyte from MI and sham mice comparison showed that upregulated H3K23succ binding peaks were enriched in the gene promoter regions (Fig. 2h). To further investigate the epigenetic effects of H3K23succ in monocytes post-MI, upregulated genes with different H3K23succ binding peaks were classified into different Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. These pathways were predominantly involved in the inflammatory response (Fig. 2i, j). Further based on hallmark gene sets and utilizing additional enrichment analysis algorithms, gene set enrichment analysis demonstrated that genes with upregulated H3K23succ binding peaks in MI mice were significantly enriched in the inflammatory response, TNF-α signaling via NF-κB (Fig. 2k), and other characteristic pathways involved in inflammation (Supplementary Fig. 3c–f). We identified 14 genes exhibiting upregulation of H3K23succ modification that were strongly associated with inflammatory processes in monocytes post-MI. To validate the observed upregulation, we performed RT-qPCR and ChIP-qPCR to confirm both the enhanced transcriptional expression and increased H3K23succ enrichment at the promoter regions of these 14 inflammatory genes--such as Cxcl10, Nos2 and Tnf (Fig. 2l, m). These results indicate that H3K23succ activates the transcription of genes encoding known inflammatory factors in monocytes post-MI. Hence, the downregulation of H3K23succ may improve cardiac function by inhibiting the transcriptional activity of inflammation-related genes, reducing the inflammatory response in the early stage of MI.
Hat1 was significantly elevated in cardiac macrophages and peripheral monocytes post-MI
The above findings prompted us to identify the upstream regulators that effectively regulate histone succinylation levels. Therefore, we hypothesize that HATs may serve as potential targets for succinylation regulation. We first identified several characterized HATs in the infarcted hearts of MI mice using bioinformatic transcriptome analysis from the GEO dataset GSE206281 and GSE775. Compared with other proteins, Hat1 was significantly upregulated in the cardiac tissues on Day 3 post-MI (Supplementary Fig. 4a, b). Moreover, Hat1 levels were higher in the infarcted area compared to the non-infarcted area during the initial phase of MI (Fig. 3a). Therefore, we investigated Hat1 expression levels in the infarcted heart at indicated time points post-MI. Similar to the transcriptomic analysis, Hat1 protein expression levels were significantly elevated in the infarcted area as opposed to the non-infarcted area. Hat1 expression was upregulated on Day 1 post-MI and continued to increase progressively until Day 3. Subsequently, Hat1 levels began to recover towards baseline on Day 7 post-MI (Fig. 3b). Similarly, immunohistochemistry demonstrated that Hat1 expression was the highest in the inflammatory-infiltrated area of the infarct region on Day 3 post-MI, with low expression in the infarct region on Days 7 and 14 (Fig. 3c). Western blotting showed that Hat1 levels were not elevated in the cardiomyocytes or fibroblasts under hypoxic conditions (Supplementary Fig. 4c, d). These results indicate that Hat1 expression was increased in inflammatory cells in the infarcted myocardium post-MI.
a Line graph showing the expression and distribution of Hat1 at different time points in MI mouse heart tissues, as revealed through reanalysis of transcriptome database (GSE775). b Immunoblot analysis of Hat1 expression in infarct zone (IZ) and remote zone (RZ) of post-MI hearts and sham controls at the indicated days (n = 4). c Representative immunohistochemical analyses of Hat1 in sham-operation and MI mice at the indicated days after MI (scale bar = 50 μm; n = 5). Hat1 expression was quantified and compared (n = 4; one-way ANOVA followed by Bonferroni multiple comparisons test). d Quantitative PCR analysis of HAT1 mRNA expression in the peripheral circulating monocytes of patients with UA and AMI (n = 8; two-tailed unpaired Student t test). e Representative Western blotting and quantification of Hat1 expression in circulating monocytes from patients with UA or AMI (n = 5; two-tailed unpaired Student t test). f Immunoblot analysis and quantification of Hat1 expression in bone marrow monocytes from MI mice at the indicated days (n = 4; one-way ANOVA followed by Bonferroni multiple comparisons test). g Flow cytometry analysis and quantification of Hat1 expression in the bone marrow monocytes on Day 1 after MI (n = 4 per group; two-tailed unpaired Student t test). h Representative Western blotting and quantification of Hat1 expression in circulating monocytes from MI mice at the indicated days (n = 4; one-way ANOVA followed by Bonferroni multiple comparisons test). i Flow cytometry analysis and quantification of Hat1 expression in the blood monocytes on Day 3 post-MI (n = 4 per group; two-tailed unpaired Student t test). j Dual immunofluorescence staining and quantification of the proportion of Hat1 and F4/80 in murine heart on Day 3 after MI (scale bar=50 μm; n = 6; two-tailed unpaired Student t test). k Flow cytometry analysis and quantification of Hat1 expression in the heart on Day 3 after MI (n = 5; two-tailed unpaired Student t test). Results are presented as the mean ± SD of independent replicates. Hat1 histone acetyltransferase 1, UA unstable angina, AMI acute myocardial infarction, DAPI 4ʹ,6-diamidino-2-phenylindole, PCR polymerase chain reaction, MFI mean fluorescence intensity. Source data are provided as a Source Data file.
Furthermore, we investigated the immune cells expressing Hat1 in a single-cell RNA sequencing data set (GSE163129) of MI mice. Seven major immune cell populations, including macrophages, monocytes, neutrophils, T cells, B cells, natural killer cells, and dendritic cells (DCs), were identified based on their characteristic markers of immune cells (Supplementary Fig. 5a, b). We observed that only Hat1 expression was specifically elevated in the monophagocytic system, including macrophages, monocytes, and DCs, whereas other HATs showed elevated or unchanged expression in multiple immune cell populations in infarcted hearts after MI (Supplementary Fig. 5c, d). Subsequently, we compared Hat1 expression in cardiac-resident (CCR2-) and infiltrated (CCR2+) macrophages post-MI. Hat1 expression was higher in CCR2+ macrophages (Supplementary Fig. 5e), and the Hat1 increase was most significant in CCR2+ macrophages on Day 3 of MI progression (Supplementary Fig. 5f). Consistent with elevated Hat1 expression, cardiac CCR2+ macrophages expanded significantly from Days 3 to 5 post-MI (Supplementary Fig. 5g). These results suggest that Hat1 is critical in cardiac-infiltrated macrophages during the inflammatory phase of MI. To validate this bioinformatics prediction, we initially observed that both protein and mRNA expression levels of Hat1 were markedly upregulated in BMDMs following stimulation with LPS and IFN-γ (Supplementary Fig. 6a, b). Notably, Hat1 exhibited the most pronounced upregulation among all succinyltransferases examined (Supplementary Fig. 6c). Next, we confirmed the inflammation-induced upregulation of HAT1 mRNA (Fig. 3d) and protein levels (Fig. 3e) in monocytes from patients with UA and AMI. Hat1 expression was most significantly upregulated in BM monocytes on Day 1 (Fig. 3f) and circulating monocytes on Day 3 (Fig. 3h) compared with other ischemia time points. Flow cytometry further confirmed that Hat1 was increased in monocytes from the BM (Supplementary Fig. 7a; Fig. 3g) and peripheral blood (Supplementary Fig. 7b; Fig. 3i) of MI model mice. Multiplex immunofluorescence co-staining of Hat1 and F4/80 confirmed that Hat1 was specifically upregulated predominantly in the nucleus of cardiac macrophages on Day 3 post-MI (Fig. 3j). Flow cytometry analysis of murine heart tissues yielded consistent results, demonstrating that Hat1 expression was significantly upregulated in cardiac macrophages isolated from MI mice at day 3 (Supplementary Fig. 7c; Fig. 3k). Furthermore, the upregulation of Hat1 was more pronounced in CCR2⁺MHCII+ macrophages than in their CCR2-MHCII- counterparts (Supplementary Fig. 6d). Our results indicate that Hat1 regulation during the inflammatory phase of MI constitutes a multistage and dynamic process. This process is initiated in the BM, persists through the peripheral circulation, and sustains high expression within macrophages residing in the cardiac microenvironment.
Hat1 knockout alleviates cardiac dysfunction and ischemic injury
The function of Hat1 in MI remains unclear. Thus, we generated Hat1 KO mice and confirmed the depletion of Hat1 in the MI hearts of these mice. Given that Hat1 is essential for processes such as DNA damage repair, Hat1−/− mice exhibit embryonic lethality. Hence, Hat1+/− (Hat1 KO) heterozygous mice were generated to evaluate the role of Hat1 in MI (Supplementary Fig. 8a, b). The remarkable reduction of Hat1 mRNA (Supplementary Fig. 8c) and protein expression (Supplementary Fig. 8d) in Hat1 KO mice was validated in both BM monocytes compared with Hat1+/+ counterparts (termed WT mice). Ischemic heart tissue similarly showed reduced Hat1 expression in the heart at 3 days after MI (Supplementary Fig. 8e). To investigate the functional role of Hat1 in MI, we subjected male Hat1 KO and WT mice to surgically induced MI at nine weeks of age. M-mode echocardiograms showed that Hat1 reduction initially improved cardiac function after MI surgery, as evidenced by increased left ventricular ejection fraction (LVEF) and left ventricular fraction shortening (LVFS). The left ventricular end-diastolic dimension (LVIDd) and left ventricular end-systolic diameter (LVIDs) were decreased (Fig. 4a, b) in Hat1 KO mice compared with WT mice after MI. TTC (Fig. 4c) and HE (Fig. 4d) staining revealed a reduction in myocardial infarct size and inflammatory cell infiltration, respectively, in Hat1 KO mice at 3 days post-MI. Furthermore, Masson’s trichrome staining indicated that Hat1 knockout conferred sustained protection against infarct expansion, as evidenced by significantly reduced fibrosis at 28 days after MI (Fig. 4e).
a, b Echocardiographic analysis of the left ventricular ejection fraction (LVEF), left ventricular fraction shortening (LVFS), left ventricular end-diastolic dimension (LVIDd), and left ventricular end-systolic diameter (LVIDs) at the indicated days after MI or sham operation (n = 6; two-way ANOVA followed by Bonferroni multiple comparisons test) in WT and Hat1 KO mice, together with representative M-mode echocardiographic images. c The TTC staining of consecutive cardiac sections from WT and Hat1 KO mice on Day 3 after MI and the quantification of infarct area size. (scale bar = 2 mm; n = 6; two-tailed unpaired Student t test). d Hematoxylin and eosin (H&E) staining demonstrates the extent of inflammatory cell infiltration (up: scale bar=1 mm, down: scale bar=50 μm; n = 6). e Masson trichrome staining of sequential heart sections from WT and Hat1 KO mice at day 28 after MI, and the quantified size of fibrotic areas (scale bar=1 mm; n = 6; two-tailed unpaired Student t test). f Flow cytometric analysis of CD45.1+ and CD45.2+ cells in the peripheral blood of CD45.1 (WT) and CD45.2 (Hat1 KO) mice transplanted with bone marrow cells from CD45.1 (WT) or CD45.2 (Hat1 KO) mice (n = 3). g, h Representative M-mode echocardiograms and quantitative analysis of cardiac function parameters (LVEF, LVFS, LVIDd, LVIDs) in bone marrow-transplanted mice at day 3 post-MI (n = 6; two-way ANOVA followed by Bonferroni multiple comparisons test). i Representative H&E staining sections of cardiac tissue from bone marrow-transplanted mice at 3 days post-MI (up: scale bar = 1 mm, down: scale bar = 50 μm; n = 6). Results are presented as the mean ± SD of independent replicates. WT wild type, Hat1 histone acetyltransferase 1, TTC triphenyl tetrazolium chloride. *P < 0.05; **P < 0.01; ***P < 0.001. Source data are provided as a Source Data file.
Hat1 expressed on bone marrow-derived cells was shown to mediate MI injury
To further investigate the functional relevance of Hat1 expression in myeloid cells infiltrating the infarcted heart, we performed bone marrow transplantation (BMT) assays using Hat1 KO (CD45.2 allele) and WT mice (CD45.1 allele) to generate chimeric mice. Eight weeks after BMT, successful hematopoietic reconstitution was confirmed by flow cytometric analysis of peripheral blood cells (Fig. 4f). Mice with confirmed BM reconstitution were then subjected to MI surgery. The heart functions were analyzed at day 3 after MI in recipient mice. The transplantation of Hat1 KO bone marrow-derived cells into WT recipients (Hat1 KO BM → WT) resulted in a significant improvement in cardiac function compared with WT recipients transplanted with WT bone marrow (WT BM → WT). In contrast, the transplantation of WT BM→Hat1 KO led to a considerable reduction in cardiac function relative to Hat1 KO mice receiving Hat1 KO bone marrow (Hat1 KO BM→Hat1 KO, Fig. 4g, h). Consistent with these findings, adoptive transfer of Hat1 KO BM → WT mice markedly reduced the infiltration of inflammatory cells compared with WT BM → WT mice. Conversely, transplantation of WT BM→Hat1 KO mice significantly enhanced inflammatory cell infiltration relative to Hat1 KO BM→Hat1 KO mice (Fig. 4i). These findings demonstrate that Hat1 in circulating monocytes and their derived macrophages plays an integral role in contributing to cardiac injury post-MI.
Absence of Hat1 ameliorated myocardial injury and reduced inflammatory macrophage infiltration
Evidence from previous studies suggests that ischemia-induced cardiomyocyte apoptosis and inflammatory responses are critically involved in myocardial ischemic injury at an early stage19,20. Thus, we performed terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling (TUNEL) of the left ventricle obtained from WT and Hat1 KO mice 3 days after MI. The number of TUNEL-positive cells around the ischemic lesions was notably decreased in Hat1 KO mice compared with that in WT mice post-MI (Fig. 5a). According to these findings, western blotting (Fig. 5b; Supplementary Fig. 9a) revealed that the pro-apoptotic molecules Bax and Cleaved Caspase-3 protein levels were decreased, and those of the anti-apoptotic molecules Bcl-2 and Caspase-3 were increased. Furthermore, the crosstalk between Hat1 KO macrophages and cardiomyocytes was investigated. Cardiomyocytes were cultured in conditioned medium with macrophage Hat1 deficiency (Supplementary Fig. 9b). Annexin V/PI double staining flow cytometry assays demonstrated that Hat1 KO medium attenuates cardiomyocyte apoptosis (Fig. 5c). Similarly, treatment with conditioned medium from Hat1‑KO BMDMs downregulated the expression of the apoptosis markers Bax and Cleaved Caspase-3 in cardiomyocytes, while also elevating levels of Bcl-2 and Caspase-3 (Supplementary Fig. 9c, d).
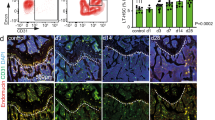
a TUNEL and α-actinin co-staining in the border regions of WT and Hat1 KO mouse hearts on Day 1 after MI. Nuclear staining with DAPI (scale bar=50 μm; n = 6; two-tailed unpaired Student t test). b Immunoblot analysis and quantification of apoptosis-related proteins including Bax, Bcl-2, Caspase-3, and cleaved Caspase-3 in peri-infarct areas of WT and Hat1 KO from infarcted hearts on Day 3 post-MI (n = 6; two-way ANOVA followed by Bonferroni multiple comparisons test). c Flow cytometry analysis of cardiomyocyte apoptosis following co-culture with LPS/IFN-γ-stimulated WT versus Hat1KO BMDMs (n = 4; two-tailed unpaired Student t test). d Flow cytometric analysis of leukocytes (CD45+), neutrophils (CD45+CD11b+Ly6G+) and macrophages (CD45+CD11b+Ly6G-F4/80+CD64+) in WT and Hat1 KO infarct tissues after MI 3 days (n = 6; two-tailed unpaired Student t test). e, f Flow cytometric analysis of pro-inflammatory (M1) macrophages (CD45⁺CD11b⁺Ly6G⁻ F4/80⁺CD64⁺CD86⁺) in infarcted heart tissues from WT and Hat1 KO mice at 3 days post-MI (n = 6; two-tailed unpaired Student t test). g, h Flow cytometric analysis of reparative (M2) macrophages (CD45⁺CD11b⁺Ly6G⁻F4/80⁺CD64⁺CD206⁺) in infarcted heart tissues from WT and Hat1 KO mice at 3 days post-MI (n = 6; two-tailed unpaired Student t test). i Flow cytometric analysis of monocyte-derived (CD45+CD11b+Ly6G-F4/80+CD64+CCR2+MHCII+) and tissue-resident (CD45+ CD11b+Ly6G-F4/80+CD64+CCR2-MHCII-) macrophage subsets in infarcted heart tissues from WT and Hat1 KO mice at 3 days post-MI (n = 5; two-tailed unpaired Student t test). j The mRNA expression levels of pro-inflammatory and reparative genes in the infarcted heart tissues of WT and Hat1 KO mice on Day 3 post-MI (n = 6; two-tailed unpaired Student t test). Results are presented as the mean ± SD of independent replicates. WT wild type, Hat1 histone acetyltransferase 1, TUNEL terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick-end labeling, DAPI 4ʹ,6-diamidino-2-phenylindole. Source data are provided as a Source Data file.
Subsequently, we investigated the effect of Hat1 loss on proinflammatory monocyte-macrophage phenotypes in MI mice. The gating strategy for circulating proinflammatory monocytes and macrophages in infarcted hearts was identified (Supplementary Fig. 10a, d, e). The results revealed that the decreased Hat1 expression suppressed the proportion and count of Ly6Chi monocytes (CD45+CD11b+Ly6G-Ly6Chi) post-MI on Day 3 (Supplementary Fig. 10b, c). Next, we examined the impact of Hat1 loss on the infiltration of cardiac leukocytes, neutrophils, and macrophages at day 3 post-MI. The obtained results demonstrated that Hat1 deficiency led to a significant reduction in leukocytes (CD45+) and neutrophils (CD45+CD11b+Ly6G-), accompanied by a reduction in both the proportion and abundance of macrophages (CD45+CD11b+Ly6G-F4/80+CD64+, Fig. 5d). Concerning tissue infiltration of macrophages in ischemic hearts, we observed that the absence of Hat1 caused a significant reduction in proinflammatory macrophages (CD45+CD11b+Ly6G-F4/80+CD64+CD86+, Fig. 5e, f) and enhanced the count of reparative macrophages (CD45+CD11b+Ly6G-F4/80+CD64+CD206+, Fig. 5g, h) compared with the WT mice on Day 3 post-MI. These changes in macrophage polarization were further supported by BMT assays. We noted that Hat1 KO BM → WT mice displayed a significant reduction in proinflammatory macrophages and a concomitant increase in reparative macrophages within the heart (Supplementary Fig. 11a, b). Moreover, compared to WT controls, Hat1 deficiency resulted in a moderate reduction in the infiltration of CCR2⁺MHCII+ macrophages at day 3 post-MI, while the abundance of CCR2⁻MHCII⁻ macrophages remained unchanged (Fig. 5i). We further assessed macrophage proliferation in the ischemic heart using Ki67 as a proliferation marker. The results revealed no significant difference in the proliferation rate, as indicated by Ki67 mean fluorescence intensity (MFI), between CCR2⁺MHCII⁺ and CCR2⁻MHCII⁻ macrophages (Supplementary Fig. 11c, d). RT-qPCR analysis yielded consistent results, demonstrating that Hat1 deficiency led to downregulation of pro-inflammatory genes (Il6, Il1β, Cd86) and upregulation of anti-inflammatory genes (Cd206, Arg1) in the ischemic myocardium at day 3 after MI (Fig. 5j). These findings indicate that the reduction of inflammation in the inflammatory phase after MI was primarily attributable to the inhibition of monocyte/macrophage infiltration and recruitment to the infarct area by Hat1 loss.
Hat1 knockdown in BM monocytes improves cardiac function after MI
We further established a BM macrophage-specific Hat1 knockdown murine model. An adeno-associated virus 9 (AAV-9) system harboring a macrophage-specific promoter with shRNA targeting Hat1 (AAV-9-F4/80-shHat1) was administered via intra-BM injection to downregulate Hat1 expression specifically to BM macrophages in mice (Supplementary Fig. 12a). Hat1 knockdown in macrophages of AAV-transduced mice was confirmed by qPCR and immunofluorescence staining (Supplementary Fig. 12b, c). Echocardiographic assessment revealed that macrophage specific Hat1 knockdown enhanced cardiac functional recovery in MI mice (Supplementary Fig. 12d). TTC staining showed reduced myocardial infarct size in Hat1 knockdown mice (Supplementary Fig. 12e); similarly, H&E staining indicated that macrophage specific Hat1 knockdown reduced inflammatory cell infiltration in the infarct zone at day 3 post-MI (Supplementary Fig. 12f). Western blot analysis (Supplementary Fig. 12g) revealed reduced levels of Bax and Cleaved Caspase-3, alongside elevated levels of Bcl-2 and Caspase-3. As anticipated, Hat1 reduction led to markedly reduced expression of pro-inflammatory macrophage markers (IL-6, IL-1β, Cd86) and concomitantly elevated expression of Cd206 and Arg1 in ischemic heart tissue at day 3 post-MI compared with controls (Supplementary Fig. 12h). Collectively, these results confirm that macrophage-specific Hat1 reduction confers a cardioprotective effect following MI.
Downregulation of Hat1 inhibited H3K23succ modification in monocytes post-MI
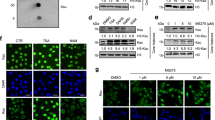
The catalytic activity of Hat1 in regulating histone succinylation in monocytes post-MI was evaluated by molecular docking, which revealed the binding modes between histone H3 and Hat1 carrying succinyl-CoA (Supplementary Fig. 13a). LigPlot+ analysis (Supplementary Fig. 13b) showed that the interaction between histone H3 and Hat1 involved the following residue pairs: Lys57 (A) - Glu54 (B), Thr33 (A) - Asn209 (B), and Arg50 (A) - Asp47 (B), with hydrogen bonds serving as the primary intermolecular forces. Molecular dynamics simulation was applied to investigate the dynamic structure changes after H3K23succ modification. Molecular dynamics simulations indicated that the succinylated H3 protein maintained overall structural stability with no significant change in compactness (Rg), suggesting no global unfolding or denaturation occurred. Locally, root mean square fluctuation (RMSF) increased significantly at H3K23 and adjacent regions, indicating enhanced backbone flexibility around this site. Additionally, a slight decrease in solvent accessible surface area (SASA) implied that succinylation may induce subtle surface rearrangements (Supplementary Fig. 13c). These calculations indicate that H3K23succ enhances local flexibility, rather than perturbing global folding, through the introduction of negative charge and a voluminous succinyl group. This increased flexibility likely promotes recognition and binding of H3K23succ at downstream gene promoters, thereby facilitating the initiation of transcriptional regulation. Next, immunoblot assays in BMDMs showed that the Hat1 decrease significantly inhibited H3K23succ levels under inflammatory conditions compared with other histone H3 succinylation sites (Fig. 6a). Subsequently, co-immunoprecipitation also demonstrated an interaction between Hat1 and H3K23succ in macrophages, which was considerably weakened by Hat1 knockdown in LPS/IFN-γ-stimulated BMDMs (Fig. 6b).
a The H3K23succ, H3K79succ, and H3K122succ immunoblots of WT or Hat1 KO BMDMs treated with LPS and IFN-γ for 24 h (n = 3; one-way ANOVA followed by Bonferroni multiple comparisons test). The blots were derived from the same experiment and processed in parallel. Total histone H3 level was set as an internal reference. b Immunoblot analysis of Hat1 and H3K23succ in protein complexes immunoprecipitated with anti-Hat1 or anti-H3K23succ antibody from lysates of WT or Hat1 KO BMDMs with LPS/IFN-γ stimulation (n = 3). c, d Immunoblots of the expression levels of Pan Ksucc in bone marrow (c) and circulating monocytes (d) sorted from WT and Hat1 KO mice post-MI (n = 4; two-tailed unpaired Student t test). e, f Immunoblots of the expression levels of H3K23succ in bone marrow (e) and circulating monocytes (f) sorted from WT and Hat1 KO mice post-MI (n = 4; two-tailed unpaired Student t test). The blots were derived from the same experiment and processed in parallel. Total histone H3 level was set as an internal reference. g, h Dual immunofluorescence staining and quantification of the proportion for Pan Ksucc (g) or H3K23succ (h) with F4/80 in the ischemic heart tissues of WT and Hat1 mice on Day 3 after MI (n = 6; Scale bar=50 μm; two-tailed unpaired Student t test). Results are presented as the mean ± SD of independent replicates. WT wild type, Hat1 histone acetyltransferase 1, LPS lipopolysaccharide, IFN-γ interferon, BMDMs bone marrow-derived macrophages; DAPI 4ʹ,6-diamidino-2-phenylindole. Source data are provided as a Source Data file.
To further validate the role of Hat1 in regulating succinylation in monocytes after MI, immunoblot analysis revealed markedly reduced global succinylation levels in monocytes isolated from bone marrow (day 1) and peripheral blood (day 3) of Hat1 KO mice (Fig. 6c, d). Consistent with the global succinylation trend, H3K23succ levels of BM and circulating monocytes were decreased (Fig. 6e, f) in Hat1 KO MI mice compared with WT MI mice. Succinylation levels were detected in cardiac-infiltrating macrophages at day 3 post-MI. As expected, reduced Hat1 expression correlated with decreased global succinylation and H3K23succ levels in ischemic heart tissues (Fig. 6g, h). Building on above findings, lentivirus-mediated knockdown of Hat1 in BMDMs (Supplementary Fig. 14a, b) reduced histone succinylation levels (Supplementary Fig. 14c). Consistently, western blot analysis revealed decreased Pan Ksucc and H3K23succ levels in monocytes from both BM and peripheral blood of AAV-9-F4/80-shHat1 MI mice compared to controls (Supplementary Fig. 14d–g). Furthermore, immunofluorescence confirmed the downregulation of both Pan Ksucc and H3K23succ in macrophages infiltrating the infarcted heart (Supplementary Fig. 14h, i). Our results suggest that Hat1 functions as a succinyltransferase responsible for regulating H3K23succ levels in monocytes and macrophages after MI at the early stage.
Hat1 regulates chromatin accessibility and H3K23succ enrichment at proinflammatory gene promoters
To further explore the underlying mechanisms governing Hat1-mediated phenotype programming in infarcted monocytes/macrophages, an assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) of circulating monocytes post-MI at 3 days was employed to investigate the involvement of Hat1 in the regulation of gene expression associated with chromatin status. Compared with AAV-9-F4/80-shNC circulating monocytes after mouse cardiac ischemia, the decreased Hat1 levels caused a significant decrease of peaks enriched near transcription start sites (TSS, ±2 kb) (Fig. 7a). Furthermore, the feature proportion concerning the intensity of the decreased differentially accessible regions from monocytes with decreased Hat1 revealed a promoter-enriching pattern, in which the decreased peaks were assigned to the promoter region (Fig. 7b). CUT&Tag assay using anti-H3K23succ antibodies also showed that the enrichment of H3K23succ near TSS (±2 kb) (Fig. 7c) was decreased in Hat1-reduced circulating monocytes from MI mice on Day 3 and that H3K23succ peaks enriched in the promoter region were similarly downregulated (Fig. 7d). RNA-seq analysis consistently validated phenotype programming at the transcriptional level. We discovered 517 downregulated and 413 upregulated differentially expressed genes in monocytes isolated from Hat1-reduced mouse blood on Day 3 post-MI (Supplementary Fig. 15a). Among these, several genes associated with the proinflammatory function and phenotype were decreased in the circulating monocytes isolated from Hat1-reduced mice compared to those from controls post-MI, whereas the expression levels of multiple reparative genes were upregulated (Fig. 7e). Correlation analyses of RNA-seq with ATAC-seq or CUT&Tag revealed significant associations between different high-throughput sequencing methods, underscoring the consistency of Hat1-mediated effects at different regulatory levels (Supplementary Fig. 15b). To identify Hat1-H3K23succ-specific genes in monocytes, we conducted a comprehensive integration and analysis of four distinct sequencing datasets: CUT&Tag from sham and MI mice, ATAC-seq, CUT&Tag, and RNA-seq from AAV-9-F4/80-shHat1/shNC mice post-MI. The analytical workflow is shown in Supplementary Fig. 15c. Subsequently, we screened for upregulated genes from CUT&Tag (sham vs. MI) and downregulated genes from ATAC-seq, CUT&Tag, and RNA-seq (MI shNC vs. MI shHat1) and identified 41 targeted genes (Fig. 7f). Thus, these 41 intersecting genes were designated as specific Hat1-H3K23succ targets in monocytes post-MI. The subsequent gene enrichment analysis revealed the active involvement of these specific genes in the regulation of cytokines and inflammatory responses (Fig. 7g). Collectively, our results define a Hat1-H3K23succ-mediated transcriptional network that modulates monocyte inflammatory responses post-MI.
a Heatmap showing the ATAC-seq peak density within ±2 kb of the transcription start sites (TSS) in circulating monocytes from AAV-9-F4/80-shNC and AAV-9-F4/80-shHat1 mice on Day 3 after MI. b Bar graph representing differential chromatin accessibility at annotated genomic regions. c Heatmaps for H3K23succ binding peaks in circulating monocytes from AAV-9-F4/80-shNC and AAV-9-F4/80-shHat1 mice after MI on Day 3. d Bar graph representing the distribution of H3K23succ sites relative to TSS in control and Hat1-reduced mice post-MI. e Volcano plot of the expression of genes in circulating monocytes isolated from AAV-9-F4/80-shNC and AAV-9-F4/80-shHat1 mice on Day 3 after MI. The labeled genes represent phenotype and repair function. f Venn diagram showing the number of overlapped genes from four datasets, including CUT&Tag (MI vs. sham, MI shHat1 vs. MI shNC), ATAC-seq (shHat1 vs. MI shNC), and RNA-seq (shHat1 vs. MI shNC); the analysis process is revealed in Supplementary Fig. 15c. g Radar plot showing enriched pathways in the overlapped genes screened from (f). h Integrative Genomics Viewer analysis of peaks density view from ATAC-seq, H3K23succ and H3K23ac binding signal from CUT&Tag in the gene locus of Aim2, Axl, Nod1, Cxcl10, Nos2, and Tnf. i The mRNA expression levels of Aim2, Axl, Nod1, Cxcl10, Nos2, and Tnf detected through quantitative polymerase chain reaction (qPCR) in WT and Hat1 KO BMDMs stimulated by LPS and IFN-γ for 24 h (n = 6; two-tailed unpaired Student t test). j ChIP and qPCR analysis of H3K23succ enrichment in the promoters of Aim2, Axl, Nod1, Cxcl10, Nos2, and Tnf in WT and Hat1 KO BMDMs stimulated by LPS and IFN-γ for 24 h (n = 4; two-tailed unpaired Student t test). Results are presented as the mean ± SD of independent replicates. NC null control, Hat1 histone acetyltransferase 1, WT wild type, BMDMs bone marrow-derived macrophages, LPS lipopolysaccharide, IFN-γ interferon, ChIP chromatin immunoprecipitation, CUT&Tag cleavage under targets & tagmentation, ATAC-seq assay for Transposase-Accessible Chromatin with High-Throughput Sequencing. Source data are provided as a Source Data file.
However, we cannot exclude the potential role of Hat1 in regulating histone acetylation to modulate proinflammatory monocytes/macrophages after MI. This notion is supported by western blot analysis showing that Hat1 knockdown downregulated acetylation levels of multiple histones, including H3K23ac, in Hat1 KO BMDMs (Supplementary Fig. 15d). To directly compare the effects of Hat1 on both modifications, we performed H3K23ac CUT&Tag in Hat1-downregulated monocytes, enabling a future integrated analysis with our existing H3K23succ data. Consistent with the H3K23succ CUT&Tag findings, H3K23ac enrichment around transcription start sites (TSS ± 2 kb) was also reduced in circulating monocytes from Hat1-downregulated MI mice at day 3 (Supplementary Fig. 15e). This reduction was particularly evident within promoter regions (Supplementary Fig. 15f). Subsequent functional enrichment analysis (GO and KEGG) revealed that genes associated with these downregulated H3K23ac peaks were primarily involved in the regulation of cell cycle, DNA replication, and inflammatory processes (Supplementary Fig. 15g, h). Moreover, the level of H3K23ac was correlated with H3K23succ signals (Supplementary Fig. 15i). Through integrated profiling of H3K23succ and H3K23ac CUT&Tag datasets, we identified 1798 genes that were co-regulated by both modifications. Cross-referencing these with the 41 previously defined revealed a core set of 15 genes where Hat1 significantly modulates both H3K23ac and H3K23succ enrichment in their promoter regions (Supplementary Fig. 15j). Based on these findings, we identified that Hat1 knockdown reduced chromatin accessibility at six inflammation-related genes. Among them, four (Aim2, Nod1, Cxcl10, and Tnf) were specifically regulated by Hat1 via H3K23succ, while two (Axl and Nos2) were co-regulated by Hat1 via both H3K23succ and H3K23ac (Fig. 7h). Functional validation by RT‑qPCR confirmed significant repression of all these six genes after Hat1 knockdown (Fig. 7i). ChIP‑qPCR corroborated the reduction of H3K23succ at Aim2, Nod1, Cxcl10, and Tnf promoters without H3K23ac change, and decreased both modifications at Axl and Nos2 (Fig. 7j; Supplementary Fig. 16a). The dual luciferase reporter assay also demonstrated that Hat1 knockdown altered the promoter activities of these target genes (Supplementary Fig. 16b). These findings support a model in which Hat1 orchestrates transcriptional tuning through dual histone modification mechanisms. Notably, Hat1 depletion perturbed H3K23succ across a broader range of inflammatory gene promoters than H3K23ac, underscoring the predominant role of its succinyltransferase activity in epigenetic inflammatory regulation.
Hat1 catalyzes H3K23 succinylation via its I243 residue
Molecular docking simulations performed with HDock predicted key amino acid residues in Hat1 that facilitate its interaction with succinyl-CoA. Among these, M241, I243, Q248, G251, A254, S281, and R415 form hydrogen-bonds with the succinyl group, and are embedded within a hydrophobic environment formed by residues such as I186, E54, K284, and L285 (Supplementary Fig. 17a–c). Therefore, we hypothesize that these residues, which are hydrogen bonded to succinyl-CoA, are required for Hat1 succinyltransferase activity. These sites exhibited a high degree of evolutionary conservation across mammals (Supplementary Fig. 17d). We therefore introduced individual mutations at each residue (M241, I243, Q248, G251, A254, S281, and R415) to perturb molecular interactions with succinyl-CoA. The Hat1-I243E mutant selectively impaired succinyltransferase activity, reducing Pan Ksucc without affecting Pan Kac levels. In contrast, Hat1-Q248A specifically decreased acetyltransferase activity (reduced Pan Kac) while maintaining succinyltransferase function (unchanged Pan Ksucc), whereas the Hat1-A254E mutation disrupted both enzymatic activities (Fig. 8a). The Hat1-I243E mutant abolished H3K23succ on histones without altering H3K23ac levels (Fig. 8b), and suppressed H3K23succ in LPS and IFN-γ-stimulated BMDMs (Fig. 8c). Co-IP assays further confirmed that the Hat1-I243E mutant exhibited markedly weakened binding to H3K23succ compared to wild-type Hat1 (Fig. 8d). To investigate the functional impact of succinyltransferase-deficient mutants, we analyzed the expression of key proinflammatory Hat1 target genes (Aim2, Axl, Nod1, Cxcl10, Nos2 and Tnf). Their transcript levels were significantly downregulated in inflammatory BMDMs expressing the Hat1-I243E mutant (Fig. 8e). Chromatin immunoprecipitation further revealed that this mutant specifically abolished H3K23succ enrichment at these promoters without affecting H3K23ac (Fig. 8f; Supplementary Fig. 18a). Consistent with this, dual-luciferase reporter assays demonstrated that the I243 mutation suppressed promoter activity of downstream genes (Supplementary Fig. 18b). These results indicate that Hat1’s succinyltransferase activity is essential for promoting the expression of its inflammatory target genes.
a HEK293T cells were transfected to express Myc-tagged wild-type Hat1 (WT1) or the indicated mutants (M241K, I243E, Q248A, G251A, A254E, S281A, and R415A). Lysine succinylation and acetylation levels were assessed by immunoblotting (n = 3). b HEK293T cells were transfected to overexpress Myc-tagged WT1 or the Hat1-I243E mutant. Levels of H3K23succ and H3K23ac were analyzed by immunoblotting (n = 3; one-way ANOVA followed by Bonferroni multiple comparisons test). The blots were derived from the same experiment and processed in parallel. Total histone H3 level was set as an internal reference. c BMDMs were transfected to overexpress Myc-tagged WT1 Hat1 or the Hat1-I243E mutant. Levels of H3K23succ were analyzed by immunoblotting (n = 3; two-tailed unpaired Student t test). The blots were derived from the same experiment and processed in parallel. Total histone H3 level was set as an internal reference. d Immunoblot analysis of Hat1 and H3K23succ in protein complexes immunoprecipitated with anti-Hat1 or anti-H3K23succ antibody from lysates of WT1 or Hat1-I243E HEK293T cells (n = 3). e The mRNA expression levels of Aim2, Axl, Nod1, Cxcl10, Nos2, and Tnf detected through quantitative polymerase chain reaction (qPCR) in WT1 and Hat1-I243E BMDMs stimulated by LPS and IFN-γ for 24 h (n = 6; two-tailed unpaired Student t test). f ChIP and qPCR analysis of H3K23succ enrichment in the promoters of Aim2, Axl, Nod1, Cxcl10, Nos2, and Tnf in WT1 and Hat1-I243E BMDMs stimulated by LPS and IFN-γ for 24 h (n = 4; two-tailed unpaired Student t test). g HEK293T cells were transfected to overexpress Flag-tagged wild-type H3.3 (WT2) or H3.3K23R mutant. Levels of H3K23succ and H3K23ac were analyzed by immunoblotting (n = 4; one-way ANOVA followed by Bonferroni multiple comparisons test). The blots were derived from the same experiment and processed in parallel. Total histone H3 level was set as an internal reference. h BMDMs were transfected to overexpress Flag-tagged WT2 or H3.3K23R mutant. Levels of H3K23succ were analyzed by immunoblotting (n = 3; two-tailed unpaired Student t test). The blots were derived from the same experiment and processed in parallel. Total histone H3 level was set as an internal reference. i The mRNA expression levels of Aim2, Axl, Nod1, Cxcl10, Nos2, and Tnf detected through qPCR in WT2 and H3.3K23R BMDMs with LPS and IFN-γ stimulation for 24 h (n = 6; two-tailed unpaired Student t test). j ChIP and qPCR analysis of H3K23succ enrichment in the promoters of Aim2, Axl, Nod1, Cxcl10, Nos2, and Tnf in WT2 and H3.3K23R BMDMs with LPS and IFN-γ stimulation for 24 h (n = 4; two-tailed unpaired Student t test). Results are presented as the mean ± SD of independent replicates. Hat1 histone acetyltransferase 1, BMDMs bone marrow-derived macrophages, LPS lipopolysaccharide, IFN-γ interferon, ChIP chromatin immunoprecipitation. Source data are provided as a Source Data file.
To elucidate the role of H3K23succ in monocyte/macrophage-mediated inflammatory responses, we generated a histone H3.3-K23R point mutant. Transfection of HEK293T cells with H3.3-K23R abolished both succinylation and acetylation at lysine 23 (Fig. 8g). Consistent with this, the same abolition of H3K23succ was observed in BMDMs following inflammatory stimulation (Fig. 8h). Furthermore, LPS- and IFN-γ-induced expression of Hat1’s targeted proinflammatory genes was significantly suppressed in H3.3-K23R BMDMs compared to H3.3-WT controls (Fig. 8i). Moreover, the H3.3-K23R mutation eliminated H3K23succ and H3K23ac enrichment at the promoters of targeted genes (Fig. 8j; Supplementary Fig. 18c). Next, we overexpressed Hat1 in HEK293T cells together with either H3.3-WT or H3.3-K23R mutants. The results showed that even under Hat1 overexpression, both H3K23succ and H3K23ac modifications were unable to be catalyzed on the H3.3-K23R mutant (Supplementary Fig. 18d, e). Subsequently, we performed ChIP-qPCR to assess the binding of Hat1 to the promoter regions of downstream inflammatory genes known to be enriched with H3K23succ, such as Aim2, Axl, and Nod1. No significant difference in Hat1 enrichment was observed at these promoters following Hat1 knockdown compared to WT controls (Supplementary Fig. 18f). Taken together, these results suggest that H3K23 residue is essential for proinflammatory responses, and Hat1 does not directly bind to the promoter of target genes but catalyzes H3K23succ (possibly in coordination with H3K23ac) to mediate chromatin openness, thereby indirectly regulating the transcriptional activation of downstream inflammatory genes. Therefore, the Hat1-H3K23succ/H3K23ac axis critically mediates epigenetic regulation of the inflammatory response following MI.
Suppressed function of Hat1 improves cardiac function after myocardial ischemia/reperfusion (I/R) injury
To preliminarily assess whether monocyte Hat1 confers cardiac protection in myocardial I/R injury, we targeted Hat1 using AAV-9-F4/80-shHat1. Echocardiography showed improved cardiac function in knockdown mice, with increased LVEF and LVFS, and decreased LVIDd and LVIDs (Supplementary Fig. 19a). Hat1 reduction also attenuated inflammatory cell infiltration (Supplementary Fig. 19b) and reduced the infarct area/AAR ratio (Supplementary Fig. 19c) on day 1 post-I/R. On day 3, proinflammatory cytokines (Il-6, Il-1β, Cd86) were downregulated in cardiac tissues, while reparative genes (Cd206, Arg1) were upregulated (Supplementary Fig. 19d). Expression of Hat1-H3K23succ-specific genes (Aim2, Axl, Nod1, Cxcl10, Nos2, Tnf) was also suppressed (Supplementary Fig. 19e). These initial findings suggest that inhibiting Hat1 in monocytes improves cardiac function and alleviate inflammation after I/R injury, indicating a potential role similar to that in permanent MI, though further studies are warranted.
Our findings indicate that epigenetic changes are crucial in monocytes/macrophages during early MI progression and provide evidence that H3K23succ is upregulated relative to inflammation in monocytes post-MI. Acting through its succinyltransferase and acetyltransferase activities, Hat1 regulates the proinflammatory response, wherein the Hat1-mediated H3K23 modification state serves as a key epigenetic mechanism driving monocyte and macrophage proinflammatory responses (Supplementary Fig. 19f). Inhibiting the function of Hat1 might be an approach to limiting the inflammatory response, ameliorating cardiac ischemic injury, facilitating cardiac tissue repair, and enhancing cardiac function post-MI.
Discussion
Myeloid cells originating from the hematopoietic system induce a sharp elevation in leukocytes, particularly monocytes and macrophages, causing excessive inflammation, which is strongly correlated with complicated outcomes after MI3,21. In this study, during the early inflammatory phase of MI, elevated histone succinylation in remote monocytes exacerbated monocyte activation and inflammatory response. Moreover, we demonstrated the significance of Hat1 in triggering pro-inflammatory programming in circulating monocytes and cardiac macrophages post-MI. Hat1 loss-of-function limits pro-inflammatory gene transcription by downregulating histone succinylation levels, reducing pro-inflammatory monocyte phenotypes, alleviating ischemic injury, and facilitating cardiac repair. Our study illustrates that inhibiting the Hat1 function represents a potential therapeutic strategy for treating MI.
Under hypoxic or inflammatory environments, disease-causing TCA cycle defects cause succinate accumulation22,23. Thus, accumulating evidence suggests that succinate is closely associated with the inflammatory response, exacerbating the progression of aortic aneurysms24,25. Similarly, our preliminary findings confirmed that patients with AMI had higher serum succinate concentrations than those with UA and that high levels of succinate are strongly associated with AMI. The hydrolysis of succinyl-CoA to succinate is catalyzed by succinyl-CoA synthetases. Succinate accumulation indicates increased production of succinyl-CoA26, which, as a protein succinylating agent, mediates protein succinylation6. Hence, we investigated the global succinylation of PBMCs in patients with AMI and UA. Consistent with succinate levels, patients with AMI had higher succinylation levels in circulating monocytes, which were significantly associated with succinate concentrations. We further discovered that the global succinylation in monocytes is related to inflammatory markers, such as NLR, TNF-α, and IL−6. Thus, we believe that the upregulated succinylation in circulating monocytes from individuals with AMI is related to excessive inflammatory activity and is pathological in MI progression, similar to succinate.
Lysine succinylation has emerged as a novel post-translational modification and has been reported in cancer and cardiovascular diseases10,27. Although most studies have explored the role and function of succinylation in nonhistone proteins, a preliminary report on succinylation also includes lysine succinylation of histones28. Histone succinylation reveals a potential role in modulating gene expression and chromatin dynamics29. However, the study of histone succinylation is unreported in MI. Our study highlights an unknown role for high levels of histone succinylation in monocytes driving MI. We also found that elevated histone succinylation continued from activation to differentiation and promoted the transcription of genes associated with inflammatory pathways in monocytes at the early stage of MI. Our results support the hypothesis that high levels of histone succinylation promote the pathological progression of MI and are not conducive to recovery after cardiac infarction. Therefore, identifying the targets that effectively inhibit histone succinylation is essential.
To identify the effective targets of histone succinylation, we screened Hat1 from HATs through the transcriptome and single-cell databases, which are specifically elevated in proinflammatory monocytes and macrophages after MI. Previous studies have reported that Hat1 can regulate various histone modifications, such as acetylation30, methacrylation31, and succinylation16. As Hat1 is specifically upregulated in proinflammatory macrophages derived from circulating monocytes compared with other HATs and may function as a succinyltransferase, we selected Hat1 as the target to regulate the levels of histone succinylation post-MI. Although Hat1 has been implicated in the pathogenesis of cancer32 and early onset aging33, its function and mechanism of action in MI are partially established. Our study showed that Hat1 regulation involves a multistage and dynamic process initiated in the bone marrow, persisting through peripheral circulation, and finally executed in the local microenvironment of the heart at the early phase of MI.
Hat1 could be activated by toll-like and TNF-α receptor-triggered calcium/calmodulin-dependent protein kinase 2 and plays a central role in maintaining the inflammatory program34. Our findings further verified the significance of Hat1 in controlling the inflammatory phenotype of monocytes, reducing the infarct size, and alleviating post-MI cardiac dysfunction. In addition, we found that reducing Hat1 levels inhibited apoptosis in the border infarct regions, possibly because we inhibited excessive inflammation in macrophages after ischemic cardiac injury35. We did not enrich the anti-inflammatory pathways in the later assays of CUT&Tag and RNA-seq; however, the infarcted cardiac tissue exhibited elevated repair gene expression after Hat1 knockdown in monocytes. BMT and Ki67 staining collectively demonstrated that the expansion of reparative macrophages following Hat1 depletion originates from functionally polarized circulating monocytes, thereby explaining their accumulation in the ischemic heart. Therefore, Hat1 inhibition is equivalent to reversing the inflammatory programming of myeloid cells at its source and consolidating it at multiple subsequent steps, ultimately leading to the phenotypic switching and attenuation of inflammation that we observed locally in the heart.
Hat1 is essential for maintaining the nuclear structure and integrity and regulating the epigenetic inheritance of heterochromatin36. Our study revealed that Hat1 regulates the chromatin accessibility of inflammatory gene promoter regions. Subsequently, we investigated that Hat1 functions as a succinyltransferase with residue I243, essential for this catalytic activity, in monocytes. Hat1 regulated the level of H3K23succ more significantly in LPS- and IFN-γ-stimulated BMDMs compared with the H3K122succ site reported in the previous study16. Our findings and those of other studies suggest that Hat1 may act as a succinyltransferase and effectively regulate histone succinylation levels. Using data from two CUT&Tag (sham vs. MI and MI AAV-9-F4/80-shHat1 vs. MI AAV-9-F4/80-shNC), ATAC-seq, and RNA-seq, we found that Hat1 controlled monocyte phenotype by regulating the enrichment of H3K23succ at the promoter regions of characteristic pro-inflammatory genes. Nevertheless, the role of Hat1 in regulating histone acetylation remains relevant. Our results indicate that Hat1 modulates H3K23ac levels and cooperates with H3K23succ to co-regulate a subset of inflammatory genes. Notably, Hat1 perturbation influenced H3K23succ enrichment across a broader spectrum of promoters than H3K23ac, underscoring the more extensive and central role of its succinyltransferase activity in transcriptional regulation. This functional divergence may be attributable to the larger molecular volume and stronger charge character of the succinyl-CoA group compared to acetyl-CoA. Our results also confirm that H3K23 serves as a key mediator of Hat1-dependent immune regulation, indicating that Hat1 modulates the transcriptional activity of downstream inflammatory genes through catalytic modification of H3K23 residues. In addition, Hat1 is known to regulate inflammatory responses by facilitating the formation of the PLZF-HDAC3-NF-κB p50 transcription complex34. Therefore, we consider it possible that Hat1-mediated H3K23succ may provide the necessary platform for the binding and function of these transcription factors to jointly regulate inflammation by changing the local chromatin accessibility or may promote succinylation of these transcription factors to promote or inhibit their function. These effects may jointly establish a sophisticated transcriptional regulation program of inflammatory genes. This study provides insights into how Hat1 coordinately regulates pro-inflammatory responses through its dual enzymatic activities. Specifically, its succinylation activity at H3K23 may regulate inflammatory genes with greater precision, either in coordination with or independently of acetylation. Our findings demonstrate that the modification status of H3K23 serves as a pivotal epigenetic nexus driving pro-inflammatory responses in macrophages, and acts as an indispensable downstream effector through which Hat1 exerts its immunomodulatory functions.
To be applicable to the clinical treatment status, we expected Hat1 to have a similar protective effect in myocardial I/R injury; however, our results have only been preliminarily validated. This is a limitation of this study. Hat1 may also affect the activation, chemotaxis, and infiltration of other immune cells, including neutrophils and lymphocytes, after MI, which we also did not explore. In addition, the effect of Hat1 on regulating nonhistone protein succinylation cannot be ignored. We explored the regulatory role of Hat1 and succinylation on specific monocyte functional proteins.
Our study revealed that increased histone succinylation depends on Hat1 levels in monocytes during early MI progression. We found the significance of Hat1 in promoting pro-inflammatory gene transcription and expression through epigenetic regulation in early remote monocytes post-MI. Our findings provide a novel perspective into the pathogenic mechanisms of monocyte activation and the inflammatory response after ischemic cardiac injury, which presents a promising therapeutic target for MI treatment.
Methods
Study population
98 patients with acute coronary syndromes (ACS) were involved between December 2023 and February 2024. Patients are between 18 and 85 years old and the medical records were complete. Acute myocardial infarction (AMI) was defined as when there is acute myocardial injury with clinical evidence of acute myocardial ischemia, a rise of cardiac troponin I with at least one value >99th percentile upper reference limit, as well as with one or more of the following markers: ischemia symptoms, new ischemic ECG changes, pathological Q waves in the ECG, evidence of new viable myocardium loss or new abnormal regional wall motion obtained from imaging, or angiography findings of an intra-coronary thrombus37. Unstable angina (UA) was defined as myocardial ischemia at rest or during minimal exercise in the absence of acute cardiomyocyte injury/necrosis. The specific criteria include the following: specific clinical features of prolonged (>20 min) angina at rest; new onset of severe angina; angina that is increasing in frequency, longer in duration or lower in threshold, or angina that occurs after a recent episode of MI38. Patients with infectious diseases, severe liver dysfunction or renal insufficiency were excluded from our study. Finally, we recruited 48 patients with AMI and 50 patients with UA.
Blood sample of ACS were collected before surgery, and mostly on the first day of admission. The human plasma was obtained from the peripheral venous blood and immediately deposited into EDTA-treated tubes. The blood was then subjected to centrifugation at 3000 g for 15 min at 4 °C temperature. The plasma was delicately segregated, deposited into 1.5 mL tubes. Human PBMCs were isolated from 5 mL of peripheral venous blood using a Ficoll-Paque solution following the manufacturer’s described protocol (P8680, Solarbio). All blood samples were collected and separated by centrifugation immediately and stored at −80 °C until analysis. This study was approved by the ethics committee of the First Affiliated Hospital of Harbin Medical University (Harbin, China) and performed according to the criteria set by the Declaration of Helsinki, and all patients provided written informed consent.
ELISA assay of human plasma
The method of ELISA assay to determine the succinate, TNF-α, IL-6, and IL-1β levels. The succinate level was measured using Succinate (Succinic Acid) Colorimetric Assay Kit (ab204718, abcam). The level of TNF-α, IL-6, and IL-1β in plasma were measured using the ELISA Kit (NE104-01, Transgen Bio; JL14113, Jianglai Bio; JL13662, Jianglai Bio), according to the instruction provided by the manufacturer.
Animals
C57BL/6J male mice aged 8-10 weeks were purchased from Liaoning Changsheng Biotechnology (Benxi, China). Mice were housed at the First Affiliated Hospital of Harbin Medical University animal facility. Hat1 conventional knockout (KO) mice were generated by disrupting the Hat1 gene using CRISPR-Cas9 gene editing at Cyagen Biosciences Inc (Suzhou, China), with Hat1+/+ littermates serving as wild-type (WT) controls. Hat1 homozygous (Hat1-/-) mouse embryos were lethal, so we used Hat1 heterozygotes (Hat1+−) for our experiments. All mice were maintained on C57BL/6 background, and male mice aged 8–10 weeks were used in this study. The cages were kept on a 12-h light/12-h dark cycle, with room temperature and humidity kept constant and monitored. Animals were given access to food and water. All animal experiments were performed according to the regulations approved by the Research Ethics Committee of the First Affiliated Hospital of Harbin Medical University (Harbin, China). The animal care and surgical procedures complied with the Principles of Animal Care outlined in the National Society for Medical Research and the Guide for the Care and Use of Laboratory Animals (NIH publication).
Myocardial infarction (MI)
Mice were anesthetized by intraperitoneal injection of 2% isoflurane and placed on mechanical ventilation. After the left thoracotomy in the third intercostal space over the left chest and exposure of heart, an 7-0 silk ligature was tied around the left anterior descending branch (LAD) near the bottom edge of the left auricular appendage. The occlusion of LAD was verified by the rapid myocardial bleaching in the area between the ligation position and the heart apex. In sham-operated mice, the ligation was at a similar location but not tied. At designated time points, mice were euthanized under anesthesia, and hearts were collected for subsequent experiments.
Myocardial I/R injury model
Myocardial I/R injury model was performed by ligating at LAD with silk ligature around fine PE-10 tubing with a slipknot. Complete occlusion of the vessel was confirmed by the presence of myocardial blanching in the perfusion bed. Mice were subjected to 45 min of LAD ischemia followed by releasing the ligature and removing the PE-10 tubing. In sham-operated mice, the ligation was at a similar location but not tied. Mice were euthanized under anesthesia on day 1 post-surgery for heart collection and subsequent analysis.
Bone marrow transplantation (BMT)
CD45.1 strain mice were obtained from Cyagen Biosciences Inc. (Suzhou, China). BMT experiments were performed between WT (CD45.1) and Hat1 KO (CD45.2) mice. Bone marrow (BM) cells were harvested from the femurs and tibias of 8-week-old male donor mice after euthanasia. Recipient male mice, also aged eight weeks, underwent lethal irradiation, and were subsequently transplanted with BM cell suspensions comprising 100% CD45.1+ or CD45.2+ cells from the respective donors. To ensure uniform irradiation doses and minimize mobility, recipient mice were placed in a pie cage and exposed to two radiation doses, each amounting to 450 rad, administered four hours apart. Following the second irradiation, each recipient mouse received an injection of 5 × 106 BM cells via the retro-orbital vein plexus. During the initial 14 days post-transplantation, these mice were housed in sterile cages and provided with food and water supplemented with antibiotics. Reconstitutions were assessed through flow cytometry analysis of peripheral blood. Eight weeks after the BMT, these mice were subjected to cardiac MI surgery.
AAV-9-F4/80-shHat1 delivery in vivo
We knocked down Hat1 in murine monocytes and macrophages by constructing an adeno-associated virus 9 (AAV-9) with F4/80-specific promoter region. Both AAV-9-F4/80-shHat1 and negative control for nonsequence-specific effects were synthesized by HanBio (Shanghai, China). The nucleotide sequence of AAV-9-F4/80-shHat1/shNC are as followings: AAV-9-F4/80-shHat1 (5′- GCTACAGACTGGATATTAA-3′). We injected 5×1010 viral genome particles of AAV-9-F4/80-shHat1 vector into 8-week-old mice via bilateral tibia bone marrow39 and performed the MI and IR surgery at 2-week after gene transfer. The injection was completed with a 10 μL microsyringe at the rate of 0.1 μL/min and then withdrawn.
Monocytes isolation from mice
Mice monocytes were isolated from peripheral blood using a mouse monocyte isolation kit (Solarbio, P5230) following the manufacturer’s described protocol. Bone marrow-derived monocytes were isolated from bone marrow using a mouse bone marrow mononuclear cell isolation kit (Solarbio, P6900), similarly following the manufacturer’s described protocol.
Triphenyltetrazolium chloride (TTC) staining
The infarct size of myocardium was analyzed by TTC staining. Mice were sacrificed at 3 days after MI. The heart was quickly excised, washed two times with cold phosphate-buffer saline (PBS), immediately frozen at -20 °C for 30 min, and sliced at 1 mm thickness. Afterward, the sections were incubated in a 1% TTC (T8877, Sigma-Aldrich) solution at 37 °C for 20 min to visualize infarcts and viable myocardium. Then the sections were fixed in 4% paraformal dehyde at 4 °C for 24 h and digitally photographed. The areas of myocardial fibrosis and infarction were measured by ImageJ software.
Evans blue/TTC double-staining
Mice were anesthetized at 1 day after IR injury, the LAD artery was reoccluded at the previous ligation, and 1 mL of 1% Evans blue (E2129, Sigma-Aldrich) was injected into the left ventricular cavity. Subsequently, we performed TTC staining, which was subjected as described above for TTC staining protocols. We assessed the left ventricular area, the area at risk (AAR, stained red) and the infarcted area (IA, stained white) by computerized planimetry and comprehensively analyzed in serial sections of each mouse, using ImageJ software as well.
Histology analysis
The cardiac specimens were immersed in a 4% paraformaldehyde solution at 4 °C for 24 h. After a series of dehydration steps using an alcohol gradient and subsequent clearing, specimens were embedded in paraffin wax. Subsequently, the sliced sections (4 μm thick) were stained with haematoxylin and eosin (H&E) to assess the infiltration of cardiac inflammatory cells. To investigate the effects of Hat1 on cardiomyocyte apoptosis, terminal deoxynucleotidyl transferase-mediated deoxyuridinetriphate nick-end labeling (TUNEL) staining was performed using HRP/ Alexa 488/Cy3 TUNEL Cell Apoptosis Detection Kit (AFIHC030, Aifang Bio) according to the manufacturer’s protocol. Following this, the sections were co-stained with Anti-Sarcomeric Alpha Actinin (ab68167, abcam) and DAPI (C1005, Beyotime Bio) to specifically label cardiomyocytes and nuclear. Images were quantified using ImageJ software. Green fluorescence-labeled apoptotic cells were counted, in order to assess the apoptotic index (number of TUNEL-positive cardiomyocytes).
Immumohistochemical (IHC) staining was performed using paraffin-embedded tissue sections. After the inhibition of endogenous peroxidase activity, the sections were incubated with primary Hat1 polyclonal antibody (11432-1-AP, Proteintech), at 4 °C overnight. Following the incubation, the appropriate Polymer-HRP-coupled secondary antibodies (PV6001, ZSGB Bio) were used, and they were applied for 1 h at room temperature. After the visualization with DAB (ZLI-9018, ZSGB Bio), the sections were counterstained with hematoxylin.
Multiplex immunofluorescence was performed using double-labeled multiplex immunofluorescence kit following the manufacturer’s described protocol (AFIHC023, Aifang Bio). After deparaffinization, heat-mediated antigen retrieval in citrate buffer pH 6.0 and blocking with 5% goat serum at room temperature for 30 min, appropriate antibodies were used to detect the specific protein expression. Primary antibodies used were Hat1 polyclonal antibody (11432-1-AP, Proteintech), mouse F4/80 Polyclonal antibody (29414-1-AP, Proteintech), Anti-Succinyl-Histone H3 (Lys23) Mouse mAb (PTM-422, PTM Bio), Anti-Succinyllysine Rabbit pAb (PTM-401, PTM Bio), and the samples were incubated with these antibodies overnight at 4 °C. The appropriate Polymer-HRP-coupled secondary antibodies (PV6001, PV6002, ZSGB Bio) were used, and they were applied for 1 h at room temperature. Afterward, the sections were counterstained with DAPI (C1005, Beyotime Bio) and cover slipped, which was followed by analysis using fluorescence microscopy. The multiplex immunohistochemistry staining procedure closely resembled traditional IHC, with the addition of a signal amplification step involving tyramide signal amplification inserted into each staining cycle. This modification enables the simultaneous detection of multiple biomarkers in the same paraffin-embedded section through successive rounds of single-label and multi-round re-staining.
Echocardiography analysis
Transthoracic echocardiography was performed using the high-resolution VIVID E9 imaging system (GE Healthcare) with a 30 MHz probe. Briefly, mice were gently anesthetized with 2% isoflurane. The cardiac images were obtained in two-dimensional mode in the parasternal long-axis section. Within this section, an M-mode cursor was placed vertically to the interventricular septum and the posterior wall of the left ventricle, at the level of the root of the papillary muscles. The left ventricular inner diameters, including the left ventricular internal diameter at end-diastole (LVIDd) and left ventricular internal diameter at end-systole (LVIDs), left ventricular ejection fraction (LVEF), and fractional shortening (LVFS) were measured and calculated from three independent cardiac cycles.
Flow cytometric analysis
Single cell suspensions were prepared from peripheral blood, bone marrow and heart tissue. Red blood cells from peripheral blood and bone marrow were lysed using ACK lysis buffer (A10492-01, Gibco) for backup use. The procedure for preparing cardiac single cell suspensions is described below. The hearts from MI mice were extensively rinsed and then removed. Use scissors to mince the heart tissue and digest it with a cocktail of 1 mg/mL type II collagenase (BS164, Biosharp), 100 U/mL hyaluronidase (H3506, Sigma-Aldrich), and 100 U/mL DNase I (10104159001, Sigma-Aldrich) for 1 h at 37 °C with gentle agitation. Following the digestion, the tissue samples were triturated and passed through a 70 µm cell strainer (Abs7008, Absin). The obtained cells were enriched, and the isolated cells were counted after erythrocyte lysis and washed with RPMI-1640 cell culture medium for further analysis.
Afterwards, single-cell suspensions were incubated with anti-CD16/32 antibody (156603, Biolegend) to prevent unspecific antibody binding. Thereafter, cells were stained with fluorochrome-labeled antibodies at 4 °C for 30 min. CD206 and Hat1 staining was performed using the True-NuclearTM Transcription Factor Buffer Set (424401, Biolegend). The samples were analyzed by Flow Cytometer (Apogee), and the data were analyzed by FlowJo V10. The primary antibodies are listed in Supplementary Data 2.
Hypoxic primary cardiomyocytes were digested with trypsin, after which the digestion was stopped and the cells were resuspended. The procedure was performed according to the instructions of the apoptosis detection kit (CA1020, Solarbio). The samples were mixed and analyzed by flow cytometry. FlowJo software was used to quantify the proportion and number of cells in various states, including early apoptosis, late apoptosis, and cell death.
HEK293T cells culture and treatment
The HEK293T cell line was kindly provided by the Stem Cell Bank, Chinese Academy of Sciences (CAS) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS) according to the manufacturer’s instructions. Cells were used within five passages for all experiments. Transfection of plasmids was performed using Lipofectamine 3000 (L3000150, Invitrogen,) following the manufacturer’s protocol. A DNA sequence encoding Hat1 was cloned into the vector (pCMV-MCS-3×Myc-Neo) to create the Myc-Hat1 plasmid. The Hat1 (M241K, I243E, Q248A, G251A, A254E, S281A and R415A) mutant was generated from the Myc-Hat1 plasmid using the QuikChange site-directed mutagenesis kit (Agilent). The plasmids were purchased from Miaoling Co. Ltd, Wuhan, China. A DNA sequence encoding H3.3 was cloned into the vector (pcDNA3.1-C-3×Flag) to create the Flag-H3.3 plasmid. The H3.3K23R mutant was generated from the Flag-H3.3 plasmid using the QuikChange site-directed mutagenesis kit (Agilent). The plasmids were purchased from Jima Co. Ltd, Suzhou, China.
Bone marrow-derived macrophages (BMDMs) culture and treatment
Bone marrow-derived macrophages (BMDMs) were isolated and differentiated into mature macrophages. BM cells were flushed from the tibias and femurs of 6-to 8-week-old male mice. Red blood cells were removed by ACK lysis buffer (A10492-01, Gibco). After beinf washed, the cell suspension was passed a 100 μm cell strainer and supplemented with 20 ng/mL macrophage colony stimulating factor (M-CSF, MCE) in RPMI 1640 medium with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin for 7 days. The culture medium was replaced every 3 days. Lentiviral particles used to knockdown Hat1 in BMDMs were purchased from GenePharma (Suzhou, China). Primary BMDMs were plated in a six-well plate, infected with the Hat1 lentivirus (shHat1), Myc-Hat1Mutant plasmid and Flag-H3.3K23R plasmid. The following shRNAs were used in this study: mouse Hat1 shRNA: 5′- GCTACAGACTGGATATTAA-3′. Mouse promoters, including Aim2, Axl, Nod1, Cxcl10, Nos2 and Tnf, which contained putative H3K23succ binding sites, were amplified and cloned into the pGL3-Basic vector (Promega) to produce firefly reporter plasmid. Different fragments of targeted gene promoters were inserted into pGL3-Basic vector to yield various firefly reporter plasmids. pRL-TK vector (constitutively expressing Renilla luciferase) was purchased from Promega. All plasmids were verified by DNA sequencing. 24 h after transfection, cells were treated with lipopolysaccharide (LPS, 100 ng/mL, Sigma-Aldrich) + IFN-γ (20 ng/mL, MCE) for 24 h or left untreated, and used to perform further experiments.
Isolation of neonatal mouse cardiomyocytes (CMs) and treatment
Hearts were aseptically isolated from neonatal mice (1–3 days old) and rinsed three times with D-Hanks solution. The tissues were then minced into approximately 1 mm³ fragments and digested with 0.25% trypsin. After digestion, the cells were collected by centrifugation at 1500 rpm for 5 minutes and resuspended in DMEM supplemented with 10% fetal bovine serum. To separate cardiac fibroblasts from CMs, differential adhesion was performed based on their distinct attachment kinetics. The resulting purified primary neonatal mouse cardiomyocytes were seeded into culture flasks or 6-well plates according to experimental requirements and maintained at 37 °C in a 5% CO₂ incubator in DMEM containing 10% FBS. Cardiomyocytes (CMs) were either subjected directly to 24 h of hypoxia or treated for 24 h with conditioned medium from inflammatory-stimulated BMDMs (WT/Hat1 KO), followed by an additional 24 h period of hypoxia.
Protein extraction and immunoblot
Whole-cell lysates were extracted using cell lysis buffer supplemented with protease inhibitor cocktail and PMSF, and histones were extracted according to the guidelines provided in the histone extraction kit manual (OP-0006, EpigenTek). Subsequently, the proteins were resolved using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, then incubated with specific antibodies. The membranes were blocked using 5% milk and incubated overnight at 4 °C with primary antibodies. The dilution doses of antibodies were according to the instructions. The next day, the membranes were then incubated with secondary antibodies for 1 h at room temperature. The band signals were visualized using Tanon Image software. The signal intensities of the target western bands were normalized with internal control protein, and then calculated as the fold change compared with the control group. The primary antibodies are listed in Supplementary Data 2.
RNA isolation and quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was extracted using Trizol reagent (Invitrogen, USA) and reverse-transcribed into complementary DNA using the Transcriptor First Strand cDNA Synthesis Kit (FSQ101, TOYOBO) following the manufacturer’s protocol. RT-qPCR was performed using Fast Start Universal SYBR®Green Master Mix (Q711-02, Vazyme) for analysis. The specific primers are listed in Supplementary Data 2.
Molecular docking
AlphaFold2 was used to predict and construct histone H3 (Uniprot ID: P68431)40. Hat1 (PDB ID: 2P0W) and succinyl-CoA (PDB ID: 5TRL) structure was used from the PDB database41. Select HDock42, the software is based on global search and mixed strategies for protein-docking, has achieved excellent performance docking score 237.22 kcal/mol. PyMOL 2.5.1was performed for graphical display and LigPlot 2.1 was used for visual display43,44.
Molecular dynamics simulation
Molecular dynamics simulations were performed using Gromacs 2019.645. The Amber14SB force field46 was applied to the protein, while the GAFF2 force field47 was used for non-standard residues. The system was solvated in a TIP3P water model within a periodic box and neutralized with sodium ions. For energy minimization, the steepest descent algorithm was employed with a maximum of 50,000 steps. Short-range nonbonded interactions were truncated at 1.4 nm for both Coulombic and van der Waals interactions. Long-range electrostatics were treated with the Particle Mesh Ewald method. The system was subsequently equilibrated under NVT and NPT ensembles, followed by a production MD simulation of 100 ns at 300 K and 1 bar. During the simulation, hydrogen bonds were constrained with the LINCS algorithm using a 2-fs integration time step. A cutoff of 1.2 nm was set for short-range electrostatics and van der Waals interactions. Temperature and pressure were maintained at 300 K and 1 bar using the V-rescale thermostat and Berendsen barostat, respectively. Both NVT and NPT equilibrations were conducted for 30 ps each. The root mean square deviation (RMSD) was calculated to assess structural convergence, with a fluctuation threshold set at 0.2 nm. The solvent accessible surface area (SASA) was monitored to evaluate changes in surface exposure of the complex during the simulation.
Immunoprecipitation assay (IP)
The total protein from the cell lysate was immunoprecipitated. Firstly, the extract was incubated with anti-Succinyl-Histone H3 (Lys23) Mouse mAb (PTM-422, PTM Bio) or Hat1 polyclonal antibody (11432-1-AP, Proteintech) for 24 h at 4 °C. The protein A/G Magnetic Beads (HY-K0202-1 mL, MCE) were added and further incubated for 3 h at 4 °C followed by centrifuged at 12,000 g for 5 min. Then, recovered the precipitate for washing, resuspended it in 30 μL SDS lysis buffer and boiled at 95 °C for 5 min, and finally analyzed the precipitate by immunoblotting with the indicated antibody.
RNA sequencing
The total RNA was extracted from 1 × 106 circulating monocytes using TRIzol reagent (n = 3 biologically independent samples). The TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) was used to construct an RNA-seq library. The RNA sequencing was performed by OE Biotechnology (Shanghai, China). Raw data reads quality was checked by FastQC and then mapped to the mouse reference genome (mm10), and quantification was performed using salmon (v1.4.0) and tximport (v1.26.1). Differential expression analysis was performed using DESeq2 (v1.38.2) to identify differential expression genes (DEGs)48. Based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) database49,50, the enrichment analysis of DEGs was conducted using clusterProfiler package (v4.6.0) to reveal their potential biological functions51. Multiple hypothesis tests were corrected by the Benjamini-Hochberg method.
The single-cell RNA sequencing (scRNA-seq) data preprocessing and analysis
The scRNA-seq dataset of mouse heart after myocardial infarction was obtained from the Gene Expression Omnibus (GEO) database (GSE163129). Initially, cells expressing at least 200 genes were selected, and the genes were expressed in at least 3 cells, and cells with a percentage of mitochondria greater than 10% were removed. Subsequently, the DoubletFinder package (v2.0.3) was employed to further identify and remove doublets52. After quality control, the integrated workflow recommended by Seurat 4 (v4.0.5) was adopted, and batch effects were mitigated and data were integrated using harmony package (v0.1.1)53,54. Next, data were clustered using the “FindNeighbor” and “FindClusters” functions, and the “RunTSNE” function was used to generate visualization plots based on the two-dimensional t-distributed stochastic neighbor embedding (t-SNE) model. Major cell clusters were assigned based on known cellular lineages predicted by marker genes within the t-SNE model. Subsequently, manual assignment of cell clusters to major cell types was performed based on established markers. Clusters showing markers for two distinct cell types were manually identified and excluded.
Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq)
ATAC-seq libraries were constructed using using Hyperactive ATAC-Seq Library Prep Kit (TD711, Vazyme Biotech, China) in accordance with the manufacturer’s instructions. NGS was leveraged to sequence the DNA (n = 3 biologically independent samples). Cell preparation was similar with CUT&Tag. Cutadapt (v1.18) was employed to remove sequencing adapters, short reads, and low-quality reads, yielding clean and high-quality data55. Subsequently, data quality was assessed using FastQC (v0.12.1). Fixed reads were then mapped to the mouse reference genome (mm10) using Bowtie2 (v2.5.1), followed by filtration using Samtools (v1.6), and the results were saved in BAM format56,57. PCR duplicates were removed using Picard (v2.0.1)58. Peak calling was performed using MACS2 (v2.2.7.1) with default parameters to identify significant genomic regions59. Finally, BED files were utilized alongside the ChIPSeeker package for peak annotation of genomic features60. To visualize the occupancy of the genome around peaks within ±2 kb of the transcription start sites (TSS), BAM files were converted to BigWig format using the bamCoverage function in deepTools (v3.5.1), and visualization was conducted using the computeMatrix and plotHeatmap functions61. For representation in genome browsers, the BigWig files were loaded into the Integrative Genomics Viewer.
Cleavage under targets & tagmentation (CUT&Tag)
CUT&Tag libraries were constructed using Hyperactive Universal CUT&Tag Assay Kit (TD903, TD904, Vazyme Biotech, China). For cell preparation, the monocytes were isolated from mice peripheral blood (Sham vs MI, MI AAV-9-F4/80-shHat1 vs AAV-9-F4/80-shNC) using the method described above. The cells (1 × 105) were coupled with concanavalin A-coated magnetic beads, followed by an overnight incubation at 4 °C with the indicated antibodies respectively. Subsequent to this, samples were incubated with a secondary antibody and Hyperactive pA/G-Transposon for 1 h. The DNA was subjected to TTBL treatment at 37 °C for 1 h, followed by extraction and purification for library amplification. Next Generation Sequencing (NGS) was leveraged to sequence the DNA (n = 3 biologically independent samples). Cutadapt (v1.18) was employed to trim sequencing adapters and filter out short, low-quality reads to generate high-quality data55. Subsequently, data quality was assessed using FastQC (v0.12.1). Processed reads were aligned to the mouse reference genome (mm10) using Bowtie2 (v2.5.1)56. Samtools (v1.6) was utilized to filter SAM format files and save them in BAM format57. Picard (v2.0.1) was utilized to remove PCR duplicates58. MACS2 (v2.2.7.1) was utilized for peak calling to identify significant genomic regions59. ChIPseeker was used to annotate peak positions in genomic features60. Additionally, deepTools (v3.5.1) was employed to convert BAM files to BigWig format for peak count normalization and visualization. Finally, these BigWig files were uploaded and viewed using the Integrative Genomics Viewer. ChIPseeker was used to annotate peak positions in genomic features. Additionally, deepTools (v3.5.1) was employed to convert BAM files to BigWig format for peak count normalization and visualization61. Finally, these BigWig files were uploaded and viewed using the Integrative Genomics Viewer.
Chromatin immunoprecipitation (ChIP)-qPCR
To prepare the samples for ChIP, 5 × 106 BMDMs/sample were cross-linked with 37% methanol-free formaldehyde for 10 min before performing a ChIP assay using SimpleChIP Plus Enzymatic Chromatin IP Kit (#9005, Cell Signaling Technology). The chromatin is then fragmented by sonication into segments of 150-900 bp in length. The fragmented chromatin was then incubated with specific primary antibodies targeting anti-Succinyl-Histone H3 (Lys23) Mouse mAb (PTM-422, PTM Bio) overnight at 4 °C with gentle rotation. After that, magnetic Beads were added into the sample and incubated for 2 h with rotation. Then the beads were sequentially washed in washing buffers with different salt concentrations and finally eluted with an elution buffer containing 1% SDS. The obtained antibody-chromatin complex sequentially underwent the steps of reverse cross-linking and DNA purification. The obtained DNA samples were subjected to RT-qPCR using the primers listed in Supplementary Data 2. The primers were designed to determine the enrichment of H3K23succ.
Dual-luciferase reporter assay
The plasmids were purchased from Miaoling Co., Ltd, Wuhan, China. Plasmid transfection for dual-luciferase reporter assay was conducted as described above. Luciferase activity was assessed using the Dual-Luciferase Reporter Assay System (E2920, Promega) following the guidelines provided by the manufacturer. In brief, cells were collected in a passive lysis buffer, and the activities of firefly and renilla luciferase in the lysate were quantified. Renilla luciferase activity (pRL-TK) served as an internal control due to variations in transfection efficiency. The luciferase activity of firefly was normalized to that of renilla to determine the promoter activity. The results were expressed as fold changes compared to the empty vector group.
Statistical analysis
Data were analyzed using SPSS (version 26.0), R (version 4.2.2) and GraphPad Prism (version 9.0.1). Categorical variables were expressed as numbers and percentages. After the Kolmogorov-Smirnov test of data distribution, continuous variables were shown as mean ± standard deviation for normally distributed data or as median (quartile) for non-normally distributed data. Between-group comparisons were performed by Student’s t test, Mann-Whitney U test, one-way ANOVA, two-way ANOVA, Kruskal–Wallis H test, χ2 test, or Fisher’s exact test, as appropriate. Odds ratios (ORs) and 95% CIs were also calculated. Restricted cubic spline analysis was applied to evaluate the association between plasma succinate levels and AMI. Receiver operating curves were performed to predict the diagnostic value of succinate in AMI. Linear regression was used to show the correlation between succinate and the global succinylation expression. Data violating core assumptions of each test were either transformed or alternative (non-parametric) tests were used, as indicated in detail in the respective figure legends. If not stated otherwise, multiplicity-adjusted (Bonferroni) P values are reported throughout, with the threshold of α set at 0.05 (two-tailed).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings described in this study are available in the article and the Supplementary Information. The CUT&Tag, ATAC-seq, and RNA-seq data generated in this study have been deposited in the Genome Sequence Archive (GSA)62 at the National Genomics Data Center63 under accession code CRA030478. Other publicly available data used in this study can be found in the Gene Expression Omnibus (GEO) under accession numbers GSE20628164, GSE16312965, and GSE77566. Source data are provided with this paper.
References
Reed, G. W., Rossi, J. E. & Cannon, C. P. Acute myocardial infarction. Lancet 389, 197–210 (2017).
Yap, J. et al. Macrophages in cardiac remodelling after myocardial infarction. Nat. Rev. Cardiol. 20, 373–385 (2023).
Dutta, P. et al. Myocardial infarction activates CCR2(+) hematopoietic stem and progenitor cells. Cell Stem Cell 16, 477–487 (2015).
Marinković, G. et al. S100A9 links inflammation and repair in myocardial infarction. Circ. Res. 127, 664–676 (2020).
Tessarz, P. & Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 15, 703–708 (2014).
Zhang, Z. et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 (2011).
Wang, Y. F. et al. Aspirin modulates succinylation of PGAM1K99 to restrict the glycolysis through NF-κB/HAT1/PGAM1 signaling in liver cancer. Acta Pharm. Sin. 44, 211–220 (2023).
Liu, L. et al. Protective role of SIRT5 against motor deficit and dopaminergic degeneration in MPTP-induced mice model of Parkinson’s disease. Behav. Brain Res. 281, 215–221 (2015).
Du, Y. et al. SIRT5 deacylates metabolism-related proteins and attenuates hepatic steatosis in OB/OB mice. EBioMedicine 36, 347–357 (2018).
Sadhukhan, S. et al. Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proc. Natl. Acad. Sci. USA 113, 4320–4325 (2016).
Bai, F. et al. Quantitative proteomics of changes in succinylated proteins expression profiling in left appendages tissue from valvular heart disease patients with atrial fibrillation. Clin. Chim. Acta 495, 345–354 (2019).
Dang, F. & Wei, W. Targeting the acetylation signaling pathway in cancer therapy. Semin. Cancer Biol. 85, 209–218 (2022).
Ai, X. & Parthun, M. R. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14, 195–205 (2004).
Capone, V. et al. HAT1: landscape of biological function and role in cancer. Cells 12, 1075 (2023).
Espíndola, M. S. et al. Epigenetic alterations are associated with monocyte immune dysfunctions in HIV-1 infection. Sci. Rep. 8, 5505 (2018).
Yang, G. et al. Histone acetyltransferase 1 is a succinyltransferase for histones and non-histones and promotes tumorigenesis. EMBO Rep. 22, e50967 (2021).
Zhang, J. et al. Accumulation of succinate in cardiac ischemia primarily occurs via canonical krebs cycle activity. Cell Rep. 23, 2617–2628 (2018).
Ammirati, E. et al. Identification and predictive value of interleukin-6+ interleukin-10+ and interleukin-6- interleukin-10+ cytokine patterns in ST-elevation acute myocardial infarction. Circ. Res. 111, 1336–1348 (2012).
Xiao, H. et al. CIRKIL Exacerbates Cardiac Ischemia/Reperfusion Injury by Interacting With Ku70. Circ. Res. 130, e3–e17 (2022).
Zheng, H. et al. CircRNA Samd4 induces cardiac repair after myocardial infarction by blocking mitochondria-derived ROS output. Mol. Ther. 30, 3477–3498 (2022).
Prabhu, S. D. & Frangogiannis, N. G. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ. Res. 119, 91–112 (2016).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Chouchani, E. T. et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515, 431–435 (2014).
Cui, H. et al. Untargeted metabolomics identifies succinate as a biomarker and therapeutic target in aortic aneurysm and dissection. Eur. Heart J. 42, 4373–4385 (2021).
Caielli, S. et al. A CD4(+) T cell population expanded in lupus blood provides B cell help through interleukin-10 and succinate. Nat. Med. 25, 75–81 (2019).
Ryan, D. G. & O’Neill, L. A. J. Krebs cycle reborn in macrophage immunometabolism. Annu. Rev. Immunol. 38, 289–313 (2020).
Ma, W. et al. OXCT1 functions as a succinyltransferase, contributing to hepatocellular carcinoma via succinylating LACTB. Mol. Cell 84, 538–551.e537 (2024).
Xie, Z. et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell Proteom. 11, 100–107 (2012).
Zorro Shahidian, L. et al. Succinylation of H3K122 destabilizes nucleosomes and enhances transcription. EMBO Rep. 22, e51009 (2021).
Xu, Q. et al. ACL and HAT1 form a nuclear module to acetylate histone H4K5 and promote cell proliferation. Nat. Commun. 14, 3265 (2023).
Delaney, K. et al. Histone lysine methacrylation is a dynamic post-translational modification regulated by HAT1 and SIRT2. Cell Discov. 7, 122 (2021).
Carafa, V. et al. RIP1-HAT1-SIRT complex identification and targeting in treatment and prevention of cancer. Clin. Cancer Res 24, 2886–2900 (2018).
Nagarajan, P. et al. Early-onset aging and mitochondrial defects associated with loss of histone acetyltransferase 1 (Hat1). Aging Cell 18, e12992 (2019).
Sadler, A. J. et al. The acetyltransferase HAT1 moderates the NF-κB response by regulating the transcription factor PLZF. Nat. Commun. 6, 6795 (2015).
Chiong, M. et al. Cardiomyocyte death: mechanisms and translational implications. Cell Death Dis. 2, e244 (2011).
Popova, L. V. et al. Epigenetic regulation of nuclear lamina-associated heterochromatin by HAT1 and the acetylation of newly synthesized histones. Nucleic Acids Res 49, 12136–12151 (2021).
Thygesen, K. et al. Fourth universal definition of myocardial infarction (2018). Eur. Heart J. 40, 237–269 (2019).
Byrne, R. A. et al. 2023 ESC Guidelines for the management of acute coronary syndromes. Eur. Heart J. 44, 3720–3826 (2023).
Chen, P. et al. Scara3 regulates bone marrow mesenchymal stem cell fate switch between osteoblasts and adipocytes by promoting Foxo1. Cell Prolif. 54, e13095 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Yan, Y., Zhang, D., Zhou, P., Li, B. & Huang, S. Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 45, W365–w373 (2017).
Mooers, B. H. M. Shortcuts for faster image creation in PyMOL. Protein Sci. 29, 268–276 (2020).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model 51, 2778–2786 (2011).
Van Der Spoel, D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005).
Maier, J. A. et al. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
He, X., Man, V. H., Yang, W., Lee, T. S. & Wang, J. A fast and high-quality charge model for the next generation general AMBER force field. J. Chem. Phys. 153, 114502 (2020).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Gene Ontology, C. et al. The Gene Ontology knowledgebase in 2023. Genetics 224, iyad031 (2023).
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima, K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–d361 (2017).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 e324 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Methods 16, 1289–1296 (2019).
Kechin, A., Boyarskikh, U., Kel, A. & Filipenko, M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24, 1138–1143 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Ramírez, F. et al. deepTools2: a next-generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Chen, T. et al. The genome sequence archive family: toward explosive data growth and diverse data types. Genom. Proteom. Bioinforma. 19, 578–583 (2021).
CNCB-NGDC Members Partners Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2025. Nucleic Acids Res. 53, D30-d44 (2025).
Han, Y. et al. Corrigendum: Analysis of time series gene expression and DNA methylation reveals the molecular features of myocardial infarction progression. Front. Cardiovasc. Med. 9, 990217 (2022).
Jung, S. H. et al. Spatiotemporal dynamics of macrophage heterogeneity and a potential function of Trem2(hi) macrophages in infarcted hearts. Nat. Commun. 13, 4580 (2022).
Tarnavski, O. et al. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol. Genom. 16, 349–360 (2004).
Acknowledgements
This study was supported by the National Natural Science Foundation of China (81871402, 82570357 to Y.T., 82170270, 82370248 to Z.D.), Scientific Research Foundation for Postdoctoral Heilongjiang Province of China (LBH-Q19037 to Y.T.), Research Project of the First Affiliated Hospital of Harbin Medical University (2021J01 to Z.D.), and Postgraduate Research & Practice Innovation Program of Harbin Medical University (YJSCX2023-42HYD to Y.G., YJSCX2023-181HYD to L.B.). The material in figures in this article is supported by Figdraw for free.
Author information
Authors and Affiliations
Contributions
Yutong G. conceived the project, designed the study, performed most of the experiments, analyzed the data, and wrote the manuscript. J.X. performed bioinformatic analysis and animal models. Z.L. performed clinical data analysis and morphological experiments. L.B. performed cell culture and immunoblot analysis. B.W. performed cell culture and RT-qPCR. B.G. performed ELISA assays. H.T. isolated neonatal mouse cardiomyocytes and flow cytometry apoptosis assay. G.L. collected human blood samples and clinical information. Y.Q. performed immunoblot analysis. L.L. performed echocardiography analysis. Z.D. provided funding and conceptual input. Y.T. supervised the study, provided funding and resources.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Cheng-chao Ruan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, Y., Xiong, J., Li, Z. et al. Histone acetyltransferase 1 promotes postinfarction inflammatory response by regulation of monocyte histone succinylation. Nat Commun 16, 11718 (2025). https://doi.org/10.1038/s41467-025-66721-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66721-9