Abstract
Quiescence (reversible cell-cycle arrest) and senescence (irreversible arrest) are challenging to distinguish due to a lack of specific biomarkers, yet both arise simultaneously after chemotherapy. While senescence suppresses tumors by limiting proliferation and recruiting the immune system, quiescent cancer cells evade future therapies and may resume proliferation. Here, we pair time-lapse imaging of cell-cycle dynamics with single-cell RNA sequencing after etoposide treatment to differentiate these states, linking heterogeneous cell-cycle phenotypes to the transcriptomic landscape. We identify diverse senescent types (senotypes) and link them to two arrest pathways — a gradual path arising after a standard mitosis-to-G0 transition, and an alternative direct path driven by a mitotic slip. Using pseudotime trajectory analysis, we find that senescent phenotypes begin to manifest early and gradually along the first trajectory, even in shallow quiescent cells. These data support a model wherein, following chemotherapy, quiescence and senescence exist on a continuum of cell-cycle withdrawal at a transcriptome-wide level.
Similar content being viewed by others
Introduction
The cellular decision to proliferate or withdraw from the cell cycle is essential for maintaining tissue health and homeostasis. When this decision goes awry, it results in diseases of hypo-proliferation, such as aging, or hyper-proliferation, such as cancer. Despite its importance, the molecular details of cell-cycle withdrawal are not fully understood. Quiescence is a state of transient and reversible cell-cycle withdrawal that occurs in response to low levels of DNA damage or lack of mitogens or nutrients1. Senescence is a state of irreversible cell-cycle withdrawal wherein cells remain metabolically active and viable but never divide again2,3. Senescence has a normal, physiological role in development, but also occurs in response to sublethal DNA damage or stress4. This arrest is initially triggered and maintained by the p53/p21 and Rb/p16 signaling axes2,3,5 and is accompanied by many cellular changes, including increases in cell size, upregulation of autophagy, changes in chromatin, and the secretion of pro-inflammatory and pro-proliferation proteins called the senescence-associated secretory phenotype (SASP)2,6,7,8.
Therapy-induced senescence (TIS) is a permanent cell-cycle withdrawal induced by exposure to radiation or chemotherapeutic drugs like etoposide and doxorubicin9,10. In the context of cancer, TIS has been identified as an alternative therapeutic strategy to cell death since these cells are irreversibly arrested, preventing the proliferation of damaged premalignant cells and initiating immune clearance through the SASP9,11. There is still debate about TIS as an end goal for therapies because some work suggests that cells may be able to evade TIS, remaining dormant for a time before returning to the cell cycle12,13,14. Therefore, understanding the molecular relationship between cells that are transiently quiescent and cells that are senescent is critically important.
Studying the relationship between these arrested states is challenging due to the lack of distinct, consistent, and specific biomarkers15, and because measuring irreversible arrest in heterogeneous populations requires single-cell, time-lapse data to define the ground-truth senescent cells. To overcome this, we recently used single-cell time-lapse microscopy and a live-cell CDK2-activity reporter to track each cell’s cumulative cell-cycle withdrawal duration across thousands of single cells recovering from etoposide, oxidative stress, or ionizing radiation16. This approach distinguished irreversibly arrested senescent cells from slow-cycling cells that spend long periods of time in quiescence between cell cycles and showed that both fates arise from the same treatment. By correlating each cell’s withdrawal duration with the intensity of eight canonical senescence markers, we found that both quiescent and senescent cells express these eight markers with a graded intensity that reflects each cell’s duration of cell-cycle withdrawal16. These findings suggested that chemotherapy-induced cell-cycle withdrawal may be a graded continuum rather than a binary decision between quiescence and senescence. However, an open question following this work is whether binary markers of quiescence vs. senescence might nevertheless exist, and whether these eight graded markers are representative of changes across the entire transcriptome. Put another way, at the transcriptome level, it is unclear whether quiescent and senescent states exist as distinct islands within a population of heterogeneous cells, or whether they exist on a continuum connected by a bridge.
Here, we address these questions by measuring the changing transcriptome in senescent cells by performing live-cell time-lapse microscopy in parallel with single-cell RNA sequencing in MCF10A mammary epithelial cells following chemotherapy. This approach avoids reliance on imperfect senescence markers because it uses long-term cell-cycle withdrawal to define senescent cells and enables transcriptome-wide analysis of gene expression changes. We uncovered a gradient of cellular states corresponding to varying probabilities of cell-cycle re-entry, supporting the quiescence-senescence continuum model across the transcriptome. Notably, quiescent cells lacked a unique signature and instead exhibited attenuated senescence-associated gene expression changes. Additionally, within a single population, we identified distinct senescence types, or senotypes17, with distinct gene expression profiles that resulted from different paths to senescence. This work contributes to a growing body of research that aims to fully understand the molecular characteristics associated with reversible and irreversible cell-cycle withdrawal.
Results
Establishment of populations with increasing fractions of senescent cells
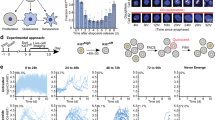
To study senescent cells at the transcriptomic level, it is essential to identify experimental conditions that yield a homogeneous senescent population as measured by a ground-truth assessment of senescence: the total lack of proliferation over time. To test whether we could alter the proportion of quiescent and senescent cells, we treated MCF10A cells with increasing doses of the chemotherapy, etoposide, a widely used inducer of senescence that causes DNA damage and cell-cycle arrest18. After treating cells for 24 h, we washed off the etoposide and allowed cells to recover in drug-free media for 6 days (Fig. 1A). On day 6, cells were fixed and stained for phospho-Rb, a marker of cell-cycle commitment19 (Fig. 1B). Higher etoposide doses resulted in fewer cycling (pRbhigh) cells. In parallel, we used time-lapse microscopy to track MCF10A cells expressing a DNA helicase B (DHB)-based CDK2-activity reporter20 (Fig. S1A) and an H2B-mTurquoise nuclear marker from day 6 to day 10 post-etoposide. CDK2 activity turns on at cell-cycle commitment, rises throughout the cell cycle, and turns off when cells withdraw from the cell cycle into quiescence or senescence. Higher etoposide doses produced a greater proportion of CDK2low non-cycling cells over the 4-day imaging period (Fig. 1C, D). We plotted the distribution of cell-cycle withdrawal durations, measured as time spent CDK2low (CDK2 activity < 0.8) for each etoposide dose and categorized cells as fast-cycling, slow-cycling, or predicted-senescent based on CDK2low durations (Figs. 1E, S1B, C). At 2.5 µM and 10 µM, we observed a mix of all three categories, while 25 µM predominantly resulted in senescent cells. We confirmed the expected expression of canonical senescent markers using immunofluorescent (IF) imaging immediately following the live-cell movies (Fig. S1D). Notably, while the average expression of canonical senescence biomarkers increased with duration of cell-cycle arrest, all cell fates showed heterogeneous expression of these markers across the population, including predicted-senescent cells. These results agreed with our previous findings that none of these biomarkers were uniquely expressed in senescent cells after etoposide or H2O216. We further confirmed these findings in MCF7 and RPE-hTERT cells, showing that senescent cells had an increased but heterogeneous expression of canonical senescence biomarkers following etoposide (Fig. S2A–C). These results showed that etoposide dose modulated the proportion of predicted-senescent cells as well as their expression of canonical senescent markers.
A Experimental timeline. B MCF10A cells were treated with 2.5 μM, 10 μM, or 25 μM etoposide for 24 h, followed by a 6 d drug-free recovery, at which point cells were stained for phospho-Rb (S807/811) to measure the fraction of cycling (pRbhigh) and non-cycling (pRblow) cells, visualized as a histogram. Log refers to natural log, and UT refers to untreated throughout this work. C MCF10A cells expressing the CDK2-activity sensor were treated as in A. Cells were filmed by fluorescent time-lapse microscopy for 96 h, from 6 d-10 d post-etoposide. Each row in the heatmap represents a single-cell trace, colored according to the colormap where yellow indicates high CDK2 activity and progression through the cell cycle, and dark blue indicates cell-cycle withdrawal into quiescence or senescence. D Percent of cells that have entered the cell cycle (CDK2 activity > 0.8) at each frame of the movie. UT refers to untreated cells (C). E Distributions of CDK2low times (CDK2 activity < 0.8) for the movie in C. Shading represents the CDK2low times corresponding to fast-cycling, slow-cycling, and non-cycling categories. Throughout the paper, the number of cells plotted and the number of biological replicates can be found in Supplementary Data 2.
Transcriptomic profiling of etoposide-treated cells reveals two paths to senescence and diverse senescent types
To understand the transcriptional changes that occurred as cells move between the different cell-cycle fates, we performed live-cell imaging and also sequenced MCF10A cells 6 d after release from a 24 h treatment with 2.5 µM, 10 µM, or 25 µM etoposide (Figs. 2A, S3A). Cells with low read counts and genes detected were filtered out (Fig. S3B, see methods), resulting in 6454, 5913, 5361, and 4878 cells profiled for UT, 2.5 µM, 10 µM, and 25 µM etoposide, respectively.
A Experimental timeline. B UMAP projection of scRNA-seq results with cells colored by etoposide dose (left), cell-cycle phase according to Seurat (middle), and clustered by gene expression similarity, where arrows depict two paths or bridges to senescence (right). C Proportion of each cluster in B by etoposide dose, and key with interpretation of the cell-cycle state of each cluster. D MCF10A cells were treated with 2.5 μM, 10 μM, or 25 μM etoposide for 24 h, followed by a 6 d drug-free recovery. Cells were stained for phospho-Rb (S807/811) and Hoechst for DNA content and plotted as a density scatter where yellow indicates a large number of cells in that region. The same number of cells is plotted for each dose. Red box indicates the 4 N/pRblow cells, a hallmark of cells that have experienced a mitotic slip. Number of cells and replicates in Supplementary Data 2. E UMAP projection colored by log-normalized expression for 10 canonically used senescent markers. F UMAP projection colored by log-normalized and scaled expression for two top single-gene markers (ranked by log2FC * (−log(p.adjusted))) from cluster 7 (quiescence-senescence transition cluster) and clusters 4, 9, 12, and 10 (senescent clusters).
We used Uniform Manifold Approximation and Projection (UMAP)21 to project all cells together in the same 2-dimensional space (Fig. 2B). Untreated cells formed the lower island of the UMAP, while the three different doses of etoposide-treated cells formed the upper island. Cells treated with 25 µM etoposide occupied a region on the far-left side of the upper island, whereas 2.5 µM and 10 µM cells were dispersed along the upper island (Figs. 2B left, S3C).
We used Seurat22,23 to categorize cells as either G1, S, or G2/M based on the expression of cell-cycle phase-specific genes (Figs. 2B middle, S3D; see methods). Seurat does not distinguish between G1-phase cells that are committed to the cell cycle and G0-phase quiescent cells. Since Seurat labels all cells that are not expressing S or G2/M markers as G1, this group contains both quiescent and senescent cells. As expected based on our live-cell imaging, 25 µM etoposide cells that we knew were predominantly senescent were labeled as G1, whereas 2.5 µM and 10 µM cells were labeled as a mix of G1, S, and G2/M (Fig. 2B middle).
Notably, untreated S and G2/M cells were transcriptionally distinct from S and G2/M etoposide-treated cells and occupied different regions of the UMAP (Fig. 2B middle), despite having similar CDK2 activities when measured by live-cell imaging (Fig. 1C). This suggested that while some cells could evade the proliferative arrest from lower etoposide doses and continue to cycle, they retained an altered transcriptional state even 6 d after the drug was washed off. Cells that cycled after etoposide treatment showed increased expression of protein chaperones (Fig. S3E) when compared to untreated cycling cells. This could be a possible adaptive mechanism allowing them to cycle with residual etoposide-induced stress, and a potential weakness that could be exploited to target cycling drug-tolerant persister cells.
We clustered cells by similarity and measured the number of cells in each cluster from each etoposide dose (Fig. 2B right, C). Optimal clustering resolution was selected based on cluster stability across various parameters (principal components and clustering resolutions) and agreement with known marker gene expression (detailed in methods and S3F–H). Most clusters in the treated island of the UMAP were composed of cells from more than one dose of etoposide, reflecting the heterogeneity in cell-cycle fate we saw in our live-cell imaging for 2.5 µM and 10 µM etoposide. 25 µM cells fell into 5 different clusters on the left side of the UMAP, suggesting transcriptional heterogeneity within the predicted-senescent cells themselves.
We identified clusters 9, 12, 4, and 10 as predicted-senescent clusters because they 1) consisted primarily of 25 µM cells that we knew from time-lapse imaging to be senescent, and 2) showed increasing cluster occupancy as a function of etoposide dose (S3C). The nearby cluster 7 contained a mix of 2.5 µM, 10 µM, and 25 µM etoposide cells, suggesting that it represented a quiescent-senescent transition cluster. Cluster 3 was also comprised of cells with a G0/G1 label but had almost no 25 µM cells and connected to the cycling cells, likely representing a cluster of quiescent cells.
Importantly, we found two different bridges between the etoposide-treated S/G2/M cycling cells and the senescent region of the UMAP (Fig. 2B). Clusters 3 and 7 made up the dominant bridge and included primarily 2.5 µM and 10 µM etoposide cells; cluster 10 formed a less populated alternative bridge and primarily consisted of 25 µM cells (arrows in Fig. 2B right). We hypothesized that these two paths represented, respectively, a standard G0/G1 arrest after completion of mitosis and a mitotic slip following G2 arrest. A mitotic slip causes a cell to enter a G0/G1-like state without mitosis and yields a population of cells with 4 N DNA content24 and elevated levels of the CDK inhibitor, p2125. To validate that the cluster 10 bridge indeed represented cells that had undergone a mitotic slip, we co-stained cells for pRb and DNA content at each dose24,25. We detected both 2 N DNA content/pRblow cells (standard completion of mitosis) and an increasing proportion of 4 N DNA content/pRblow (mitotic slippage) cells with increasing etoposide dose (Fig. 2D). Identification of cells that have experienced a mitotic slip via scRNA-seq was notable because this represented a direct path from cycling (clusters 1 and 5) to senescence (cluster 10), without transiting through quiescence. We also confirmed the existence of 4 N and >4 N arrested cells in both RPE-hTERT and MCF7 released from etoposide (Figs. S2, S4A).
We next visualized the expression of ten canonical senescence biomarkers across all cells (Fig. 2E). While specific marker expression patterns varied, the mRNA levels of canonical markers of senescence failed to specifically identify senescent cells, with the exception of CDKN1A (p21), which showed a marked but graded increase in all senescent clusters. To determine if this was unique to etoposide treatment in MCF10A cells, we tested this same panel of markers in two published single-cell RNA sequencing data sets of cells induced to senescence. The first data set from Wechter et al. included WI38 fibroblasts aged to replicative senescence (RS), treated with 50 μM etoposide, or exposed to ionizing radiation (IR)26. The second data set from Chan et al. included WI38 cells at multiple population doublings, including RS27. Both data sets revealed heterogeneous expression patterns for the panel of markers, mirroring our dataset (Fig. S4B–E). For some markers, such as GLB1 mRNA, a lack of correlation between senescence and GLB1 mRNA levels is expected since this gene encodes senescence-associated β-galactosidase, a marker typically assessed by an enzymatic activity assay. Interestingly, other markers, such as IL6, performed better in the RS WI38 cells than in MCF10A, suggesting that some markers may be more effective in specific contexts.
While multiple studies of senescence heterogeneity in different cell types or senescence induction methods exist, the extent to which individual senescent cells within the same population are transcriptionally different is less well studied. To determine the specific differences in expression profiles of our senescent clusters, we performed differential gene expression (DGE) analysis on each senescent cluster compared to all other cells. DGE analysis revealed specifically upregulated markers for cluster 7, cluster 4, cluster 9, and cluster 12 that could be used to identify these clusters (Figs. 2F, S5A right). Cluster 9 was particularly well marked by upregulation of PURPL, a p53-induced lncRNA with emerging roles in senescence and cancer28,29,30,31, and by GRID2, an ionotropic receptor involved in glutamate transfer. Cluster 12 was particularly well marked by increased expression of many lncRNAs. However, cluster 10 genes showed less cluster-specific upregulation. This analysis thus revealed several genes whose distinctive expression can be used to flag the senescent clusters we identified. By contrast, the top downregulated genes were much less specific to each cluster and instead were downregulated in G0/G1 cells generally (Fig. S5A left, B).
Pathway analysis reveals biological processes altered in senescent cells
We next performed gene set enrichment analysis (GSEA)32 using the Gene Ontology (GO) Biological Processes gene set annotations33 (see methods) to determine if the most differentially expressed (DE) genes for each senescent cluster belonged to a shared pathway. We found multiple significantly (p.adjusted < 0.05) upregulated and downregulated pathways for clusters 7, 4, 9, and 10 (Figs. 3A, S6). Cluster 12 had no significant up- or downregulated pathways, since many of the cluster 12 DE genes were lncRNAs (S5A) and not annotated in specific pathways.
A Number of pathways significantly up- or down-regulated in each cluster according to Gene Set Enrichment Analysis (GSEA) (p.adjusted < 0.05). B Significant pathways from A were grouped by general biological process (see methods, Supplementary Data 1). C GSEA for cluster 7 (quiescence-senescence transition cluster) and clusters 4 and 9 (senescent clusters). Top 10 pathways (by p.adjusted) for each cluster are displayed. Gene ratio for each pathway (proportion of genes in each pathway that are significantly DE compared to the total number of genes in that pathway) plotted on the x-axis. Dots colored by the normalized enrichment score for each pathway. Dot size reflects adjusted p-value.
We plotted the gene ratio and normalized enrichment score (NES) for the top 10 (ranked by p.adjusted) up- and down-regulated pathways for each cluster, ignoring cell-cycle related pathways that we expected to change (Fig. 3B, S6). Many pathways had similar GO terms and overlapping leading-edge genes, so we grouped them based on general cellular process (see methods, Supplementary Data 1, Fig. 3C, S6B). As expected, we found that pathways related to cell-cycle progression were the largest group of down-regulated pathways for all senescent clusters (Figs. 3C, S6B). Interestingly, the second and third largest groups of down-regulated pathways were related to the processing/transport of RNA and translation. Importantly, we detected changes in these pathways in all senescent clusters.
Despite having DNA damage from the etoposide treatment, clusters 9, 4, and 7 had decreased expression of genes associated with DNA repair pathways. This is likely because many DNA repair genes are E2F-target genes that are only expressed upon cell-cycle commitment. All three clusters also showed down-regulation of DNA/chromatin organization-related pathways, indicating a shared feature of senescent cells. Cluster 9, the farthest away from cycling cells and the cluster with the highest p21 expression, had the most varied down-regulated pathway types, including translation, RNA processing, telomere maintenance, and energy/metabolism pathways, with translation related-pathways showing the strongest change.
We expected to see an increase in pro-inflammatory pathways characteristic of the SASP7,8 in all senescent clusters, but we only saw this in clusters 4 and 7. Interestingly, cluster 4 upregulated cytokine- and interferon-responsive immune pathways such as antimicrobial humoral response and defense response to bacterium, consistent with the inflammatory arm of the SASP. In contrast, cluster 7 upregulated wound-healing and epidermal-differentiation pathways enriched for pro-growth and matrix-reorganizing SASP factors. Notably, cluster 9 lacked SASP-related pathways and was instead defined by an upregulation of many synapse-related pathways, including regulation of trans-synaptic signaling and regulation of membrane potential, with leading-edge genes that included many ion channels and metabolite-sensing transmembrane receptors (Supplementary Data 1).
Altogether, these data reveal that senescent cells exhibit gene expression changes associated with diverse biological processes. While some processes are shared, the extent to which they each manifest varies across clusters, leading to four senescent types, or senotypes, within irreversibly arrested populations.
G0/G1 quiescent cells following etoposide represent a transitional transcription state between cycling and senescent cells
If the continuum model of cell-cycle withdrawal is correct, then 1) quiescent cells would not have a unique gene expression program when compared to senescent cells, and 2) quiescent cells would begin to show gene expression changes associated with late senescent phenotypes, even in G0 cells that are in close proximity to cycling cells.
To test if quiescent cells have a unique expression signature, we performed two DGE analyses. First, we compared quiescent cluster 3 to all other cells. The top up- and down-regulated DE genes for cluster 3 were not uniquely expressed in cluster 3 and instead showed altered expression in neighboring clusters as well (Fig. S7A). Specifically, upregulated genes in cluster 3 also showed elevated expression in clusters 2 and 7, while downregulated genes in cluster 3 showed decreased expression in clusters 7, 4, 9, and 12. Pathway analysis of these DE genes revealed only two significantly upregulated pathways in cluster 3, establishment of organelle localization and enamel mineralization (Supplemental Data 1), and the significantly down-regulated pathways were cell-cycle related (Fig. S7B), suggesting that the primary feature of quiescent cells when compared to all other cells was their lack of cycling markers.
In a second analysis, we specifically compared quiescent and senescent cells by performing DGE analysis on quiescent cluster 3 vs. senescent clusters 4, 9, and 12, as well as on quiescence-senescence transition cluster 7 vs. senescent clusters 4, 9, and 12. The most differentially expressed genes (Fig. 4A) and pathways (Fig. 4B) in cluster 3 vs. senescent cells were shared with cycling cells (cluster 2) and were some of the top pathways that defined our senescent cells, such as SASP, translation, and RNA-processing related pathways. These findings suggested that quiescent cells in the etoposide-released context are defined by a lack of cycling genes when compared to all cells and an attenuated senescent program when compared to senescent cells, thus representing a transitional transcriptional state between cycling and senescence.
A Top 5 up- and down-regulated genes for cluster 3 (shallow quiescence) vs. senescent cells (clusters 4, 9, 12) and cluster 7 (quiescence-senescence transition) vs. senescent cells. Color corresponds to the average scaled expression. B GSEA pathway analysis for results from the DGE analysis in A. Top 8 pathways (ranked by p.adjusted) are shown. Cluster 7 had no significantly downregulated pathways. For full pathway names, see Supplementary Data 1. C Distribution of module scores by cluster for leading-edge genes in two selected up- and down-regulated pathways for quiescent cluster 3 vs. senescent cells. D UMAP with cells colored by progress along pseudotime trajectory. Trajectory branches are visualized separately as Trajectory 1 and Trajectory 2. E Etoposide dose distribution of cells over pseudotime. F Log-normalized expression of select top senescent markers over pseudotime. Cells colored by cluster. Loess fit visualized in blue. G Left: Module scores for grouped biological processes over pseudotime (see methods). Scores for one select up- and down-regulated pathway group over pseudotime are shown as single-cell data. Cells colored by cluster. Loess fit in blue. Right: Heatmap of all pathway groups over pseudotime. Color represents the Loess fit value for each module score vs. pseudotime scatter plot for the pathways listed along the y-axis. Module scores were rescaled for visualization on the same scale.
Within the quiescence-senescence transition cluster 7, some cells may more readily re-enter the cell cycle when compared to other senescent clusters based on the etoposide dose distribution in this cluster. Interestingly, pathways upregulated in cluster 7 relative to senescent clusters included epithelium migration, tissue migration, and cell-adhesion related pathways (Figs. 4B, S7C), pathways that have been shown to promote metastasis and epithelial-mesenchymal transition (EMT) in dormant cancer cells34,35,36. If such cells were to re-enter the cell cycle, they could promote metastasis in a cancer context.
scRNA-seq reveals a transcriptomic gradient along the quiescence-senescence continuum
Clustering analysis defines clusters and pseudobulks the cells, sometimes masking heterogeneity within clusters. We scored cells based on their expression of leading-edge genes (see methods) for pathways differentially expressed in quiescent vs. senescent cells and found significant overlap in the distribution of scores for top pathways between adjacent clusters (Fig. 4C). We reasoned that this overlap between clusters might have been due to continuous, graded changes in gene expression between cells on one side of the cluster compared to the other.
To test this idea, we performed pseudotime trajectory analysis using Monocle337,38 (see methods), a computational method used to infer the progression of single cells through a biological process. Our goal was to identify transcriptional changes that occurred as a function of progress through G1-labeled clusters. We therefore subsetted the G1 etoposide-treated cells and selected the cells closest to proliferating clusters as our root. Cells were then ordered and colored based on their progression along the learned trajectory (Figs. 4D, S7D). Some cells with a G1 label were in the G1 phase of the cell cycle instead of being arrested, and thus, by including all G1-labeled cells, the resulting pseudotime trajectory spanned all depths of quiescence into our most senescent cells. The trajectory branched partway, resulting in two different regions of late pseudotime, which were split into two trajectories for clarity during analysis (Fig. 4D). We measured the distribution of each etoposide dose along pseudotime (Fig. 4E). Progress along pseudotime correlated with a decreasing proportion of 2.5 µM and an increasing proportion of 25 µM cells. Given the declining likelihood of cell cycling along this trajectory, progress along pseudotime measured progression toward senescence.
We examined whether the top DE genes from our previous analyses were graded along pseudotime (Figs. 4F, S7E). Indeed, CDKN1A and PSCA showed a graded increase over pseudotime, dipping slightly in cells that had progressed the farthest (Fig. 4F). To extend this analysis beyond individual genes, we identified senescence-associated pathways in our data by selecting the significantly up- and down-regulated pathways for cluster 4 and cluster 9. We then used the leading-edge genes (most DE, see methods) from each of these pathways to calculate an expression score in every cell. Once the score was calculated, we plotted the score vs. pseudotime and performed a Loess fit to find the average score for each pathway across pseudotime. The Loess fit values for all pathways were then rescaled between 0 and 1 for visualization on the same color scale (Fig. S7F bottom).
Since many pathways were changed in the senescent cells, we combined pathways from the same general cellular functions as in Fig. 3, such that the score was based on the leading-edge genes from all pathways grouped in that general cellular function (Figs. 4G left, S7F top). To visualize all grouped pathways together over pseudotime, we generated a heatmap where the color corresponded to the fit and rescaled score (Fig. 4G right). All downregulated pathways gradually declined over pseudotime, with NTP biosynthesis, metabolism, and translation-related pathways going back up in late pseudotime along Trajectory 2 but not Trajectory 1. The upregulated pathways showed the reverse behavior, with gradual increases across pseudotime. The varied behavior in later pseudotime reflected the heterogeneity of the expression of these phenotypes within the different senescent clusters.
We then sought to test if the leading-edge genes from these grouped biological processes following etoposide were changed in WI38 cells at various PDLs from Chan et al. and WI38 cells aged to RS, exposed to IR, or treated with etoposide from Wechter et al. We plotted the distributions of expression scores based on our leading-edge genes of each biological category for cells in the published data sets from Fig. S4. Importantly, only cells labeled G1 by Seurat were included in this analysis since many of the conditions contained a mix of G1/S/G2M cells (Fig. S4A, B). Excluding S and G2M cells prevented large differences in expression of these genes driven by a cycling subpopulation and allowed us to test if these biological processes were different between quiescent and senescent cells, as our etoposide sequencing suggested. We found the leading-edge genes of all of the biological process groups except for epithelial cell differentiation-related pathways were changed in the same way (all up or all down) in the WI38 senescent cells as in our dataset (Fig. S8). The average expression of all of these processes in WI38 cells at various PDLs up to RS was strikingly graded. Expression of the leading-edge genes for these processes even at low PDLs changed gradually, with strong changes in these genes upon RS (PDL_50) (Fig. S8C). Together, these results showed that senescent phenotypes began to manifest early in pseudotime, even in cells with the potential to re-enter the cell cycle. Further, quiescence represented a transition state where cells exhibited expression signatures between those of proliferating and senescent cells.
Protein biosynthesis rate is a key difference between quiescent and senescent cells
An outstanding question is how senescent cells permanently lose the ability to re-enter the cell cycle when compared to quiescent cells faced with the same stressor. We found that pathways associated with RNA-processing and translation comprised the two largest pathway groups downregulated in senescent clusters, after cell-cycle related pathways (Figs. 3B, S9A). These pathways were also the most downregulated in senescent cells when compared to quiescent cells directly (Fig. 4B, C). RNA-processing and translation are essential for efficient protein biosynthesis, which is in turn essential for the cell cycle, particularly in cancer cells39,40. Therefore, a decrease in RNA-processing and translation can impinge directly on the cell cycle.
We hypothesized that a decrease in protein biosynthesis might be predictive of an inability to re-enter the cell cycle. To test this, we treated MCF10A cells with 2.5 µM, 10 µM, or 25 µM etoposide for 24 h, followed by a 6-day drug-free recovery. At the end of the 6 d, we treated cells for 30 min with O-propargyl-puromycin (OPP), which incorporates into actively translating proteins and measures global translation rate at the single-cell level. We detected a dose-dependent decrease in both the average and the integrated OPP intensity (Figs. 5A, S9B, respectively) in senescent cells, indicating reduced protein biosynthesis.
A MCF10A cells were treated with 2.5 μM, 10 μM, or 25 μM etoposide for 24 h, followed by a 6 d drug-free recovery. Cells were treated with OPP for 30 min prior to fixation. Each cell’s OPP intensity was normalized to the cell area and plotted as histograms for each etoposide dose. Number of cells and replicates in Supplementary Data 2. B Experimental timeline (top). MCF10A cells expressing the CDK2-activity reporter and a DHFR-mCherry translation reporter were imaged by live-cell microscopy for 96 h from 6d–10d after etoposide washout. TMP was added at the start of the gray shading to induce the DHFR-mCherry reporter, and the mCherry signal (each cell normalized to its average signal before TMP) was plotted for fast-cycling, slow-cycling, or predicted-senescent cells for each dose of etoposide. Dark line is the mean for each group, shading is the 95% confidence interval. Gray shading indicates the frames used to calculate the mCherry signal slope plotted in (C). Number of cells and replicates in Supplementary Data 2. C A line was fit to the initial slope of mCherry accumulation after TMP addition. The average slope for each cell-cycle behavior group for each dose is shown. Error bars represent 95% CI. PS is predicted-senescent. Number of cells and replicates in Supplementary Data 2. D All significantly downregulated pathways from the RNA-processing and translation pathway groups in Fig. 3C for senescent clusters 4 and 9 were broken down into more specific pathway groups. E Average scaled expression for select leading-edge genes by cluster from groups of pathways in (D). F Heatmap of all GO biological processes from the RNA-processing and translation pathway groups over pseudotime. Colormap represents the Loess fit value for each module score over pseudotime. Module scores were rescaled for visualization on the same scale. For full pathway names, see Supplementary Data 1. G Cells were treated with 2.5 μM, 10 μM, or 25 μM etoposide for 24 h followed by a 6 d drug-free recovery. OPP was multiplexed with RNA FISH for three SASP genes and imaged. RNA FISH puncta number and total OPP per cell were quantified and plotted as a single-cell density scatter. Red line is drawn at the 95th percentile of untreated cells. Number of cells and replicates in Supplementary Data 2.
To determine if translation rate could predict if cells were quiescent or senescent, we used a dihydrofolate reductase (DHFR)-trimethoprim (TMP) protein stabilization system fused to mCherry, where the rate of mCherry accumulation upon TMP treatment reflects protein biosynthesis rate (Fig. S9C)41,42. We treated cells with 10 µM or 25 µM etoposide for 24 h and then tracked them from 6-10 d post-recovery (Fig. 5B top) to measure CDK2 activity and mCherry accumulation rate (Figs. 5B, S9D–F). We found that senescent cells had a lower protein biosynthesis rate than slow-cycling cells that spend long periods in quiescence, in a dose-dependent manner (Figs. 5B–C, S9D–F), identifying reduced translation as a distinguishing factor between quiescent and senescent cells. Importantly, we verified this difference in MCF7 cells by inducing senescence with 10 µM or 25 µM etoposide for 24 h, followed by drug washout and live-cell imaging from 6–10 d. We measured translation rate for quiescent and senescent cells using post-hoc OPP staining linked to each cell’s cycling history. Indeed, fast-cycling cells had a higher translation rate than slow-cycling cells, which had a higher translation rate than predicted-senescent cells, even within the same etoposide dose group (Fig. S9G–J). We then tested the expression levels of the GO BP Ribosome Biogenesis and RNA Processing pathways in WI38 cells labeled by Seurat as G1 after RS, IR, and etoposide (Fig. S10A–F) and also saw decreased expression in these pathways upon senescence induction, indicating that these findings are likely applicable in other senescence contexts.
Protein biosynthesis is impaired in senescence, particularly in cluster 9
Protein biosynthesis relies on many important biological processes upstream. To understand which steps of this process could have been driving a decrease in protein biosynthesis following senescence induction, we examined the DE pathways in our senescent clusters identified in Fig. 3. We further broke down the translation and RNA-processing related pathways for clusters 4, 9, 10, and 7 by similarity (Fig. 5D, Supplementary Data 1). We noted downregulation of biological processes that could affect protein biosynthesis at multiple steps, including rRNA-related pathways, tRNA-related pathways, and ribosome biogenesis and assembly (Fig. 5E). Examples of the strongly down-regulated genes included NPM1, EIF4A1, and exosome complex components. The largest number of pathways was related to general RNA-processing and localization. We confirmed that these same genes were strongly downregulated in other senescence contexts by plotting their expression levels in WI38 cells induced to senescence that were labeled by Seurat as G1 (Fig. S10G). In the WI38s approaching RS, we found that many of these genes were most strongly downregulated in PDL_50, the RS cells, suggesting these genes may play an important role in maintaining cell-cycle arrest by decreasing upstream protein biosynthetic processes like RNA processing and ribosome biogenesis. Thus, declining translation likely occurs both in cellular aging and chemotherapy-induced senescence contexts.
These data suggested that protein biosynthesis was decreased in senescent cells due to a multi-step failure, where no one step is solely responsible. In addition to the direct changes in RNA-processing, ribosome biogenesis, and translation initiation, we also measured a decrease in NTP biosynthesis and metabolism-related pathways (Fig. 4G, S11A) in cells further along pseudotime. Knockout of key regulators of NTP biosynthesis can cause cell-cycle arrest43, and NTP biosynthesis is coupled with protein biosynthesis in some cancers44. Thus, changes in these processes could also indirectly affect protein biosynthesis and cell-cycle progression.
Senescent cells that express SASP factors have higher protein biosynthesis than cells that do not
The decrease in protein biosynthesis was interesting because translation has been shown to be especially important for maintaining the SASP45,46,47. We knew from our clustering and pseudotime analysis that pseudotime Trajectory 2 ended in cells that showed greater expression of SASP pathways than Trajectory 1. To test whether Trajectory 2 retained expression of pathways associated with translation and RNA processing, we used the same analysis strategy as in Fig. 4, plotting the expression score of leading-edge genes over pseudotime, rescaled between 0 and 1. However, here we plotted this score for every individual GO biological process associated with translation or RNA processing (Figs. 5F, S11B). We found that a majority of pathways associated with ribosome biogenesis, ribosome assembly, and translation initiation initially declined but then increased in late pseudotime along Trajectory 2, whereas these pathways only declined along Trajectory 1, as hypothesized.
To experimentally test this phenotype in cells, we multiplexed RNA FISH for three SASP factors, IL6, CXCL8, or CXCL10, with the OPP translation assay in single cells released from 2.5 µM, 10 µM, or 25 µM etoposide 6 d prior (Fig. 5G). We found a dose-dependent increase in expression of all three SASP factors. In agreement with our scRNA-seq results, only a subset of the cells in each sample showed increased expression of the SASP factors, even at 25 µM where nearly all cells were senescent, supporting our finding of distinct senotypes. This SASP expression was highest in the cells with the greatest translation rate as measured by OPP. We also confirmed this relationship in MCF7 and RPE-hTERT cells induced to senescence with etoposide (Fig. S11C). Expression of the SASP can be detrimental if cells are not properly cleared48,49. Therefore, future work investigating the pathways that were expressed differentially between Trajectory 1 and Trajectory 2 could offer new insights into suppressing the SASP without suppressing cell-cycle arrest.
Finally, mammalian Target of Rapamycin (mTOR) is a master regulator of protein biosynthesis. We therefore also tested whether mTOR was inactive in cells with low protein biosynthesis (Trajectory 1 or 2 N/pRblow cells) or if mTOR was active yet insufficient to drive robust translation. We treated cells with 10 µM or 25 µM etoposide for 24 h and allowed them to recover for 6 d. We then stained for phosphorylation of 4EBP1, an mTOR substrate, in 2 N/pRblow cells and found that mTOR activity showed a slight decrease with increasing etoposide dose and was slightly lower in 2 N/pRblow cells than 4 N/pRblow cells (Fig. S11D). We co-stained these cells with OPP and found that this small decrease in phospho-4EBP1 was correlated with a decrease in OPP (Fig. S11E). We also plotted each cluster’s module score for the hallmark MTORC1 signaling pathway (Fig. S11F) and found that MTORC1 signaling was upregulated in etoposide-treated cycling cells relative to untreated cells, but was less induced in senescent cells. This increased MTORC1 signaling in the cycling etoposide persister cells likely enhances their ability to cycle50. Further, we plotted the mRNA expression of negative regulators of mTOR in each cluster (Fig. S11F) and found that cluster 9, which had low protein biosynthesis and lower SASP expression, showed higher expression of mTOR negative regulators (e.g., TSC1/2), suggesting that mTOR might have been inhibited to some extent in cells arresting along the 2 N trajectory.
Expression of the SASP is higher in cells that have undergone a mitotic slip
Because the SASP can alter the surrounding tissue microenvironment and is thought to be a driver of age-related pathologies48,49, there is significant interest in characterizing the SASP at the single-cell level. It was surprising that SASP expression varied by senescent cluster, with cluster 4 containing the most upregulated pathways associated with the SASP (Fig. 3B). We plotted the score for leading-edge SASP-related genes from our data set over pseudotime (Fig. 6A) and saw that Trajectory 2, which was connected to the cluster of cells undergoing mitotic slippage, showed a graded and steady increase in expression. By contrast, Trajectory 1 showed an initial increase followed by a decrease in SASP-related genes, in agreement with our cluster analysis. We also used an existing SASP gene list from the Molecular Signatures Database (MSigDB)32,51 and scored cells based on their expression of these SASP factors (Fig. 6B). When visualizing the existing SASP gene list on our UMAP, we saw that cells near cluster 4 and cluster 10, the mitotic slip cluster, scored the highest for SASP expression.
A Module score for leading-edge genes from SASP-related biological processes over pseudotime. Loess fit in blue. Colored by cluster as in Fig. 4F. B Left: Distributions of module scores for REACTOME_SENESCENCE_ASSOCIATED_SECRETORY_PHENOTYPE_SASP gene set by cluster. Right: UMAP colored by module score for the SASP gene set. C Cells were left untreated (UT) or treated with 10 μM or 25 μM etoposide for 24 h before being washed and allowed to recover for 6 d. Number of RNA FISH puncta for three SASP genes vs. DNA content is plotted as a single-cell density scatter. Horizontal line is the 95th percentile of untreated puncta for each SASP factor. Vertical line is the saddle point between 2 N and 4 N DNA content (measured by Hoechst dye). Number of cells and replicates in Supplementary Data 2. D Single-cell scaled expression levels for 21 SASP genes expressed in all senescent clusters (Supplementary Data 3). Quiescent cluster 3 and quiescence-senescence-transition cluster 7 are shown for contrast. E Module scores for the same genes shown in D, by cluster.
To test experimentally whether cells that arrested via a mitotic slip were the senescent cells with the highest SASP expression, we performed RNA FISH for IL6, CXCL8, and CXCL10 in MCF10A (Fig. 6C) and MCF7 cells (Fig. S12A). We found that cells with 4 N DNA content, most of which had undergone a mitotic slip at 10 µM and 25 µM etoposide (Fig. 2D), had the highest expression of SASP RNAs (Figs. 6C, S12A). While cells arrested by the G0/G1 trajectory showed upregulation of some SASP-related pathways (Fig. 3, cluster 7), these pathways were related more to wound-healing, cell migration, and extracellular matrix organization rather than the cytokine/chemokine secretion and defense response-related pathways that we saw for cells that had undergone a mitotic slip (Fig. S12B, C). Notably, the pro-growth proteins secreted as part of these pathways have been linked to the survival, invasiveness, and epithelial-mesenchymal transition (EMT) of surrounding cells34,35,36 rather than immune clearance. Indeed, when we plotted the hallmark epithelial mesenchymal transition and GO cell migration gene sets (Fig. S12D), we saw increased levels in senescent clusters, especially cluster 7, the quiescent-senescent transition cluster. These data suggest that not only are there different senotypes in our population, but the type of senescence arising in each cell is tied, at least in part, to its initial mode of arrest.
A subset of SASP genes is expressed in all types of senescence
Because different clusters showed differing expression of SASP genes, we tested whether a subset of SASP genes was expressed in all senescent clusters. We found 21 SASP genes that were universally upregulated (see methods) in every senescent cluster at every dose of etoposide (Fig. 6D). Quiescent cluster 3 was used for contrast. We plotted the expression levels of these SASP factors in single cells and confirmed that all senescent clusters showed expression of these factors (Fig. 6E). Interestingly, many of these specific genes were double-stranded (ds) RNA-sensing proteins (OASL, OAS2) or interferon-inducible (IFI) antiviral genes (IFIT1, IFIT2, IFI6) downstream of these RNA-sensing proteins. We confirmed that this shared SASP was not dose-dependent in MCF10As (Fig. S13A) and found increased expression of this shared SASP list in WI38 cells induced to senescence by RS, IR, and etoposide (Fig. S13B). Notably, the shared SASP was correlated with CDKN1A at the single-cell level in the WI38 cells (Fig. S13B), suggesting that greater stress across multiple senescence-inducing treatments correlated with expression of these 21 SASP genes. While the upstream stresses driving the SASP can be varied, these data suggest that dsRNA accumulation might be a common stress driving SASP factor expression across senescent types.
A gene set to identify TIS cells
The lack of clear biomarkers to distinguish between quiescent and senescent cells is a persistent problem in the field. While we identified 21 SASP genes that were upregulated in all senescent clusters, these genes were occasionally also expressed in other clusters. We therefore generated a gene set based on genes represented in all treated senescent clusters that did not overlap with treated quiescent cells (see methods, Fig. S14A) and scored cells based on their expression of this gene set (Figs. 7A, S14A). Some of the top markers for senescent cells showed elevated expression in untreated spontaneously quiescent CDK2low cells20, and we therefore removed them to generate a senescence-specific list of genes, dubbed Etoposide Induced Senescence (EIS) Boulder. EIS Boulder includes both broadly expressed genes shared across senescent clusters and cluster-specific top markers. This composition allows the gene set to robustly capture multiple senotypes, while buffering against variability or low expression of individual top genes. Some of the best general, single-marker genes include IFI6, IFI27, ISG15, DDX60, and RGS2. Importantly, the expression of genes in this list was not dose-dependent or driven by a changing proportion of 25 µM cells, since it was able to identify senescent cells across all doses (Fig. S14B, C). The genes in our gene set were generally highly expressed in senescent clusters, showed low or no expression in other clusters, and included several new senescent biomarkers (Fig. 7B). We tested EIS Boulder in WI38 cells induced to senescence under various conditions and found that cells induced to senescence scored higher in expression of this gene set (Fig. 7C, D). We only included cells labeled G1 by Seurat in this analysis; therefore, the increasing score in WI38 cells at various PDLs suggested that our gene set helped distinguish quiescent from senescent cells in non-etoposide contexts. Finally, because there was no ground truth senescent population identified in the WI38 data sets, we plotted our senescent score vs. CDKN1A (our best single marker of senescence) in single cells and found a strong correlation for both WI38 data sets (Fig. 7E).
A Left: Distribution of module scores for cells from each cluster based on our senescent gene set, dubbed Etoposide Induced Senescence (EIS) Boulder (Supplementary Data 3). Right: UMAP with cells colored by module score for EIS Boulder. B Average scaled expression of all genes in EIS Boulder by cluster. C Left: UMAP of scRNA-sequencing data of WI38 fibroblasts cultured until replicative senescence (population doubling level (PDL) > 50, RS), exposed to 10 Gray (Gy) ionizing radiation and grown for an additional 10 d (IR), or treated with 50 μM etoposide for 7 d (ETO7d) from Wechter et al. UMAPs colored by condition or Seurat cell-cycle phase annotation. Right: UMAP colored by log-normalized expression of CDKN1A (p21) or module score for EIS Boulder. D Left: UMAP of scRNA-sequencing data of WI38 fibroblasts cultured to various PDLs from Chan et al. UMAPs colored by condition or Seurat cell-cycle phase annotation. Right: UMAP colored by log-normalized expression of CDKN1A (p21) or module score for EIS Boulder. E Distributions of expression scores of the EIS Boulder gene set for the data sets in C and D with only Seurat-labeled G1 cells plotted. F Single-cell expression scatter plots of the EIS Boulder gene set vs log-normalized CDKN1A expression for the data sets in C and D. Dots colored by condition and Loess fit in blue. Gray shading represents a 95% confidence interval. Only Seurat-labeled G1 cells plotted.
Discussion
The relationship between quiescence and senescence remains incompletely understood (Fig. 8A), in large part due to the lack of specific and consistent biomarkers. Here, we paired scRNA-seq with single-cell time-lapse microscopy to identify ground-truth senescent cells based on a functional definition of senescence: the long-term lack of proliferation. A limitation of our study is the reliance on in vitro cell culture models of senescence that may not completely capture the complexity of senescence in vivo. This is an ongoing limitation in the field of senescence that arises from the lack of reliable biomarkers to identify senescent cells in vivo, and the need for ground-truth identification of senescent cells (e.g., by time-lapse imaging). Our approach of combining longitudinal single-cell tracking paired with scRNA-seq allows us to distinguish senescent cells from cells in a transient quiescence, providing uniquely detailed insights into the dynamic and heterogeneous nature of senescence that should be validated with in vivo studies where possible. Our data revealed several findings that we discuss below.
A Left: Canonical model of a switch-like transition between quiescence and senescence. We did not include an arrow from quiescence to senescence because it has not been clear in the field whether senescent cells can arise from existing quiescent cells or only from proliferating cells. Middle: Simple continuum model of cell-cycle withdrawal where quiescence and senescence represent different levels on the same continuum of withdrawal. Right: Model of cell-cycle arrest supported by this work. In the G0/G1 path, cells complete mitosis and first enter a shallow quiescence (G0). In this path, quiescent cells represent a reversible intermediate between proliferation and senescence and show a graded expression of senescence genes. In an alternative path, cells skip mitosis and arrest with 4 N DNA content. These cells transit directly to a senescent state, bypassing the quiescence deepening observed along the G0/G1 path.
Related to the cell cycle itself, we found that cycling cells after etoposide treatment retain a transcriptionally encoded memory from the initial etoposide stress, even after 6 d. Further work characterizing the gene expression signatures in cells that resume cycling after chemotherapy treatment relative to untreated cycling cells could expose vulnerabilities to eliminate this population. We also found two distinct arrest paths, or bridges, from the cell cycle to senescence: a mitosis-to-G0 path and a mitotic-slip path. While cells taking the first path progress from G0/quiescence to senescence in a graded manner, cells arresting by the mitotic slip path appear to move directly from cycling to senescence based on the fact that they move straight from the G2/M cluster to a senescent cluster (Fig. 2B). This mitotic slip path was primarily occupied by 10 and 25 µM cells, suggesting that higher levels of damage may drive cells towards this path over the mitosis-to-G0 arrest path. Cells that are in S phase when they are first treated with etoposide may also be more susceptible to this arrest path52.
Related to senescence, we identified multiple transcriptomic clusters of senescent cells, or senotypes. Thus, senescence heterogeneity arises not only from different treatments or cell types, as previously reported53,54,55,56, but also within the same population of cells undergoing the same treatment. Although these senotypes were clustered discretely for analysis and showed cluster-to-cluster differences, there was a continuous distribution of cells across the senescent region of our UMAP, and our clusters did not show strong separation by Silhouette Score (Fig. S3G). This suggests that the gene expression distinctions between these senotypes are graded, with single senescent-cell transcriptomes varying across the entire senescent transcriptomic space. Prior work, including ours, noted variability in senescence markers under identical conditions16,57. Our findings suggest that this variability reflects both incomplete senescence induction as well as the presence of senescent cells lacking expression of certain canonical markers.
Interestingly, these different senotypes arose following the two distinct arrest paths. Mitotic slipping cells passing from cluster 10 to cluster 4 expressed higher levels of SASP, inflammation, and protein biosynthesis pathways compared to cells taking the mitosis-to-G0 path, which showed higher expression of pro-growth and EMT-related pathways, such as wound healing. In the context of cancer, the pro-inflammatory SASP can recruit the immune system to initiate clearance, whereas the pro-growth arm of the SASP can drive surrounding cells to proliferate7,8. Further work is needed to understand how these different senotypes expressing unique pro-inflammatory signatures may affect surrounding cells. Additionally, further work is needed to understand how broadly applicable these senotypes are in other contexts, as these findings were primarily based on MCF10A cells. The SASP requires a basic level of cellular function for coordinated expression, synthesis, and secretion of the program. Cluster 9 cells experiencing a high level of stress and impaired protein biosynthesis—marked by high p21, low RNA processing/translation, metabolic stress, and distance from cycling cells—likely lack this capacity. Indeed, cluster 9 cells showed reduced SASP-associated pathways, whereas SASP-expressing cells in cluster 4 had higher expression of genes linked to translation, RNA processing, NTP biosynthesis, and metabolism. This aligns with a recent study using multiplexed immunofluorescence imaging, which identified distinct senescent cell trajectories, including a subset characterized by increased protein biosynthesis and sustained high SASP levels in response to senescence-inducing stress58. In contrast, larger senescent cells displayed lower SASP expression. These results underscore the importance of distinguishing between senescent subpopulations of cells to develop targeted strategies for reducing the SASP, or to push cells along a low-SASP senescence trajectory.
We identified reduced protein synthesis as a key difference between senescence and quiescence. It has been shown that changes in RNA processing and translation are a part of the DNA damage response59,60,61 and that reduced mTOR and translation signaling are also associated with senescence62,63. Here, we reveal that these gene changes span a range of processes involved in the processing of mRNA, tRNA, rRNA, and lncRNAs, ribosome biogenesis and assembly, and direct translational control. Importantly, the differential expression of these pathways between quiescent and senescent cells places renewed importance on protein biosynthesis as a uniquely useful predictor of future cell-cycle re-entry, with a lack of protein biosynthesis as a causal determinant of irreversible cell-cycle withdrawal.
We defined a core gene set, EIS Boulder, shared across all senescent clusters that distinguishes them from non-senescent clusters, along with a few shared upregulated SASP genes (Fig. 6D, E) that are expressed in all of our senescent cells. These gene sets may enable transcriptional-level identification of senescent cells, and thus should be validated in further cell types and in other modes of senescence induction. Future studies will be needed to elucidate the mechanistic role, if any, of these markers in senescent cell biology.
Lastly, we found that our senescent population was connected to the treated, cycling cells by a continuous UMAP bridge. Using pseudotime trajectory analysis of the mitosis-to-G0 path, we found that rather than a switch-like change, multiple senescent phenotypes begin to manifest early and gradually, even in shallow quiescent cells. Thus, in the context studied here, quiescence represents a path through which cells move from proliferation to senescence, and cells positioned along this continuum express gradually varying levels of senescence markers.
This model represents an emerging view of the relationship between quiescence and senescence, where quiescence and senescence are not viewed as binary, distinct states, but instead exist on a continuum with declining probabilities of cell-cycle re-entry (Fig. 8). This model is consistent with previous work from our lab examining eight protein-based senescence biomarkers16. However, it was nevertheless surprising to see that the graded model of cell-cycle arrest extends across every senescence pathway we identified here, providing transcriptome-wide support for this model. A handful of other studies also support the continuum model. One study that used bulk RNA-seq found that prolonged serum starvation of fibroblasts, a reversible quiescence-inducing treatment, led to some transcriptional changes that are typical of senescence, such as lysosome biogenesis and autophagy64. While we also observed changes in some of these genes, they were less pronounced. Not all senescent phenotypes may change in every arrested condition, warranting further study to understand their overlap and trajectories. Stallaert et al. used multiplexed immunofluorescence to examine arrest induced by serum starvation, etoposide, and oxidative stress57, concluding that senescence represents a G1-like arrested state that eventually converges on a single senescent state following different arrest trajectories. Importantly, their study also found two paths to senescence via either a G1-like arrest or a G2/M path, mirroring our two-paths-to-arrest finding. This work was limited to cell cycle-related proteins and a few canonical senescent markers, potentially explaining why their data appeared to converge on a single senescent phenotype. In our study here, with measurements across the entire transcriptome, we found multiple senotypes. Additionally, the study by Chan et al. in WI38 cells approaching replicative senescence27 found that senescent-like changes manifest early, before replicative senescence was reached, suggesting that our model likely extends to replicative aging as well. Future work delineating the specific genes and processes that are universally changed in irreversibly arrested cells is warranted.
At what point along this continuum of withdrawal do cells become irreversibly arrested, and why? We found that irreversibly arrested cells showed perturbations in multiple genes and pathways that likely impair cell-cycle entry and progression in various ways. This suggests that no single, switch-like expression change drives the irreversibility of cell-cycle withdrawal. Instead, as cells progress along the continuum, multiple different senescent traits that deepen arrest intensify, creating an increasingly higher barrier to cell-cycle re-entry, which eventually becomes insurmountable.
Methods
Cell lines and culture media
MCF10A (ATCC CRL − 10317) cells were obtained from ATCC and grown in DMEM/F12 supplemented with 5% horse serum, 20 ng/ml EGF, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, and 100 μg/mL of penicillin and streptomycin. RPE-hTERT (ATCC CRL-4000) were obtained from ATCC and grown in DMEM/F12 supplemented with 10% FBS, 1x Glutamax, and 100 μg/mL of penicillin and streptomycin. MCF7 (ATCC HTB-22) were obtained from ATCC and grown in RPMI supplemented with 10% FBS, 1x Glutamax, and 100 μg/mL of penicillin and streptomycin. Cells were imaged in phenol red-free full growth media during live-cell movies. Cells were cultured and imaged in a humidified incubator at 5% CO2 and 37 °C. Cells were maintained in the CU Boulder Biochemistry Cell Culture Facility (BCCF; RRID:SCR_018988).
Drug treatments
MCF10A cells were split 1:10 (from a confluent plate) into a plastic 10 cm culture plate before being treated with 2.5, 10, or 25 μM etoposide the following day for 24 h. Cells were then washed twice with PBS before being returned to full-growth media. Full-growth media was refreshed every 3 d during the drug recovery period. 24 h prior to imaging, the etoposide-released cells were trypsinized and replated onto a collagen-coated (1:50 dilution in water) (Advanced BioMatrix, No. 5005) 96-well glass-bottom plate (Cellvis Cat. No. P96 − 1.5H-N). Cells were plated at 1000, 2000, 3000, and 4000 cells per well for untreated, 2.5, 10, or 25 μM etoposide, respectively. Cells were plated at 3000 cells per well for immunofluorescent imaging.
Immunofluorescence imaging
Cells were seeded onto a collagen-coated (1:50 dilution in water) (Advanced BioMatrix, No. 5005) 96-well glass-bottom plate (Cellvis Cat. No. P96-1.5H-N) 24 h prior to fixation. Cells were fixed for 15 min with 4% PFA in PBS, then permeabilized at room temperature with 0.1% TritonX for 15 min. Cells were then washed with PBS and blocked with 3% Bovine Serum Albumin (BSA) for 1 h at room temperature. Primary antibodies were incubated overnight in 3% BSA at 4 °C, and secondary antibodies were incubated for 1–2 h in 3% BSA at room temperature. Nuclei were labeled with Hoechst at 1:10,000 in PBS for 10 min. The whole cell was stained with succinimidyl ester 488 at 1:10,000 in PBS for 30 min. Cells were washed with 100 μL per well of PBS between every step. All images were obtained using a 10X, 0.4 numerical aperture objective on a Nikon TiE microscope. To visualize IL8, we blocked secretion for 6 h with 5 µg/mL Brefeldin A before fixing and staining. For the OPP translation assay, cells were treated according to the manufacturer’s protocol (Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis Assay Kit, ThermoFisher C10456), with cells incubated with OPP reaction component A for 30 min prior to fixation.
RNA FISH imaging
Cells were seeded onto a collagen-coated (1:50 dilution in water) (Advanced BioMatrix, No. 5005) 96-well glass-bottom plate (Cellvis Cat. No. P96-1.5H-N) 24 h prior to fixation. Cells were fixed for 15 min with 4% PFA in PBS. IL6 (VA6 − 12712-VC), CXCL8 (VA6-13192-VC), and CXCL10 (VA6-13729-VC) mRNA were visualized according to the manufacturer’s protocol (ViewRNA ISH Cell Assay Kit, ThermoFisher QVC0001), with cells permeabilized for 30 min and mRNA probes hybridized for 4 h at 40 °C. RNA FISH imaging was performed on a Revvity Opera Phenix using a 20X objective at the BioFrontiers Institute’s Advanced Light Microscopy Core (RRID: SCR_018302; supported by NIH grant 1S10OD025072).
Time-lapse microscopy
Cells were plated 24 h prior to imaging, and full-growth media was replaced with phenol red-free full-growth media. Images were taken for each fluorescent channel every 12 min at two sites per well that were spaced 2 mm apart. Total exposure across all fluorescent channels was kept below 800 ms. Cells were imaged in a humidified, 37 °C chamber at 5% CO2. All images were obtained using a 10X, 0.4 numerical aperture objective on a Nikon TiE microscope. The tracking code is available for download here: https://github.com/scappell/Cell_tracking.
Image processing and quantification
Image processing and cell tracking were performed using MATLAB Mathworks 2017b as previously described65. Quantification of 53BP1 puncta was measured as previously described66. Nuclear signals (phospho-Rb, 53BP1, p21, and Lamin B1) were quantified from a nuclear mask (median nuclear intensity). Cytoplasmic signals (LAMP1 and IL8) were quantified from a cytoplasmic mask (median cytoplasmic intensity) derived from succinimidyl ester total protein stain. RNA FISH puncta detection was performed using Revvity’s Harmony High-Content Imaging and Analysis Software. All puncta inside the cytoplasmic mask belonging to each nucleus were counted.
Antibodies and reagents
Antibodies against pRb (S807/811) D20B12 XP (8516), LAMP1 D2D11 XP (9091), p21 Waf1/Cip1 (12D1) (2947), p4EBP1 (S65) (174A9) (9456), and Lamin B1 (D9V6H) (13435) were purchased from CST and used at 1:500, 1:1000, 1:250, 1:500, and 1:1000 dilutions, respectively. Antibodies against 53BP1 (612523), and IL-8 (550419) were purchased from BD and were all used at dilutions of 1:1000. All secondary antibodies, Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Cyanine3 (A10521), Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Cyanine3 (A10520), Goat anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (A-21236), Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (A-21245) were purchased from Thermo Scientific and used at 1:1000 dilutions. IL6 FISH mRNA probe set (VA6 − 12712-VC), CXCL8 FISH mRNA probe set (VA6-13192-VC), and CXCL10 mRNA probe set (VA6-13729-VC) were purchased from Thermo Scientific. The ViewRNA ISH Cell Assay Kit was purchased from Thermo Scientific (QVC0001). CF 488 A succinimidyl ester (SCJ4600018) was purchased from Sigma and used at a 1:10,000 dilution. Hoechst 33342 was purchased from Thermo Scientific (H3570) and used at a 1:10,000 dilution. Etoposide (E1383) and Brefeldin A (B7651) were purchased from Sigma. Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis Assay Kit (C10456) was purchased from Thermo Scientific.
Single-cell RNA sequencing
Single-cell RNA sequencing (scRNA-seq) libraries were generated using the 10x Genomics Chromium platform with the 3’ mRNA capture kit. Single-cell suspensions were prepared following the 10x Genomics “Cell Preparation for Single Cell Protocols” guide with a viability of >95%. Single-cell encapsulation, barcoding, cDNA amplification, library preparation, and sequencing were performed by the University of Colorado Anschutz Genomics Shared Resource facility (RRID: SCR_021984). Cells were encapsulated into droplets using the Chromium Controller, where reverse transcription occurred within individual gel beads-in-emulsion (GEMs). Following barcoding and cDNA amplification, sequencing libraries were constructed according to the manufacturer’s protocol. Libraries were sequenced on an Illumina NovaSeqX to a depth of 81,810 reads per cell for untreated, 74,214 reads per cell for 2.5 µM, 81,254 reads per cell for 10 µM, and 76,973 reads per cell for 25 µM.
scRNA-seq data processing
Publicly available single-cell RNA-seq data sets for Chan et al. and Wechter et al. were downloaded from GEO (GSE175533 and GSE226225, respectively). Data from Wechter et al. were downloaded as processed count matrices and were analyzed using the same analysis pipeline described below. Cells with fewer than 3000 genes and more than 15% mitochondrial gene expression were excluded to remove low-quality cells. Data from Chan et al. were downloaded as processed count matrices with low-quality cells already removed and were analyzed using the same analysis pipeline described below.
scRNA-seq data were processed and analyzed using the Seurat22,23 (v5.1.0) package in R (v4.4.1). Raw sequencing reads were pre-processed using CellRanger (v7.1.0) and aligned to the Human (GRCh38) 2020-A reference genome. Gene-cell count matrices were generated, and low-quality cells were filtered out based on the number of detected genes, unique molecular identifiers (UMIs), and mitochondrial gene percentage. Cells with fewer than 3,500 genes were excluded to remove low-quality cells. Cells with greater than 15% mitochondrial gene expression were also removed. Data were log-normalized using Seurat’s NormalizeData() function with a scale factor of 10,000.
Feature selection and dimensionality reduction
Highly variable genes (HVGs) were identified using FindVariableFeatures(), and data were scaled using ScaleData(). Principal component analysis (PCA) was performed using RunPCA(). The optimal number of PCs (30) was determined using an elbow plot and JackStraw analysis22,23 and used in downstream dimension reduction.
Clustering and visualization
Cells were clustered using the shared nearest neighbor (SNN) graph-based clustering algorithm implemented in Seurat’s FindNeighbors() and FindClusters() functions, with a final resolution of 0.8. Clustering was performed using the top 30 principal components (PCs) derived from the 2000 most variable genes. We selected 2000 variable genes as this number was sufficient to capture the major biological variation in our dataset. Specifically: (1) Elbow plots generated using the top 2000, 6000, and all genes exhibited similar shapes, with the plots for larger gene sets being left-shifted. This indicated that the main variance structure could be captured efficiently with fewer features. (2) The cluster structures resulting from using 2000 versus more genes were highly similar, suggesting that additional features did not contribute substantially to cluster resolution.
The final clustering resolution of 0.8 was selected based on iterative testing across a range of dimensions (PCs) and clustering resolutions (Fig. S3G). Cluster stability was evaluated in two ways: (1) using the clustree R package to visualize cluster stability across resolutions, and (2) by visually inspecting cluster structure at each resolution (Fig. S3H). We assessed cluster quality by performing a parameter sweep across different combinations of dimensions and resolutions and computing the average Silhouette Score for each. As expected for a continuous dataset, Silhouette Scores were consistently low and showed little variation across parameters, reflecting the gradual transitions between cell states.
To further refine our resolution, we iteratively clustered the data across a range of resolutions (0.4 to 1.4; Fig. S3H) and assessed whether known marker genes for distinct cell states (e.g., cyclins, E2F targets, mitotic genes, and tumor suppressors such as CDKN1A/p21 and CDKN1B/p27) were appropriately resolved at each resolution. Clustering resolutions > 0.4 to 0.6 appropriately resolved known cell states. Importantly, clustering resolutions from 0.8 to 1.4 consistently preserved the cluster structure within the senescent population, which was the focus of our downstream analyses.
Finally, we performed differential gene expression (DGE) analysis to ensure that cluster-defining genes for each cluster were not confined to a small subset of cells within each cluster and that neighboring clusters did not share the same set of top markers. Importantly, our clustering analysis was followed by pseudotime trajectory inference to better capture the continuous nature of the data and to address the limitations of discrete clustering.
Uniform Manifold Approximation and Projection (UMAP)21 was used for dimensionality reduction and visualization with RunUMAP(). Cell-cycle scoring was performed using Seurat’s CellCycleScoring() function, which assigns cell-cycle phase predictions based on predefined gene sets for G2/M and S phases. Any cells that are not categorized as G2/M or S are labeled G1.
Differential expression analysis
Differentially expressed genes (DEGs) between clusters were identified using FindMarkers() with the Wilcoxon rank-sum test. Multiple testing correction was performed using the Bonferroni method to adjust p-values. The expression of canonical markers was visualized using FeaturePlot(), VlnPlot(), and DotPlot(). Expression values shown in all single-gene UMAPs and heatmaps represent z-scored gene counts, which were normalized using NormalizeData() and subsequently scaled and centered with ScaleData().
Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA) was performed using the clusterProfiler package67 in R to identify enriched biological processes. Enrichment significance was assessed using a permutation-based, weighted Kolmogorov–Smirnov–like test implemented in clusterProfiler, with Benjamini–Hochberg correction for multiple testing. The ranking metric was calculated as: Ranking Score = avg_log2FC × −log10 (adj_pvalue), where avg_log2FC represents the average log2 fold change in gene expression, and adj_pvalue is the Benjamini-Hochberg adjusted p-value. This ranking method prioritizes genes that are both highly differentially expressed and statistically significant. Enrichment analysis was conducted using the gseGO() function, specifying the org.Hs.eg.db annotation package for human genes and the Biological Process (BP) ontology. The results were visualized using the gseaplot() function to display the leading enriched pathway and the dotplot() function to summarize the top enriched pathways. All analyses were performed in R using the following packages: clusterProfiler, org.Hs.eg.db, and ggplot2 for visualization.
Identifying shared SASP genes
We first took the 118 leading-edge genes for SASP-related pathways in our senescent clusters (clusters 4, 9, 10, 12) and then for each senescent cluster, we selected the top 50 of the 118 original genes. We then found the overlap between these lists, resulting in 21 SASP genes that were universally upregulated in every senescent cluster at every dose of etoposide (Fig. 6D). The complete list is in Supplementary Data 3.
Identifying the etoposide-induced senescent gene set
To make a gene set to identify etoposide-induced senescent cells, we combined our senescent clusters (clusters 4, 9, 10, 12) and performed DE expression analysis on our combined senescent cells vs. all other cells sequenced. We selected the top 100 genes (based on Ranking Score = avg_log2FC × −log10 (adj_pvalue)) and combined these with the top 50 genes from DE analysis of cluster 4 alone vs. all other cells, since this cluster was less robustly identified. We then found the unique genes from this combined list. Finally, any genes from this list that were also elevated in untreated G0/G1 cells were removed. The complete list is in Supplementary Data 3. A list of the top 200 DE genes for senescent cells vs. all other sequenced cells, ranked by 1) adj_pvalue alone and 2) avg_log2FC × −log10 (adj_pvalue) without any additional filtering, is also provided in Supplementary Data 3.
Expression score calculation
Gene module scores were computed using AddModuleScore() for predefined gene sets, and results were visualized with FeaturePlot() and DoHeatmap().
Pseudotime analysis
Pseudotime trajectory inference was performed using Monocle337,38, with preprocessed Seurat data converted via as.cell_data_set(). Trajectories were learned with learn_graph() and cells were ordered along pseudotime using order_cells().
Statistics & reproducibility
Statistical analyses for single-cell RNA sequencing are detailed in the Single-cell RNA sequencing section of the Methods. Statistical tests for imaging data are described in the corresponding figure captions. No statistical method was used to predetermine sample size. For single-cell RNA sequencing, sample size was determined by the number of captured cells passing quality control, with a target of approximately 5000 cells per condition, as described in the Methods. For live-cell imaging, the sample size was limited by the number of cells that could be plated in 96-well plates, and all observable single cells were imaged. All fixed-cell imaging experiments included at least three replicate wells, with nine independent fields of 10X magnification imaged per well. No data were excluded from the analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All raw sequencing data and processed count matrices have been deposited in GEO under accession code GSE297393. The raw and tracked live-cell imaging data are available upon request to the corresponding author. Publicly available single-cell RNA-seq data sets for Chan et al. and Wechter et al. were downloaded from GEO, GSE175533 and GSE226225, respectively. Source data are provided with this paper.
References
Marescal, O. & Cheeseman, I. M. Cellular mechanisms and regulation of quiescence. Dev. Cell 55, 259–271 (2020).
He, S. & Sharpless, N. E. Senescence in health and disease. Cell 169, 1000–1011 (2017).
Gorgoulis, V. et al. Cellular senescence: defining a path forward. Cell 179, 813–827 (2019).
Huang, W., Hickson, L. J., Eirin, A., Kirkland, J. L. & Lerman, L. O. Cellular senescence: the good, the bad and the unknown. Nat. Rev. Nephrol. 18, 611–627 (2022).
Mijit, M., Caracciolo, V., Melillo, A., Amicarelli, F. & Giordano, A. Role of p53 in the regulation of cellular senescence. Biomolecules 10, 420 (2020).
Di Micco, R., Krizhanovsky, V., Baker, D. & d’Adda di Fagagna, F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 22, 75–95 (2021).
Wang, B., Han, J., Elisseeff, J. H. & Demaria, M. The senescence-associated secretory phenotype and its physiological and pathological implications. Nat. Rev. Mol. Cell Biol. 25, 958–978 (2024).
Coppé, J.-P., Desprez, P.-Y., Krtolica, A. & Campisi, J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev. Pathol. 5, 99–118 (2010).
Gewirtz, D. A., Holt, S. E. & Elmore, L. W. Accelerated senescence: an emerging role in tumor cell response to chemotherapy and radiation. Biochem. Pharmacol. 76, 947–957 (2008).
Ewald, J. A., Desotelle, J. A., Wilding, G. & Jarrard, D. F. Therapy-induced senescence in cancer. J. Natl Cancer Inst. 102, 1536–1546 (2010).
Wang, L., Lankhorst, L. & Bernards, R. Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer 22, 340–355 (2022).
Romanov, S. R. et al. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature 409, 633–637 (2001).
Prasanna, P. G. et al. Therapy-Induced Senescence: Opportunities to Improve Anticancer Therapy. J. Natl Cancer Inst. 113, 1285–1298 (2021).
Schmitt, C. A. et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109, 335–346 (2002).
Sharpless, N. E. & Sherr, C. J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 15, 397–408 (2015).
Ashraf, H. M., Fernandez, B. & Spencer, S. L. The intensities of canonical senescence biomarkers integrate the duration of cell-cycle withdrawal. Nat. Commun. 14, 4527 (2023).
Walker, K. A., Basisty, N., Wilson, D. M. & Ferrucci, L. Connecting aging biology and inflammation in the omics era. J. Clin. Invest. 132, e158448 (2022).
Baldwin, E. L. & Osheroff, N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. Anticancer Agents 5, 363–372 (2005).
Hatakeyama, M. & Weinberg, R. A. The role of RB in cell cycle control. In Progress in Cell Cycle Research. 1, (eds. Meijer, L., Guidet, S. & Tung, H. Y. L.) https://doi.org/10.1007/978-1-4615-1809-9_2 (Springer US, Boston, MA, 1995).
Spencer, S. L. et al. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013).
McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 3, 861 (2018).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Cornwell, J. A. et al. Loss of CDK4/6 activity in the S/G2 phase leads to cell cycle reversal. Nature 619, 363–370 (2023).
Johmura, Y. et al. Necessary and Sufficient Role for a Mitosis Skip in Senescence Induction. Mol. Cell 55, 73–84 (2014).
Wechter, N. et al. Single-cell transcriptomic analysis uncovers diverse and dynamic senescent cell populations. Aging 15, 2824–2851 (2023).
Chan, M. et al. Novel insights from a multiomics dissection of the Hayflick limit. eLife 11, e70283 (2022).
Rossi, C. et al. PURPL and NEAT1 Long Non-Coding RNAs Are Modulated in Vascular Smooth Muscle Cell Replicative Senescence. Biomedicines 11, 3228 (2023).
Han, S. et al. PURPL represses autophagic cell death to promote cutaneous melanoma by modulating ULK1 phosphorylation. Cell Death Dis. 12, 1–13 (2021).
Li, X. L. et al. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep. 20, 2408–2423 (2017).
Hartford, C. C. R. et al. Context-dependent function of long noncoding RNA PURPL in transcriptome regulation during p53 activation. Mol. Cell Biol. 42, e0028922 (2022).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. 102, 15545–15550 (2005).
The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2020).
Aouad, P., Quinn, H. M., Berger, A. & Brisken, C. Tumor dormancy: EMT beyond invasion and metastasis. Genesis 62, e23552 (2024).
Smith, B. N. & Bhowmick, N. A. Role of EMT in metastasis and therapy resistance. J. Clin. Med. 5, 17 (2016).
Brabletz, S., Schuhwerk, H., Brabletz, T. & Stemmler, M. P. Dynamic EMT: a multi-tool for tumor progression. EMBO J. 40, e108647 (2021).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Kovalski, J. R., Kuzuoglu-Ozturk, D. & Ruggero, D. Protein synthesis control in cancer: selectivity and therapeutic targeting. EMBO J. 41, e109823 (2022).
White-Gilbertson, S., Kurtz, D. T. & Voelkel-Johnson, C. The role of protein synthesis in cell cycling and cancer. Mol. Oncol. 3, 402–408 (2009).
Rong, Y., Darnell, A. M., Sapp, K. M., Vander Heiden, M. G. & Spencer, S. L. Cells use multiple mechanisms for cell-cycle arrest upon withdrawal of individual amino acids. Cell Rep. 42, 113539 (2023).
Han, K. et al. Parallel measurement of dynamic changes in translation rates in single cells. Nat. Methods 11, 86–93 (2014).
Delfarah, A. et al. Inhibition of nucleotide synthesis promotes replicative senescence of human mammary epithelial cells. J. Biol. Chem. 294, 10564–10578 (2019).
Cunningham, J. T., Moreno, M. V., Lodi, A., Ronen, S. M. & Ruggero, D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell 157, 1088–1103 (2014).
Herranz, N., Gallage, S. & Gil, J. TORn about SASP regulation. Cell Cycle 14, 3771–3772 (2015).
Herranz, N. et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat. Cell Biol. 17, 1205–1217 (2015).
Laberge, R.-M. et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 17, 1049–1061 (2015).
Chaib, S., Tchkonia, T. & Kirkland, J. L. Cellular senescence and senolytics: the path to the clinic. Nat. Med 28, 1556–1568 (2022).
Tchkonia, T., Palmer, A. K. & Kirkland, J. L. New horizons: novel approaches to enhance healthspan through targeting cellular senescence and related aging mechanisms. J. Clin. Endocrinol. Metab. 106, E1481–E1487 (2021).
Nangia, V. et al. MAPK and mTORC1 signaling converge to drive cyclin D1 protein production to enable cell cycle reentry in melanoma persister cells. Sci. Signal. 18, eadw3231 (2025).
Gillespie, M. et al. The Reactome Pathway Knowledgebase 2022. Nucleic Acids Res. 50, D687–D692 (2022).
Hsu, C.-H., Altschuler, S. J. & Wu, L. F. Patterns of early p21 dynamics determine proliferation-senescence cell fate after chemotherapy. Cell 178, 361–373.e12 (2019).
Cohn, R. L., Gasek, N. S., Kuchel, G. A. & Xu, M. The heterogeneity of cellular senescence: insights at the single-cell level. Trends Cell Biol. 33, 9–17 (2023).
Cohn, R. L., Gasek, N. S., Kuchel, G. A. & Xu, M. The heterogeneity of cellular senescence: insights at the single-cell level. Trends Cell Biol. 33, 9–17 (2022).
Hernandez-Segura, A. et al. Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 27, 2652–2660.e4 (2017).
Kirschner, K., Rattanavirotkul, N., Quince, M. F. & Chandra, T. Functional heterogeneity in senescence. Biochem Soc. Trans. 48, 765–773 (2020).
Stallaert, W. et al. The molecular architecture of cell cycle arrest. Mol. Syst. Biol. 18, e11087 (2022).
Sessions, G. A., Loops, M. V., Diekman, B. O. & Purvis, J. E. Multiplexed single-cell imaging reveals diverging subpopulations with distinct senescence phenotypes during long-term senescence induction. GeroScience 47, 3891–3905 (2025).
Paronetto, M. P., Miñana, B. & Valcárcel, J. The ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol. Cell 43, 353–368 (2011).
Lü, X., de la Peña, L., Barker, C., Camphausen, K. & Tofilon, P. J. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 66, 1052–1061 (2006).
Halim, V. A. et al. Doxorubicin-induced DNA damage causes extensive ubiquitination of ribosomal proteins associated with a decrease in protein translation*. Mol. Cell Proteom. 17, 2297–2308 (2018).
Payea, M. J., Anerillas, C., Tharakan, R. & Gorospe, M. Translational control during cellular senescence. Mol. Cell Biol. 41, e00512–e00520 (2021).
Weichhart, T. mTOR as regulator of lifespan, aging and cellular senescence. Gerontology 64, 127–134 (2018).
Fujimaki, K. et al. Graded regulation of cellular quiescence depth between proliferation and senescence by a lysosomal dimmer switch. Proc. Natl. Acad. Sci. USA 116, 22624–22634 (2019).
Gookin, S. et al. A map of protein dynamics during cell-cycle progression and cell-cycle exit. PLOS Biol. 15, e2003268 (2017).
Arora, M., Moser, J., Phadke, H., Basha, A. A. & Spencer, S. L. Endogenous replication stress in mother cells leads to quiescence of daughter cells. Cell Rep. 19, 1351–1364 (2017).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Acknowledgments
We thank current and past members of the Spencer Laboratory for general discussion and insight over the course of the work, and Theresa Nahreini and the cell culture facility for cell sorting (RRID:SCR_018988). The Aria Fusion FACS sorter is supported by NIH grant S10OD021601. The RNA FISH imaging was performed at the BioFrontiers Advanced Light Microscopy Core directed by Joe Dragavon (RRID: SCR_018302). The Revvity Opera Phenix is supported by NIH grant 1S10OD025072. Single-cell capture, library generation and sequencing were performed at the University of Colorado Anschutz Medical Campus Genomics Shared Resource (RRID: SCR_021984), which is supported by the Cancer Center Support Grant (P30CA046934). We thank Dr. James DeGregori for providing facilities for sample preparation at CU Anschutz. This work was supported by an NIH T32 GM142607 (to B.F.), an NIH F31 CA284877 (to B.F.), an NIH Director’s New Innovator Award 1DP2CA238330-01 (to S.L.S), and an R01 R01AG082942 (to S.L.S). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
B.F. and S.L.S. designed the research; H.A., V.P., and B.F. conducted the research; B.F. analyzed the data; B.F. and S.L.S. conceived the project; B.F. and S.L.S. wrote the paper; S.L.S. supervised the project.
Corresponding author
Ethics declarations
Competing interests
S.L.S. has a sponsored research agreement with Genesis Therapeutics, had a past sponsored research agreement with Pfizer Inc., is on the scientific advisory board of Meliora Therapeutics, has been a paid consultant for Astex Therapeutics and Guidepoint Global, and has received honoraria/speaking fees from Blueprint Therapeutics, Ensem Therapeutics, Genentech Inc, Iambic Therapeutics, Nereid Therapeutics, Odyssey Therapeutics, and Pfizer Inc. A joint patent has been filed by the Regents of the University of Colorado and Pfizer Inc. related to CDK2 inhibitors (PCT/IB2021/052894). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fernandez, B., Passanisi, V.J., Ashraf, H.M. et al. Single-cell RNA sequencing reveals a quiescence-senescence continuum and distinct senotypes following chemotherapy. Nat Commun 17, 169 (2026). https://doi.org/10.1038/s41467-025-66836-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66836-z