Abstract
DNA topology is critical for regulating transcription and maintaining cellular homeostasis. Z-DNA is a left-handed DNA helix in regions with high transcriptional activity. Its physiological function remains poorly understood. Here, we demonstrate that oncoprotein MYC induces the formation of Z-DNA by recruiting the chromatin remodeler FACT, independent of RNA Polymerase II activity. FACT facilitates Z-DNA formation by remodeling H2A/H2B dimers within intact nucleosomes. Additionally, the phosphorylation of FACT regulates its liquid-liquid phase separation, promoting its efficient recruitment to chromatin by MYC. Through a genome-wide analysis and characterization of engineered Z-DNA promoters, we found that Z-DNA directly facilitates the loading of RNA Polymerase II, thereby promoting transcriptional activity. This study elucidates the molecular mechanisms of Z-DNA dynamics and emphasizes its functional importance in transcriptional regulation, providing insights into the role of left-handed DNA structures in chromatin biology and MYC-driven cancer.
Similar content being viewed by others
Introduction
The dynamic architecture of chromatin plays a fundamental role in regulating gene expression1,2. Within this architecture, various DNA structures, beyond the conventional B-DNA double helix, have emerged as significant regulators of transcriptional activity3,4,5. Among these non-conventional DNA structures, Z-DNA, a left-handed DNA helix, has garnered particular interest due to its association with regions of high transcriptional activity6,7,8. Despite its well-documented formation under supercoiling stress, the precise mechanisms underlying Z-DNA induction and its functional consequences on transcription remain poorly understood9.
The existence of Z-DNA was an unexpected discovery, initially identified during the crystallization of the first synthetic DNA6,10. While the biological significance of Z-DNA was initially overestimated, it has since been relatively underappreciated3,11. A pivotal moment in our understanding of left-handed nucleic acid was the identification of the binding domain, known as the Zα domain, located in ADAR112. In-depth studies by various scientists have highlighted the roles of ADAR1 and other Z-DNA-binding proteins, such as ZBP1, in connecting left-handed nucleic acid with proteins, especially regarding human health and disease13,14,15,16. The expression of ADAR1 can be induced by interferon, which negatively regulates immune responses triggered by dsRNA17,18. It is suggested that the transition from B-form to Z-form DNA occurs when processive enzymes, such as polymerases and helicases, create regions of underwound DNA during their activity19. When formed through these enzymatic processes, the formation of Z-DNA is considered a passive byproduct of transcription.
The oncogenic transcription factor MYC serves as a crucial regulator of gene expression and cellular proliferation, influencing a broad range of biological processes20,21,22. Alterations in MYC activity are frequently observed in various cancers23. MYC proteins are components within a network of interacting transcription factors, which act as regulators of gene transcription24,25. By directly influencing the duration of transcriptional activity, MYC amplifies gene expression in various genes and cell types26. Additionally, MYC regulates RNA Polymerase II (RNAP II) binding to promoters, facilitates promoter escape, induces pause release, and resolves obstacles caused by R-loops and DNA damage27,28,29. MYC is also recognized for its ability to remodel chromatin structures, which in turn elevates gene expression30,31. MYC can also regulate gene transcription by recruiting various co-factors, such as histone modifiers, topoisomerases, and chromatin remodeling complexes, to specific chromatin regions32,33,34,35. This recruitment facilitates the transcriptional activation of target genes, which are often involved in critical cellular functions such as cell cycle progression, metabolism, and apoptosis31,36,37,38. Despite the extensive characterization of MYC’s role in gene regulation, the interplay between MYC activity and the formation of non-B-DNA structures, such as Z-DNA, remains relatively underexplored. Understanding how MYC influences or interacts with these non-B-DNA structures could provide valuable insights into the complexity of gene regulation39.
In this study, we employ a combination of cellular and biochemical techniques to demonstrate that MYC recruits the FACT complex to chromatin, promoting the formation of Z-DNA independently of RNAP II activity. FACT facilitates Z-DNA formation by remodeling intact nucleosomes through specific interactions with H2A/H2B dimers. Furthermore, the phosphorylation of FACT is essential for regulating its liquid-liquid phase separation (LLPS), which enables its efficient recruitment to chromatin by MYC. Importantly, Z-DNA serves as a functional inducer for RNAP II loading and transcriptional activation, supported by genome-wide analysis and investigations of engineered Z-DNA-containing super core promoters (Z-SCPs), highlighting its role in regulating gene expression beyond mere structural characteristics. By delineating the molecular interplay between MYC and Z-DNA, our work advances the understanding of how Z-DNA directly contributes to transcriptional regulation, with potential implications for targeting MYC-driven transcriptional programs in cancers.
Results
MYC induces Z-DNA formation in a FACT-dependent manner
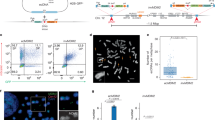
To investigate the relationship between gene expression and Z-DNA formation, we overexpressed the oncoprotein MYC in U2OS cells. MYC overexpression functions not by activating silent gene clusters, but rather as a nonlinear amplifier for active genes by binding to those already undergoing transcription33,36. Furthermore, MYC enhances overall transcriptional output by modifying the residence times of transcription machinery components27,35. Previous reports have shown that Z-nucleic acid (Z-NA) can be specifically recognized by the Z22 monoclonal antibody17,40,41. We then purified this antibody and analyzed its specificity using the CBL0137-treated cells in comparison to the vehicle (DMSO)-treated cells. CBL0137, originally designed as a FACT inhibitor, has been reported to be directly incorporated into B-formed DNA, thereby inducing the formation of Z-DNA17. Our results demonstrated that the Z22 antibody we employed could specifically recognize the signals within CBL0137-treated cells (Supplementary Fig. 1a, b), thereby confirming its specificity for Z-DNA. Utilizing this Z22 antibody, we observed an increased signal of Z-NA, which specifically recognizes Z-NA17,40,41, in the nucleus (Fig. 1a, b). In addition, the observed Z-NA levels were strongly correlated with MYC expression across different cells (Fig. 1c). To further confirm this increase in Z-NA signal, we also overexpressed MYC in other cell lines, including HeLa, HEK293T, Astrocyte, and HGC27. The expression of exogenous MYC proteins showed comparable levels to the endogenous MYC proteins (Supplementary Fig. 1c). The Z-NA signals were all elevated by the overexpression of MYC (Supplementary Fig. 1d, e). Additionally, the induction of Z-NA was rapidly reversed by the prompt degradation of MYC within 1 h (Supplementary Fig. 1f–h). To determine whether the Z-NA signal originated from DNA, RNA, or DNA-RNA hybrids, we treated MYC-overexpressing cells with DNase I, RNase H, and RNase A, respectively. Only DNase I treatment effectively reduced the Z-NA signal (Fig. 1d, e), confirming that the MYC-induced Z-NA was left-handed Z-DNA. DNase I treatment generates DNA breaks, which alleviates negative superhelical stress. This process facilitates the transition from Z-DNA to the more energetically stable B-DNA, thereby establishing a thermodynamically favorable conformation under physiological conditions.
a IF results showing the localization and expression levels of MYC and Z-NA. HA-tagged MYC was overexpressed in U2OS cells and stained with HA antibodies 3 days after transfection. Z-NA was detected using Z22 antibodies. Nuclei were stained with DAPI. Scale bar, 10 µm. b Z22 fluorescence intensity quantification results for each cell in (a). The data were represented by the scatter dots and mean values (WT, N = 50 cells, HA-MYC, N = 65 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. c The correlation between HA-MYC and Z22 fluorescence intensity among each cell (N = 50 cells) in (a). Data were analyzed using simple linear regression (two-sided test), with no adjustments for multiple comparisons. The exact p values were indicated in the figure. Source data are provided as a Source Data file. d IF results showing the Z-NA signals after treatment with various nucleases. U2OS cells expressing HA-MYC for 3 days were fixed, permeabilized, and then treated with 0.2 U/μL DNase I, 1 mg/mL RNase A, and 0.2 U/μL RNase H, respectively, at 37 °C for 3 h. The efficiencies of DNase I, RNase H, and RNase A treatments were verified by DAPI, S9.6, and Acridine Orange staining, respectively. Cells treated with PBS served as the negative control. Scale bar, 10 µm. e Z22 fluorescence intensity quantification results of each cell in (d). The data were represented by the scatter dots and mean values (WT, N = 51 cells; PBS control for DNAse I treatment, N = 52 cells; DNAse I, N = 52 cells; PBS control for RNAseH treatment, N = 64 cells; RNAseH, N = 54 cells; PBS control for RNAseA treatment, N = 64 cells; RNAseA, N = 63 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. f IF results showing the MYC and Z-NA signals in H526 (low MYC expression), H211 and H82 cells (high MYC expression) of human SCLC lines. Scale bar, 10 µm. g Z22 fluorescence intensity quantification results for each cell in (f). The data were represented by the scatter dots and mean values (H526, N = 53 cells; H211, N = 52 cells; H82, N = 53 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. h IF results showing the MYC and Z-NA signals in RPS1 (low MYC expression), RPP-mTmG and RPP-GSDME-KO cells (high MYC expression) of mouse SCLC lines. Scale bar, 10 µm. i Z22 fluorescence intensity quantification results for each cell in (h). The data were represented by the scatter dots and mean values (RPS1, N = 53 cells; RPP-mTmG, N = 55 cells; RPP-GSDME-KO, N = 55 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. j Z22 fluorescence intensity quantification results of U2OS cells treated with different RNAP II inhibitors. Cells expressing HA-tagged MYC were treated with 1 μM BMH−21, 1 μM Flavopiridol, 1 μM THZ1, 1 μg/mL Actinomycin-D, and 10 μg/μL α-amanitin for 6 h. The data were represented by the scatter dots and mean values (Vector, N = 61 cells; DMSO, N = 57 cells; BMH-21, N = 69 cells; Flavopiridol, N = 52 cells; THZ1, N = 67 cells; Actinomycin-D, N = 74 cells; α–amanitin, N = 68 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. k IF results showing the Z-DNA signals in HA-MYC overexpressing cells 3 days after SSRP1 or SPT16 depletion. NT non-target control. Scale bar, 10 µm. l Z22 fluorescence intensity quantification results for each cell in (k). The data were represented by the scatter dots and mean values (WT, N = 59 cells; shNT shSSRP1-1, N = 69 cells; shSSRP1-2, N = 60 cells; shSPT16-1, N = 60 cells; shSPT16-2, N = 61 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. m SSRP1 bound with MYC and SPT16. FLAG-tagged SSRP1 was purified by FLAG beads in HEK293T cells. Proteins from input and IP samples were analyzed by Western blotting using the indicated antibodies. The assay was performed in three independent biological replicates with similar results. Source data are provided as a Source Data file. n Endogenous MYC bound with SSRP1 and SPT16. MYC antibody was used to pull down endogenous MYC and bound FACT proteins in HEK293T cells. Proteins from input and IP samples were analyzed by Western blotting using the indicated antibodies. The assay was performed in three independent biological replicates with similar results. Source data are provided as a Source Data file. o IF results showing DAPI and GFP merged signals after GFP1-10-tagged MYC and GFP11-tagged SSRP1 or SPT16 were expressed in U2OS cells together or respectively. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results.
Since MYC overexpression was utilized to detect increased Z-DNA formation, we are also interested in investigating whether physiologically elevated MYC levels in tumor cells could similarly promote Z-DNA formation. We utilized human and mouse small cell lung cancer (SCLC) cell lines with varying physiological MYC expression to investigate Z-DNA formation. Specifically, we analyzed H526 cells (low MYC expression) as well as H211 and H82 cells (high MYC expression) of human SCLC lines (Supplementary Fig. 1i). Similarly, for mouse SCLC lines, we included RPS1 cells (low MYC expression), RPP-mTmG and RPP-GSDME-KO cells (high MYC expression) (Supplementary Fig. 1j). Increased MYC expression consistently correlated with heightened Z22 nuclear signal in both human and mouse SCLC cell lines, suggesting that elevated MYC expression promotes Z-DNA formation under physiological conditions (Fig. 1f–i).
Previous research has proposed that, during transcription, RNAP II unwinds the DNA double helix and generates positive supercoiling tension. The unwinding process, which is characterized by negative supercoiling, can promote the transition to the higher-energy Z-DNA conformation by alleviating torsional stress in the DNA helix42,43. To investigate this further, we inhibited RNAP II activity after MYC overexpression. The Z-DNA signal remained unchanged upon treatment with various RNAP II inhibitors (Fig. 1j and Supplementary Fig. 1k). These RNAP II inhibitors effectively repressed the RNAP II functionality (Supplementary Fig. 1l). While the endogenous MYC protein exhibited a reduction due to its inherent short lifespan, the exogenously expressed MYC protein remained stable. This stability was likely attributed to the accumulation of exogenous MYC mRNA, which sustains ongoing translation (Supplementary Fig. 1l, m). Furthermore, the Z-DNA signal remained unchanged following treatment with the histone deacetylase inhibitor Trichostatin A after MYC overexpression (Supplementary Fig. 1n, o). To further test whether the treatment with RNAP II inhibitors could affect Z-DNA signal, we also inhibited RNAP II in wild-type U2OS cells and detected no induction of Z-DNA signal (Supplementary Fig. 1p–r). These results indicate that the formation of Z-DNA is independent of RNAP II activity. We then considered whether chromatin remodeling factors44, which alter the chromatin structure to facilitate transcription, were involved in the formation of MYC-induced Z-DNA. To test this hypothesis, we depleted various chromatin remodeling factors in cells that overexpressed MYC (Supplementary Fig. 2a, b). Among the chromatin remodelers tested, we found that depleting the FACT complex, which is composed of SSRP1 and SPT1645, reduced MYC-induced Z-DNA formation without affecting MYC protein levels (Fig. 1k, l and Supplementary Fig. 2c). Notably, altering either of the FACT components changed the protein levels of the other one (Supplementary Fig. 2d, e), as previously reported46,47. Furthermore, we depleted SSRP1 in two human cell lines with high MYC expression (H211 and H82) and observed that the Z22 signals were abolished (Supplementary Fig. 2f–h). This provides additional evidence for the involvement of the FACT complex in MYC-induced Z-DNA formation under physiological MYC expression levels.
Moreover, we observed that MYC can bind to the FACT complex within cells, as evidenced by co-immunoprecipitation (Fig. 1m, n) and split-GFP tag methods (Fig. 1o). To further investigate whether ectopically expressed MYC interacts with MAX, we expressed either WT MYC or a MAX-binding-deficient mutant, MYC-ΔbHLHZip, which lacks the basic-helix-loop-helix-leucine zipper (bHLHZip) domain. As expected, WT MYC co-immunoprecipitated with MAX, whereas the mutant failed to do so (Supplementary Fig. 2i–k). Additionally, the MYC-ΔbHLHZip mutant exhibited markedly diminished interaction with FACT. Notably, Z-DNA formation was induced only by WT MYC and not by MYC-ΔbHLHZip. Together, these findings indicate that MYC induces Z-DNA formation in a FACT-dependent manner.
FACT facilitates the formation of Z-DNA by remodeling H2A/H2B
Since FACT is a chromatin remodeler that functions by binding with nucleosomes, we then investigated whether this process would facilitate the formation of Z-DNA. To investigate this, we incubated the recombinant FACT complex, which was purified from insect SF9 cells, with reconstituted nucleosomes and magnesium. This resulted in a mobility shift in the wrapped DNA on a native gel, indicating that FACT binds to the nucleosome (Fig. 2a). More importantly, this complex was recognized by the Z-NA antibody, resulting in an additional shift of the DNA. In addition, overexpression of SSRP1 or SPT16 induced the Z-NA signals in multiple cell types, including U2OS, HeLa, HEK293T, Astrocyte, and HGC27 (Supplementary Fig. 3a, b). The expression of exogenous SSRP1 or SPT16 proteins showed comparable levels to their corresponding endogenous proteins (Supplementary Fig. 3c, d). Moreover, the Z-NA signals induced by SSRP1 or SPT16 overexpression were reduced by DNase I treatment, thereby directly confirming the formation of Z-DNA (Fig. 2b, c and Supplementary Fig. 4a, b). We also observed that SSRP1 and SPT16 formed puncta in cells, with both proteins being enriched, as detected by the split-GFP tag (Supplementary Fig. 4c). Since the FACT complex may also play a role in DNA replication, the formation of puncta and Z-DNA could result from disturbances in the cell cycle. To investigate this, we synchronized cells to the G1/S phase and then overexpressed SPT16. To synchronize the U2OS cells at the G1/S phase boundary, we pre-treated cells with 4 mM thymidine for 6 h. Subsequently, the cells were infected using a viral expression system to facilitate the expression of SPT16 while maintaining the presence of 4 mM thymidine. Throughout the expression period of SPT16, the cells were continuously cultured with thymidine to ensure synchronization. Under these conditions, SPT16 still formed puncta and induced Z-DNA formation (Supplementary Fig. 4d, e).
a EMSA assay showing the formation of Z-DNA by FACT. Nucleosomes, FACT, and Z22 antibody were incubated at molar ratios of 3:1:1 (nucleosome:FACT:Z22) in 20 μL of reaction buffer (10 mM HEPES pH 7.5 and 10 mM MgCl2). A 6% Native-PAGE gel was stained with GelRed to show the gel shifting of DNA by FACT and Z22 antibody. The assay was performed in three independent biological replicates with similar results. b IF results showing the localization and expression level of SSRP1-GFP and Z-NA after treatment with various nucleases. U2OS cells expressing SSRP1-GFP for 3 days were fixed, permeabilized, and then treated with 0.2 U/μL DNase I, 1 mg/mL RNase A, and 0.2 U/μL RNase H, respectively, at 37 °C for 3 h. The efficiencies of DNase I, RNase H, and RNase A treatments were verified by DAPI, S9.6, and Acridine Orange staining, respectively. Cells treated with PBS served as a negative control. Scale bar, 10 µm. c Z22 fluorescence intensity quantification results of each cell in (b). The data were represented by the scatter dots and mean values (WT, N = 56 cells; PBS control for DNAse I treatment, N = 62 cells; DNAse I, N = 61 cells; PBS control for RNAseH treatment, N = 60 cells; RNAseH, N = 60 cells; PBS control for RNAseA treatment, N = 60 cells; RNAseA, N = 60 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. d IF results showing the localization and expression level of WT and mutant SSRP1-GFP, endogenous SPT16, and Z-DNA. GFP-tagged SSRP1 and mutants were transfected into U2OS cells for 3 days. NLS, nuclear localization signal. Scale bar, 10 µm. e Z22 fluorescence intensity quantification results for each cell in (d). The data were represented by the scatter dots and mean values (Vector, N = 62 cells; SSRP1-GFP, N = 102 cells; SSRP1ΔCTD-GFP, N = 79 cells; SSRP1ΔCTD-GFP-NLS, N = 60 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. f EMSA assay showing the formation of Z-DNA by nucleosome or H3/H4 wrapped with DNA. Nucleosomes or H3/H4 wrapped with DNA, FACT, and Z22 antibody were incubated at molar ratios of 3:1:1 (nucleosome:FACT:Z22) in 20 μL of reaction buffer (10 mM HEPES pH 7.5 and 10 mM MgCl2). A 6% Native-PAGE gel was stained with GelRed to show the gel shifting of DNA by FACT and Z22 antibody. The assay was performed in three independent biological replicates with similar results. g In vitro pull-down results showing the binding between FACT and the indicated substrates. SSRP1 antibody was used to pull down FACT and histones. The assay was performed in three independent biological replicates with similar results. Source data are provided as a Source Data file.
The FACT complex may facilitate nucleosome reorganization via the “dimer eviction model”. This model posits that the FACT complex utilizes its histone chaperone function to actively displace a single H2A-H2B dimer from the nucleosome48,49,50. To analyze how the flipping of H2A/H2B affects FACT-mediated induction of Z-DNA, we expressed C-terminal deletions of SSRP1 or SPT16, which have been reported to lose their binding to H2A/H2B51. These deletions resulted in the cytoplasmic localization of SSRP1 and SPT16, likely due to a nuclear localization signal (NLS) in C-terminal regions (Fig. 2d and Supplementary Fig. 4f–h). We subsequently added an NLS to relocate these mutated proteins to the nucleus. While these mutant proteins still formed puncta in the cells, they lost the ability to induce Z-DNA formation (Fig. 2d, e and Supplementary Fig. 4h, i).
We next examined the effect of removing H2A/H2B from intact nucleosomes on Z-DNA formation. To test this, we wrapped DNA with H3/H4 tetramers to reconstitute H3/H4 tetrasomes, which were then incubated with the FACT complex. As expected, the FACT complex could still bind to H3/H4 tetrasomes, causing a mobility shift in the wrapped DNA on a native gel (Fig. 2f). However, additional incubation with a Z-NA antibody did not alter this mobility, indicating that FACT could not induce Z-DNA formation without H2A/H2B. Moreover, FACT bound to H3/H4 tetrasomes with lower affinity compared to intact nucleosomes (Fig. 2g). These results suggest that FACT induces Z-DNA formation primarily through remodeling H2A/H2B dimers within intact nucleosomes.
FACT puncta are formed through liquid-liquid phase separation (LLPS)
We observed that FACT can form puncta (Fig. 2b and Supplementary Fig. 4a), so we further analyzed their condensate behavior. We found that, when incubated with physiological concentrations of magnesium (about 1 mM), recombinant FACT spontaneously formed small circular droplets (Fig. 3a, b and Supplementary Fig. 5a). A higher concentration of FACT resulted in larger droplet formation (Supplementary Fig. 5b, c). These droplets were sensitive to disruption with 1,6-hexanediol (Fig. 3c, d). Additionally, the puncta of SSRP1 and SPT16 exhibited movement and fusion in cells over time (Fig. 3e). We also observed that, after photobleaching, the fluorescence signals of SSRP1 exhibited slow recovery, while those of SPT16 demonstrated rapid recovery, regardless of whether SSRP1 and SPT16 were expressed together or separately (Fig. 3f–h and Supplementary Fig. 5d–g). More importantly, 10-min exposure to 2% and 4% 1,6-hexanediol suppressed SSRP1 and SPT16 puncta formation, respectively (Fig. 3i). These results collectively suggest that FACT forms puncta through LLPS. Relevant literature indicates that FACT predominantly associates with partially unwrapped nucleosomes, rather than those that are fully wrapped. However, under high concentrations, FACT could still bind with fully wrapped nucleosomes52,53. It is also known that nucleosomes with identical DNA sequences tend to cluster in the presence of magnesium54. Our research demonstrated that FACT formed LLPS in magnesium-containing environments. Thus, magnesium consequently elevated the local concentration of both nucleosomes and FACT. This increase may facilitate the detection of their mutual binding. To directly assess the influence of magnesium, we titrated magnesium concentration in our binding assays and observed that higher magnesium concentrations led to a marked increase in the shifted band due to FACT binding (Supplementary Fig. 5h).
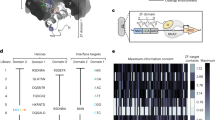
a Microscopy results showing the FACT condensates in different concentrations of MgCl2. FACT (15 µM) mixed with MgCl2 at different concentrations (0.25, 0.5, 1, 2, and 4 mM from left to right) was analyzed for the formation of droplets at 75 mM NaCl and 25 mM Tris-HCl pH 7.5. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results. b Condensate area quantification results of each droplet in (a). The data were represented by the scatter dots and mean values (0.25 mM, N = 140 droplets; 0.5 mM, N = 180 droplets; 1 mM, N = 187 droplets; 2 mM, N = 178 droplets; 4 mM, N = 149 droplets). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. c Microscopy results showing the FACT condensates treated with 1,6-Hexanediol. FACT (15 µM) mixed with 1,6-Hexanediol at different concentrations (0, 0.5%, 1%, 2%, and 4% from left to right) was analyzed for the formation of droplets at 0.5 mM MgCl2, 75 mM NaCl, and 25 mM Tris-HCl pH 7.5. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results. d Condensate area quantification results for each droplet in (c). The data were represented by the scatter dots and mean values (0, N = 130 droplets; 0.5%, N = 124 droplets; 1%, N = 29 droplets; 2%, N = 31 droplets; 4%, N = 243 droplets). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. e Fluorescence results of live U2OS cells transfected with SSRP1-GFP and SPT16-GFP. The live cell imaging was conducted 72 h following the transfection of U2OS cells with SSRP1-GFP or SPT16-GFP. Representative images showing a condensate at different time points. Scale bar, 10 µm. Arrows indicate the merging sites. The assay was performed in three independent biological replicates with similar results. f FRAP assay of U2OS cells transfected with SPT16-mCherry and SSRP1-GFP together. Representative images showing a condensate before and at different time points after photobleaching. Scale bar, 10 µm. Arrows indicate the photobleaching sites. Relative intensity quantification of SSRP1-GFP (g) and SPT16-mCherry (h) in representative bleached and unbleached droplets. The data were represented by the mean ± SD (N = 3 independent replicates). Source data are provided as a Source Data file. i Microscopy results showing the SSRP1 condensates treated with 2% 1,6-Hexanediol and SPT16 condensates treated with 4% 1,6-Hexanediol at the indicated time points in U2OS cells. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results. j The disordered regions of SSRP1 predicted by NovoPro (upper panel) and the illustration showing the protein domains of SSRP1 (lower panel). Source data are provided as a Source Data file. k IF results showing the localization and expression levels of WT and mutant SSRP1-GFP, endogenous SPT16, and Z-NA in U2OS cells. Scale bar, 10 µm. l Condensate number quantification results for each cell in (k). The data were represented by the scatter dots and mean values (SSRP1-GFP, N = 50 cells; SSRP1ΔNTD-GFP, N = 55 cells; SSRP1ΔIDR1-GFP, N = 52 cells; SSRP1ΔIDR2-GFP, N = 49 cells; SSRP1ΔMD-GFP, N = 50 cells; SSRP1ΔIDD-GFP, N = 52 cells; SSRP1ΔHMG-GFP, N = 51 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. m Z22 fluorescence intensity quantification results for each cell in (k). The data were represented by the scatter dots and mean values (WT, N = 58 cells; SSRP1-GFP, N = 59 cells; SSRP1ΔNTD-GFP, N = 56 cells; SSRP1ΔIDR1-GFP, N = 54 cells; SSRP1ΔIDR2-GFP, N = 56 cells; SSRP1ΔMD-GFP, N = 54 cells; SSRP1ΔIDD-GFP, N = 55 cells; SSRP1ΔHMG-GFP, N = 54 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. n The deletions of IDD and NTD domains of SSRP1 disrupted the binding of SSRP1 with MYC and SPT16, respectively. FLAG-tagged WT SSRP1 and mutants were purified by FLAG beads in HEK293T cells. Proteins from input and IP samples were analyzed by Western blotting using the indicated antibodies. The assay was performed in two independent biological replicates with similar results. Source data are provided as a Source Data file.
To investigate the domains responsible for the formation of FACT condensate and Z-DNA, we created single deletions of all domains in SSRP1/SPT16 and assessed their effects. The predicted intrinsically disordered region (IDR) is located at the C-terminal end of SSRP1, including IDD and CTD (Fig. 3j). The intrinsically disordered domain (IDD) within this region played a critical role in the formation of SSRP1 condensates (Fig. 3k, l and Supplementary Fig. 5i). Additionally, the N-terminal domain (NTD), which exhibits less internal disorder, was also important for the formation of SSRP1 condensates. When the HMG domain, which facilitates the binding between SSRP1 and SPT16, was truncated, SPT16 and Z-DNA were observed as a circular arrangement at the surface of SSRP1 condensates. This observation supports the notion that SSRP1 and SPT16 form FACT complexes to contribute to the formation of Z-DNA.
The above results showed that truncating the IDR domain of SSRP1 resulted in the loss of the protein’s ability to form condensates. In contrast, single domain truncation of SPT16 had a minimal effect on its condensation properties. Only the dimerization domain (DD) of SPT16, which interacts with SSRP1 to form a heterodimer, was essential for the formation of SPT16 condensates (Supplementary Fig. 5j–m). Nonetheless, the promotion of Z-DNA formation by FACT remained unaffected by these domain truncations in SSRP1/SPT16, except for the truncation of the IDR2 in SPT16 (Fig. 3m and Supplementary Fig. 5n). Notably, the segment corresponding to the IDR2 domain in SPT16 is longer than that of the CTD domain. This difference resulted in its diminished capacity to promote Z-DNA formation, similar to the effects observed with the CTD truncation. We conducted further experiments to assess the critical domains involved in the interaction between FACT and MYC. The truncation of the IDD domain in SSRP1 reduced its association with MYC (Fig. 3n). Consistent with previous reports, the N-terminal domain (NTD) of SSRP1 was essential for its binding with SPT16. Moreover, the DD domain of SPT16 played a pivotal role in its interactions with both MYC and SSRP1 (Supplementary Fig. 5o). These findings indicate that FACT puncta are formed through LLPS in cells, with the IDD of SSRP1 being essential for its LLPS.
Phosphorylation regulated by the CK2-PP2AC axis controls FACT LLPS
Given that post-translational modifications often change the dynamic of FACT complex55,56,57, we explored how such changes affect its modulation of LLPS. The FACT complex may undergo diverse post-translational modifications, including phosphorylation, PARylation, acetylation, ubiquitylation, and glycosylation. We then employed the following inhibitors: CX-4945 (phosphorylation inhibitor), Olaparib (PARylation inhibitor), Tannic acid (deacetylation inhibitor), LEN (ubiquitination inhibitor), and OSMI (O-GlcNAc glycosylation inhibitor). Of these, only CX-4945 significantly inhibited the LLPS of SSRP1 (Fig. 4a and Supplementary Fig. 6a), and this effect was observed in a concentration-dependent manner (Supplementary Fig. 6b, c).
a Condensate number quantification results of U2OS cells treated with 10 µM CX-4945, Olaparib, Tannic acid, LEN, and OSMI for 24 h, respectively. CX-4945, a CK2 inhibitor that suppresses protein phosphorylation; Olaparib, a PARP inhibitor that blocks PARylation (poly-ADP ribosylation); Tannic acid, an HDAC inhibitor that increases acetylation; Lenalidomide (LEN), a CRBN E3 ubiquitin ligase modulator that reduces ubiquitination; OSMI-1, an OGT inhibitor that reduces O-GlcNAc glycosylation. The data were represented by the scatter dots and median value (DMSO, N = 45 cells; CX-4945, N = 41 cells; Tannic acid, N = 38 cells; LEN, N = 45 cells; Olaparib, N = 58 cells; OSMI, N = 42 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. b IF results showing the localization and expression level of endogenous SSRP1, SPT16, and Z-NA after CK2 overexpression in U2OS cells. Empty vectors were used as negative controls. Scale bar, 10 µm. c Condensate number quantification results for each cell in (b). The data were represented by the scatter dots and mean values (SSRP1-CT, N = 51 cells; SSRP1-CK2, N = 60 cells; SPT16-CT, N = 51 cells; SPT16-CK2, N = 60 cells; Z22-CT, N = 50 cells; Z22-CK2, N = 60 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. d IF results showing the localization and expression level of WT and mutant SSRP1-GFP, endogenous SPT16, and Z-NA in U2OS cells. The S-to-A mutation was used to disrupt phosphorylation, and the S-to-E mutation was used to mimic phosphorylation of SSRP1. Scale bar, 10 µm. e Condensate number quantification results for each cell in (d). The data were represented by the scatter dots and mean values (SSRP1-GFP, N = 50 cells; SSRP1-S510A-GFP, N = 52 cells; SSRP1-S657A/S688A-GFP, N = 51 cells; SSRP1-S510A/S657A/S688A-GFP, N = 51 cells; SSRP1-S510E-GFP, N = 50 cells; SSRP1-S657E/S688E-GFP, N = 50 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. f Schematic illustration of phosphorylation mimic of SSRP1 at S510 by genetic code expansion. The red tRNA represented aminoacyl-tRNA in the engineered E. coli. g Western blotting results showing the detection of pS510-SSRP1 in reconstituted proteins in vitro. Proteins from wild-type and reconstituted phosphorylation mimics of pS510-SSRP1, which were purified from E. coli, were analyzed by Western blotting using homemade antibodies. The assay was performed in two independent biological replicates with similar results. Source data are provided as a Source Data file. h Western blotting results showing the detection of pS510-SSRP1 in vitro. Phosphorylated levels of SSRP1 at the S510 site of WT SSRP1, mutant SSRP1, and SSRP1 treated with CIP/denatured CIP were analyzed by homemade antibodies. FLAG-tagged WT and mutant SSRP1 were purified by FLAG beads in HEK293T cells. FLAG immunoprecipitated (IPed) SSRP1 was further treated with 250 U/mL of CIP or denatured CIP at room temperature for a duration of 30 min. Denatured CIP, heat-inactivated at 85 °C for 10 min. Proteins from input and IP samples were analyzed by Western blotting using the indicated antibodies. The assay was performed in two independent biological replicates with similar results. Source data are provided as a Source Data file. i Western blotting results showing the level of pS510-SSRP1 after CK2 overexpression. FLAG-tagged SSRP1 was purified by FLAG beads in HEK293T cells. Proteins from input and IP samples were analyzed by Western blotting using the indicated antibodies. The assay was performed in two independent biological replicates with similar results. Source data are provided as a Source Data file. j Microscopy results showing the FACT condensates with WT or S510A mutant SSRP1 in vitro. Proteins were purified from SF9 cells. WT and mutant FACT were mixed with 0.5 mM MgCl2 to form droplets at 75 mM NaCl and 25 mM Tris-HCl pH 7.5. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results. k Condensate area quantification results for each droplet in (j). The data were represented by the scatter dots and mean values (WT, N = 225 droplets; SSRP1-S510A, N = 72 droplets). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. l Microscopy results showing the FACT condensates with CIP treatment in vitro. FACT was purified from SF9 cells and was further treated with 250 U/mL of CIP or denatured CIP at room temperature for a duration of 30 min. Denatured CIP, heat-inactivated at 85 °C for 10 min. FACT treated with CIP or denatured CIP were mixed with 0.5 mM MgCl2 to form droplets at 75 mM NaCl and 25 mM Tris-HCl pH 7.5. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results. m Condensate area quantification results for each droplet in (l). The data were represented by the scatter dots and mean values (CT, N = 259 droplets; CIP, N = 57 droplets; Denatured CIP, N = 161 droplets). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. n The level of pS510-SSRP1 in FACT complex in vitro. FACT and FACT-S510A were purified from SF9 cells. Purified FACT was further treated with 250 U/mL of CIP or denatured CIP at room temperature for a duration of 30 min. Denatured CIP, heat-inactivated at 85 °C for 10 min. Upper panel, the input of the FACT was shown by Coomassie Brilliant Blue (CBB) staining. Lower panel, phosphorylated level of SSRP1 at S510 detected by Western blotting. The assay was performed in three independent biological replicates with similar results. o SSRP1 condensate number quantification results for each cell after 36 different phosphatases were expressed in SSRP1-GFP overexpression U2OS cells. The data were represented by the mean values for each condition (N = 49–59 cells). p value was determined by unpaired two-sided t-test. p IF results showing the localization and expression level of endogenous SSRP1, SPT16, and Z-NA after PP2AC was knocked down in U2OS cells. Two independent shRNAs were generated to knock down PP2AC. NT non-target control. Scale bar, 10 µm. q SSRP1 condensate number quantification results for each cell in (p). The data were represented by the scatter dots and mean values (NT, N = 53 cells; shPP2AC #1, N = 51 cells; shPP2AC #2, N = 49 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. r Mutations at S510 of SSRP1 disrupted the binding between SSRP1 and MYC. FLAG-tagged SSRP1 was purified by FLAG beads in HEK293T cells. Proteins from input and IP samples were analyzed by Western blotting using the indicated antibodies. The assay was performed in two independent biological replicates with similar results. Source data are provided as a Source Data file. s Overexpression of CK2 increased the binding between FACT and MYC. FLAG-tagged SSRP1 was purified by FLAG beads in HEK293T cells. Proteins from input and IP samples were analyzed by Western blotting using the indicated antibodies. The assay was performed in two independent biological replicates with similar results. Source data are provided as a Source Data file. t Knocking down PP2AC increased the binding between FACT and MYC. FLAG-tagged SSRP1 was purified by FLAG beads in HEK293T cells. Proteins from input and IP samples were analyzed by Western blotting using the indicated antibodies. The assay was performed in two independent biological replicates with similar results. Source data are provided as a Source Data file. u IF results showing the relocation of FACT by MYC. HA-tagged MYC and GFP-tagged SSRP1 or SPT16 were overexpressed in U2OS cells, either separately or together. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results.
CX-4945 is an inhibitor of CK2, which consists of CK2α and CK2β. We then overexpressed the CK2 complex (Supplementary Fig. 6d) and observed that endogenous SSRP1 and SPT16 formed puncta (Fig. 4b, c). In line with this result, Z-DNA was predominantly formed at these puncta. Previous reports showed that CK2 catalyzed the phosphorylation of SSRP1 at S510, S657, and S68856. We then investigated various S-to-A and S-to-E mutations of SSRP1. Our analysis revealed that only the mutation with S510A resulted in a significant reduction in condensate formation (Fig. 4d, e). Considering that the S510E mutation had little effect on condensate formation, the changes in electronic charge resulting from phosphorylation are important for FACT LLPS.
The above findings indicate that CK2-mediated S510 phosphorylation of SSRP1 plays a crucial role in the phase separation of FACT. To strengthen this notion, we generated a specific antibody against the S510 phosphorylation of SSRP1. This antibody recognized the reconstituted phosphorylation mimic of pS510-SSRP1 protein (Fig. 4f, g), but not CIP (calf intestinal alkaline phosphatase)-treated SSRP1 or the S510A mutant of SSRP1 (Fig. 4h). Overexpression of CK2 indeed increased the levels of pS510-SSRP1 (Fig. 4i). Furthermore, the SSRP1-S510A mutation repressed the formation of FACT condensates in vitro (Fig. 4j, k). Consistently, the treatment with CIP, but not with denatured CIP, reduced the formation of FACT condensates and the S510 phosphorylation of SSRP1 (Fig. 4l–n).
After identifying the kinase responsible for the phosphorylation and LLPS of SSRP1, we next aimed to determine the phosphatase catalyzing its dephosphorylation. To this end, we overexpressed a library of 36 different phosphatases and observed their effects on SSRP1 condensates. Among the phosphatases we examined, PP2AC was primarily found to inhibit the formation of SSRP1 condensates (Fig. 4o and Supplementary Fig. 6e–g). Consistently, treatment with the PP2AC inhibitor (LB-100) increased the endogenous FACT puncta (Supplementary Fig. 6h, i). In addition, the depletion of PP2AC enhanced the level of pS510-SSRP1 and condensation of endogenous FACT (Fig. 4p, q and Supplementary Fig. 6j), although the resulting FACT condensates were smaller compared with those observed with CK2 overexpression. Collectively, these data suggest that the CK2-PP2AC axis regulates the S510 phosphorylation of SSRP1, thereby controlling the LLPS of FACT.
Phosphorylation elevates FACT’s recruitment by MYC
After observing that the phosphorylation of SSRP1 influenced its condensation, we sought to understand how phosphorylation impacts its role in nucleosome binding and the induction of Z-DNA. FACT primarily interacts with nucleosomes, which serve as its direct substrate. We found that the interaction between FACT and nucleosomes remained unchanged when SSRP1 carried an S510A mutation (Supplementary Fig. 6k). Additionally, this binding is not affected by CIP treatment of FACT (Supplementary Fig. 6l). Moreover, neither the SSRP1-S510A mutation nor CIP treatment altered the Z-DNA formation induced by FACT and nucleosome binding (Supplementary Fig. 6m, n).
Since the phosphorylation of SSRP1 does not impact its ability to bind nucleosomes or facilitate the formation of Z-DNA, we next explored whether the phosphorylation altered its interaction with MYC. Our results revealed that the SSRP1-S510A mutation decreased the binding between MYC and FACT (Fig. 4r). Furthermore, the binding between FACT and MYC increased when the levels of pS510-SSRP1 were elevated through CK2 overexpression or PP2AC depletion (Fig. 4s, t). We also observed that MYC disrupted the condensate formed by FACT, causing FACT to relocate to the MYC-enriched region (Fig. 4u). These findings indicate that phosphorylation plays a crucial role in regulating FACT condensation and its association with MYC, thereby preparing FACT for recruitment to chromatin by MYC.
Z-NA binding domain of ADAR1 is sufficient to repress MYC-induced Z-DNA formation
Accumulation of Z-DNA or Z-RNA in cells may trigger PANoptosis and immune response activation9,18. We examined whether overexpression of MYC affects the expression of ADAR1 or ZBP1, which are the main proteins recognizing Z-NA in response to its accumulation. Our results indicated that the protein levels of ADAR1 and ZBP1 were not affected by MYC overexpression (Fig. 5a). Moreover, the phosphorylation status of RIPK3, a key downstream event in PANoptosis execution, was also unchanged. The overexpression of either component of the FACT complex showed minimal impact on the protein levels of ADAR1 and ZBP1, as well as on the phosphorylation levels of RIPK3 (Fig. 5b). These findings suggest that while MYC and FACT promote Z-DNA formation, they do not directly induce PANoptosis by Z-DNA.
a Western blotting result showing the indicated protein levels after the FLAG-tagged MYC was overexpressed for 3 days in U2OS cells. Empty vectors were used as negative controls. The assay was performed in three independent biological replicates with similar results. Source data are provided as a Source Data file. b Western blotting result showing the indicated protein levels after FLAG-tagged SSRP1 and FLAG-tagged SPT16 were overexpressed in U2OS cells for 3 days, respectively. Empty vectors were used as negative controls. The assay was performed in three independent biological replicates with similar results. Source data are provided as a Source Data file. c IF results showing the changes of Z-DNA in MYC overexpressing cells by ADAR1 expression. FLAG-tagged MYC and HA-tagged ADAR1, including both wild-type and mutant forms, were overexpressed in U2OS cells. Empty vectors were used as negative controls. Scale bar, 10 µm. d Nuclear Z22 fluorescence intensity quantification results for each cell in (c). The data were represented by the scatter dots and mean values (WT, N = 51 cells; Vector, N = 52 cells; P150, N = 89 cells; P110, N = 60 cells; Zαβ, N = 59 cells; E912A, N = 54 cells; N173A/Y177A/P193A, N = 59 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. e IF results showing the changes of Z-DNA in SSRP1 overexpressing cells by ADAR1 expression. GFP-tagged SSRP1 and HA-tagged ADAR1, including both wild-type and mutant forms, were overexpressed in U2OS cells. Empty vectors were used as negative controls. Scale bar, 10 µm. f Nuclear Z22 fluorescence intensity quantification results for each cell in (e). The data were represented by the scatter dots and mean values WT, N = 67 cells; Vector, N = 66 cells; P150, N = 59 cells; P110, N = 65 cells; Zαβ, N = 78 cells; E912A, N = 65 cells; N173A/Y177A/P193A, N = 56 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. g Nucleocytoplasmic fractionation assay showing the indicated protein levels after FLAG-MYC and HA-ADAR1-Zαβ overexpression in HEK293T cells. Empty vectors were used as negative controls. The assay was performed in three independent biological replicates with similar results. Source data are provided as a Source Data file.
Since ADAR1 is responsible for the removal of Z-NA in cells to prevent programmed cell death induced by ZBP118, we hypothesized that ADAR1, although not activated, could reduce the induced Z-DNA levels. We overexpressed the full-length ADAR1 (ADAR1-P150) and observed that the Z-DNA induced by MYC was largely eliminated (Fig. 5c, d and Supplementary Fig. 7a). In contrast, the ADAR1 isoform (ADAR1-P110) that lacks the Z-DNA binding domain did not have this effect. Similarly, FACT-induced Z-DNA was also abolished by ADAR1-P150, but not by ADAR1-P110 (Fig. 5e, f and Supplementary Fig. 7b, c). We also noticed that ADAR1-P150 overexpression induced Z-NA formation in the cytoplasm, likely because ADAR1-P150 can bind with RNA to induce Z-RNA formation. Furthermore, the chromatin loading of MYC and FACT remained unchanged when ADAR1-P150 was overexpressed (Supplementary Fig. 7d), indicating that the removal of Z-DNA was unlikely due to decreased MYC/FACT recruitment to chromatin.
Based on the above findings, we further explored the minimal functional domains of ADAR1 required to eliminate Z-DNA induced by MYC. We created three mutants: a Z-NA binding domain protein (ADAR1-Zαβ), a deaminase-dead mutant (ADAR1-E912A), and a Zα domain mutant (ADAR1-N173A/Y177A/P193A) to evaluate their effects on Z-DNA elimination. The Z-NA binding domain alone, or the deaminase-dead mutant, was sufficient to suppress the formation of MYC- or FACT-induced Z-DNA without causing Z-NA formation in the cytoplasm (Fig. 5c–f and Supplementary Fig. 7b, c). The absence of Z-NA formation in the cytoplasm is likely due to a reduced ability to bind RNA in these two mutants. Supporting this, the ADAR1-N173A/Y177A/P193A mutant, which cannot bind Z-NA, was unable to suppress the formation of MYC- or FACT-induced Z-DNA. Similar to ADAR1-P150, ADAR1-Zαβ kept the chromatin loading of MYC and FACT unchanged (Fig. 5g). Collectively, these findings indicate that MYC-driven Z-DNA formation is resolved by ADAR1, and the Z-NA binding domain of ADAR1 is sufficient for this resolution.
The formation of Z-DNA induced by MYC is essential for regulating gene expression
Z-DNA formation is typically associated with various cellular processes, including transcription, changes in chromatin structure, and DNA modifications. As a result, it can be challenging to investigate the specific function of Z-DNA without influencing these other factors. Notably, we found that ADAR1-Zαβ specifically repressed the formation of Z-DNA without affecting MYC or FACT. This unique observation allows us to separate the role of Z-DNA formation from other contributing factors within the cell.
To investigate the role of Z-DNA in regulating global gene expression, we conducted transient transcriptome sequencing (TT-seq) analysis on empty-vector overexpressed wild-type (WT) cells, as well as cells with overexpression of MYC, ADAR1-Zαβ, and both MYC and ADAR1-Zαβ. We used an internal spike-in to normalize the sequencing results since MYC, an oncoprotein, can lead to a global increase in gene expression. Indeed, MYC largely enhanced gene expression levels (Fig. 6a). In contrast, cells expressing ADAR1-Zαβ exhibited reduced gene expression compared to WT cells and mitigated the expression changes induced by MYC. Consistent with this, the rates of RNA synthesis were also reduced by ADAR1-Zαβ in both WT and MYC-overexpressing cells (Fig. 6b and Supplementary Fig. 8a).
a The normalized distribution profiles of TT-seq signals spanning 5 kb around TSS showing the average levels in Vector, ADAR1-Zαβ overexpressing, MYC overexpressing, and MYC + ADAR1-Zαβ overexpressing U2OS cells. Sense (+) and antisense (−) transcripts associated with NCBI RefSeq TSS were shown. b Boxplots showing the synthesis rates. The p values were calculated by a two-sided unpaired Wilcoxon rank-sum test. Centerline, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range. Vector, N = 13,464 genes, Zαβ, N = 13,016 genes, MYC, N = 13,256 genes, MYC + Zαβ, N = 13,931 genes. The exact p values were indicated in the figure. Source data are provided as a Source Data file. The normalized distribution of MYC (c), Z22 (d), RNAP II (e), and SSRP1 (f) levels spanning 3 kb around the TSS of genes identified by NCBI RefSeq. The distribution profiles were presented at the top, while the heatmaps were displayed below. MYC, RNAP II, and SSRP1 distributions were detected using CUT&Tag with corresponding antibodies. Z-DNA was detected by Z22 antibodies using ChIP-seq. RRPM reference-adjusted reads per million. g Integrative Genomics Viewer (IGV) tracks presenting TT-seq signals and the enrichment of MYC, Z-DNA, RNAP II, and SSRP1. h Correlations between changes in RNAP II and Z-DNA. Signals were analyzed at RNAP II peaks identified in Vector U2OS cells. FC fold-change. R, the correlation coefficient, was assessed by Pearson product-moment correlation. The p value was calculated by a two-sided paired t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. i Same as in (h), except for changes at RNAP II peaks identified in MYC overexpressing U2OS cells. The p value was calculated by a two-sided paired t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. j IF results showing the localization and expression levels of FLAG-MYC, RNAP II, and Z-DNA after MYC and ADAR1-Zαβ expression in U2OS cells. FLAG-tagged MYC and HA-tagged ADAR1-Zαβ were overexpressed in U2OS cells. Empty vectors were used as negative controls. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results. k IF results showing the localization and expression levels of SSRP1-GFP, RNAP II, and Z-DNA after SSRP1 and ADAR1-Zαβ expression in U2OS cells. GFP-tagged SSRP1 and HA-tagged ADAR1-Zαβ were overexpressed in U2OS cells. Empty vectors were used as negative controls. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results.
To further assess how chromatin was altered, we profiled Z-DNA, MYC, SSRP1, and RNAP II on a genome-wide scale. MYC overexpression enhanced the enrichment of SSRP1, Z-DNA, and RNAP II on chromatin (Fig. 6c–g). ADAR1-Zαβ largely diminished the Z-DNA signal in WT and MYC-overexpression cells, while the levels of SSRP1 and MYC were not down-regulated. More importantly, RNAP II was reduced by ADAR1-Zαβ in WT and MYC-overexpression cells. In MYC-overexpression cells, the ADAR1-Zαβ slightly increased MYC loading at the TSS, suggesting that RNAP II depletion is not due to reduced MYC levels.
The above results indicate that Z-DNA formation facilitates the loading of RNAP II, whereas the depletion of Z-DNA, as mediated by ADAR1-Zαβ, leads to a reduction in RNAP II loading. To further investigate this association, we analyzed the correlation between changes in Z-DNA and RNAP II levels in response to ADAR1-Zαβ expression. We found a positive correlation between these changes in both WT and MYC-overexpressing cells (Fig. 6h, i). In addition, the genes that displayed diminished levels of Z-DNA, a phenomenon attributed to the expression of ADAR1-Zαβ, were associated with a decrease in transient expression (Supplementary Fig. 8b, c). Since the recruitment of SSRP1 may alter the nucleosome integrity to regulate gene expression, we also analyzed the distributions of nucleosomes by MNase-seq. The results showed that ADAR1-Zαβ had little effect on the nucleosome positioning regulated by FACT (Supplementary Fig. 8d).
In light of these findings, we further investigated the regulatory mechanisms by which Z-DNA influences RNAP II with its positional context within cellular environments. We found that RNAP II was enriched at regions associated with MYC/FACT, in conjunction with the presence of Z-DNA (Fig. 6j, k and Supplementary Fig. 8e). Notably, ADAR1-Zαβ was observed to diminish the enrichment of RNAP II at these MYC/FACT regions while concurrently reducing the Z-DNA levels. Collectively, these data suggest that MYC-induced Z-DNA enhances the recruitment of RNAP II to promote gene expression.
Z-DNA is a direct facilitator of RNAP II loading and gene expression
After demonstrating the potential of Z-DNA to enhance gene expression by promoting RNAP II loading, we aimed to investigate whether Z-DNA can directly activate gene expression. To this end, we synthesized a series of Z-DNA-containing super core promoters (Z-SCPs) based on SCPs58. Each promoter contains an upstream TATA box, a downstream TFIID-binding element, a Poly(A) site, and a variable sequence prone to form Z-DNA flanking the transcription start site (TSS). The Z-DNA-forming sequence is characterized by alternating purine and pyrimidine bases (Fig. 7a). These Z-SCPs were effectively expressed in cells when transfected as linear double-stranded DNA (dsDNA) (Supplementary Fig. 9a, b).
a The illustration showing the design of Z-SCPs. b Schematic illustration of the hybridization products between two complementary linear or circular ssDNA. c CD spectra of the Z − B chimera hybrids. The structures of CC and CL Z-SCPs were confirmed by CD spectroscopy. The spectra were recorded from 215 nm to 320 nm. Source data are provided as a Source Data file. d IF results showing the formation of Z-DNA by Z-SCPs in U2OS cells. CC Z-SCPs and CL Z-SCPs were transfected into U2OS cells for 24 h. Scale bar, 10 µm. The assay was performed in three independent biological replicates with similar results. e The normalized read profiles of RNA-seq for transcripts across CC and CL Z-SCPs. f Schematic illustration of the desthiobiotin-modified CC and CL Z-SCPs. g CD spectra of the desthiobiotin-labeled Z − B chimera hybrids. The structures of modified CC and CL Z-SCPs were confirmed by CD spectroscopy. The spectra were recorded from 215 nm to 320 nm. Source data are provided as a Source Data file. In vitro pull-down assay showing the binding between desthiobiotin-modified Z-SCPs #1 (h) and #2 (i) with RNAP II, FACT, MYC, and ADAR1. Proteins were detected with the indicated antibodies by Western blotting (upper panel). Z-SCPs levels were analyzed through a 6% Native-PAGE gel (lower panel) stained with GelRed. The assay was performed in three independent biological replicates with similar results. Source data are provided as a Source Data file. j IF results showing the changes of Z-DNA in Z-SCPs-transfected U2OS cells by ADAR1-P150 expression. Empty vectors were used as negative controls. Scale bar, 10 µm. k Condensate number quantification results for each cell in (j). The data were represented by the scatter dots and mean values (N = 51 cells). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file. l RT-qPCR results showing the expression levels of CC Z-SCPs with or without ADAR1-P150 overexpression in U2OS cells. The mRNA expressions were normalized to β-ACTIN and subsequently adjusted to the levels of the transfected DNA, which was further normalized by the genomic DNA. The value in the corresponding vector-transfected cells was set as 1. The data were represented by the mean ± SD (N = 3 independent replicates). p value was determined by unpaired two-sided t-test. The exact p values were indicated in the figure. Source data are provided as a Source Data file.
Next, we synthesized topologically constrained circular DNA59,60. When two single-stranded DNA (ssDNA) circles (CC) are annealed, the Z-DNA-prone sequences snap into left-handed Z-DNA in one half of the circle, while the other half remains in the right-handed B-DNA form. As controls, we generated flexible B-DNA circles by first annealing a circular ssDNA with a linear one (CL) and then ligating the linear strand (Fig. 7b and Supplementary Fig. 9c). Under physiological conditions, the designed CC Z-SCPs successfully formed a Z-DNA structure, as demonstrated by Circular Dichroism (CD) spectroscopy (Fig. 7c). These CC Z-SCPs exhibited a positive Cotton effect around 270 nm and a negative effect at 290 nm, which are characteristic signatures of Z-DNA10. Furthermore, the CC Z-SCPs, but not the CL Z-SCPs, could be recognized by Z-NA antibodies (Supplementary Fig. 9d). The formation of Z-form DNA in the designed region was further confirmed by sequential digestion with nuclease S1 and restriction enzymes (Supplementary Fig. 9e, f)61. We then transfected these Z-SCPs into cells and observed that only the CC Z-SCPs formed Z-DNA structures, as confirmed by Z-NA antibody recognition (Fig. 7d). More importantly, the CC Z-SCPs induced higher gene expression compared to their corresponding CL Z-SCPs (Supplementary Fig. 9g). The expression level was normalized to the internal β-ACTIN expression and further normalized to the transfected DNA level.
We then conducted RNA-seq to further analyze the expression of the Z-SCPs. Traditional mapping methods typically use a linear sequence as a reference. However, because the Z-SCP sequence is circular and short, using a linearized reference would reduce mapping efficiency at the ends of the sequence. To improve mapping accuracy and overall read distribution across the entire sequence, we generated a reference by duplicating the Z-SCPs. After mapping the reads to this new reference sequence, we merged and normalized the reads to a uniform Z-SCP length, allowing us to assess the distribution of reads across the Z-SCPs more effectively. The expressed RNA was primarily aligned between the designed TSS and poly(A) site. Moreover, the CC Z-SCPs that maintained a Z-DNA conformation induced higher RNA expression levels compared to the CL Z-SCPs (Fig. 7e and Supplementary Fig. 9h). Interestingly, the detected RNA reads were mapped slightly upstream of the designed TSS in both CC and CL Z-SCPs. This observation aligns with previous results that transcription was not initiated at a single fixed TSS but rather from multiple adjacent nucleotide loci in cells62,63.
We also investigated the role of topoisomerases in Z-DNA formation of Z-SCPs. Treatment with TOP1 or TOP2 inhibitors exhibited no detectable impact on the Z-DNA formation of these Z-SCPs (Supplementary Fig. 9i–k), suggesting that these enzymes are dispensable for Z-SCP structural dynamics. The efficacy of the TOP2 inhibitor was evidenced by a notable increase in γ-H2A.X levels. Additionally, the effect of the TOP1 inhibitor was corroborated by both an elevation in γ-H2A.X and a decrease in TOP1 protein levels (Supplementary Fig. 9j). To further investigate the impact of Z-DNA on RNAP II loading, we labeled these Z-SCPs with desthiobiotin (Fig. 7f and Supplementary Fig. 9l). The desthiobiotin-labeled CC (CC-Bio) Z-SCPs effectively formed a Z-DNA structure (Fig. 7g and Supplementary Fig. 9m). These Z-SCPs were then coupled with streptavidin beads to pull down proteins from cell lysates (Supplementary Fig. 9n). The CC-Bio Z-SCPs enriched more RNAP II than the CL-Bio Z-SCPs (Fig. 7h, i). Importantly, we detected an interaction with ADAR1-P150, which demonstrated a higher affinity for CC-Bio Z-SCPs. In contrast, no significant binding was detected between Z-SCPs and FACT, H3, or MYC. To investigate the functional consequences of this interaction, we then overexpressed ADAR1-P150 in CC Z-SCPs-transfected cells, monitoring Z-DNA formation and transcription of Z-SCPs. Consistent with our previous findings (Supplementary Fig. 7a), the overexpression of ADAR1-P150 induced cytoplasmic Z-NA formation (Fig. 7j). In parallel, the Z-NA antibody indicated a substantial reduction in CC Z-SCPs foci due to the overexpression of ADAR1-P150 (Fig. 7j, k). Consequently, the expression of the four tested CC-ZCPs was repressed following ADAR1-P150 overexpression (Fig. 7l and Supplementary Fig. 9o). Together, these data suggest that Z-DNA directly facilitates RNAP II loading, thereby increasing gene expression.
Discussion
Our study provides a comprehensive exploration of the interplay between the MYC protein, FACT complex, and Z-DNA formation, revealing a nuanced mechanism of transcriptional regulation mediated by DNA dynamics. Our findings reveal how MYC collaborates with chromatin remodeling to induce Z-DNA formation, facilitating the recruitment of RNAP II and promoting gene expression. This offers new insight into the active role of Z-DNA in transcriptional regulation.
The induction of Z-DNA by MYC underscores its critical role in transcriptional activation. Previous studies have shown that MYC overexpression is associated with transcriptional upregulation32,33. Our work extends these findings by demonstrating that the effect of MYC overexpression on transcriptional upregulation relies on Z-DNA formation mediated by FACT. This dependency suggests a synergistic relationship where MYC recruits FACT to chromatin, facilitating chromatin remodeling and subsequent Z-DNA formation. Importantly, our observations indicate that Z-DNA formations can persist independently of RNAP II activity, challenging the conventional view that Z-DNA is just a byproduct of transcription. Our genome-wide analyses demonstrate that Z-DNA facilitates the recruitment of RNAP II, thereby promoting transcriptional activation. In addition, we demonstrate the intrinsic capacity of Z-DNA to directly elevate gene expression through engineered Z-SCPs. With minor modifications, these Z-SCPs can be utilized to examine the direct effects of Z-DNA on any chromatin-binding factors. These results unequivocally establish Z-DNA as a functional inducer of RNAP II loading and transcriptional initiation, providing direct evidence of its role beyond its structural properties. Our data highlight Z-DNA as an active regulatory structure, not merely a passive response to transcriptional tension.
FACT primarily functions in transcription activation by alleviating nucleosomal barriers49,64. During the transcription elongation phase, FACT interacts with RNAP II, helping to destabilize nucleosomes ahead of the transcription machinery while preserving histone integrity. This process reduces polymerase stalling, allowing for the efficient elongation of mRNA transcripts. Moreover, FACT has been implicated in releasing paused RNAP II65,66. By destabilizing nucleosomes located near promoter-proximal regions, FACT facilitates the transition of RNAP II into productive elongation67. Interestingly, FACT also plays a role in transcription repression by stabilizing nucleosomes68. It maintains chromatin architecture, which leads to RNAP II pausing during transcription. FACT has been reported as a sensor of DNA torsional stress to recognize Z-DNA52. In that study, CBL0137 was predominantly utilized as the FACT inhibitor to demonstrate its role in repressing Z-DNA formation. However, a subsequent study demonstrated that CBL0137, as a small molecule, can be directly intercalated into B-form DNA, thereby inducing the transition to Z-form DNA17. Our observations indicate that FACT induces the formation of Z-DNA to facilitate RNAP II loading. In the previous report69, FACT failed to interact with the nucleosome fully wrapped with DNA. The assay is conducted in a magnesium-free environment. However, we performed the EMSA assay in the presence of magnesium. Magnesium facilitates the preferential interaction and assembly of identical nucleosome species54. Furthermore, magnesium may induce structural changes within the nucleosomes70. These magnesium-induced changes in the nucleosome could enhance the association of the FACT complex with the wrapped nucleosome. These findings provide a fresh perspective on the role of FACT and help clarify its dual and seemingly paradoxical function in both the activation and repression of transcription.
Two major models exist in the literature regarding FACT-mediated nucleosome reorganization48. The “dimer eviction model” proposes that the FACT complex, through its histone chaperone activity, actively displaces a single H2A/H2B dimer from a nucleosome, thereby enhancing DNA accessibility for transcriptional machinery49,50. Conversely, the “global accessibility/non-eviction model” posits that H2A/H2B dimer displacement induced by FACT is a nonessential byproduct of its action and is not crucial for nucleosome reorganization71,72. Critically, our findings indicate that, in the presence of magnesium, FACT can engage nucleosomes fully wrapped with DNA. This supports the hypothesis that FACT actively displaces a single H2A/H2B dimer from a nucleosome. Furthermore, FACT may tether the evicted H2A/H2B dimer to the hexasome structure before reinserting it to restore a complete nucleosome. The eviction and reinsertion process may induce DNA-binding and bending torsion, leading to Z-DNA formation. Under this condition, the presence of H2A/H2B is critical for the induction of Z-DNA by FACT.
It is important to note that in vitro structural studies utilizing hexasome as the starting material do not accurately reflect the dynamics of fully wrapped nucleosomes bound by the FACT complex45. In such structures, the FACT subunit SPT16 anchors to nucleosomal DNA and tethers H2A/H2B, while SSRP1 also contributes to DNA binding and can transition between two conformations depending on the presence of a second H2A/H2B dimer. This observation aligns with our results, suggesting that these protein complex structures likely represent the intermediate states of FACT binding to nucleosomes during H2A/H2B eviction. Future investigations focusing on the structural and conformational changes occurring in the presence of nucleosomes and the FACT complex may elucidate the detailed mechanisms underlying this process and how Z-DNA formation is induced.
Our study also shows that the phosphorylation, regulated by the CK2-PP2AC pair, controls FACT LLPS. These FACT condensates are critical for the dynamic localization of FACT within the nucleus and its interaction with MYC. However, the phosphorylation status of SSRP1 and the formation of LLPS are not necessary for Z-DNA formation. This separation of FACT condensation from its Z-DNA induction function offers insight into the complex roles of FACT in transcriptional regulation. As a key chromatin remodeler, FACT regulates nucleosome structure and gene transcription. The loading of FACT onto chromatin is a vital step in activating its function, and this process must be tightly controlled. Once phosphorylated, FACT is more likely to form condensates, which increase the local concentration of FACT, facilitating MYC binding and the relocation of FACT to the chromatin. The observation that phosphorylation acts as a molecular switch governing FACT condensate assembly and MYC dynamics, while MYC binding appears to disrupt the FACT condensate, can be attributed to the increased local concentration of SSRP1/SPT16 facilitated by FACT condensation. This heightened concentration likely enables MYC to readily engage with available SSRP1/SPT16 and subsequently relocate them to the MYC-binding loci.
PP2AC is a highly conserved protein found across various species, playing a crucial role in numerous cellular processes, including signal transduction, cell proliferation, and neuronal function. Dysregulation of PP2AC has been implicated in several types of cancer and neurodegenerative diseases. Previous research has shown that PP2AC affects gene expression by targeting multiple signaling pathways73. Moreover, PP2AC directly modulates transcription by dephosphorylating the C-terminal domain (CTD) of RNAP II74. In this context, PP2AC is part of a complex known as the integrator-PP2A complex (INTAC), which operates as a noncanonical holoenzyme. Moreover, different phosphatases also regulate the dephosphorylation of RNAP II CTD to fine-tune and control gene transcription. This raises an intriguing question: how might transcription-associated phosphatases participate in regulating FACT?
Our research shows that ADAR1 eliminates Z-DNA formed by MYC and FACT, thereby decreasing Z-DNA accumulation. The observation that the ADAR1’s Z-NA binding domain is sufficient for this resolution highlights a precise approach for regulating Z-DNA levels without disrupting other chromatin-associated processes. Furthermore, MYC and FACT levels on chromatin remain unchanged by the Z-NA binding domain of ADAR1. This indicates that ADAR1 targets Z-DNA specifically, rather than the upstream processes responsible for its formation. Since Z-DNA is induced by various factors, ADAR1’s role in specifically repressing Z-DNA is crucial for distinguishing the function of Z-DNA from a cause-and-consequence paradox. Our data, along with several previous reports18,75,76, reveal that ADAR1 suppresses the formation of the Z-form structure in the nucleic acid base pairs. ADAR1 deaminates RNA, disrupting the double-stranded structures formed when RNA molecules anneal or form self-hairpins, consequently resolving the Z-form RNA structure. Although the resolution of Z-RNA by ADAR1 is well understood, its role in resolving Z-DNA is less clear. It is possible that ADAR1 recruits additional components, such as helicases, to help unwind DNA to facilitate this process. The ADAR1-P150 protein is located in the cytoplasm and is initially transported into the nucleus, subsequently re-exported via a nuclear export signal (NES) near the N-terminal region. This process, together with the nuclear localization signal (NLS) possessing nuclear import activity, facilitates the shuttling characteristics of the ADAR1 protein77. We did not observe a significant accumulation of ADAR1 within the nuclei, likely due to the translocation of ADAR1 into the nuclei to eliminate Z-DNA and subsequently return to the cytoplasm following Z-DNA clearance.
The detection of Z-DNA involves various methods to recognize Z-form nucleotides in cells. Antibodies are one approach to identifying Z-DNA40. However, the particular monoclonal antibody used for this purpose was raised against a GC-rich sequence, which may lead to biased recognition favoring GC-rich sequences. Furthermore, under normal physiological conditions, the amount of Z-DNA detected by immunofluorescence is low, likely due to the low basal level of Z-DNA or the limited affinity of the Z22 antibody for Z-DNA. Additionally, Z-NA reader proteins, such as the Zα domain of ZBP1 or ADAR1, have also been employed8. These domains can induce Z-NA formation in vitro, despite their protein structures potentially leading to biased recognition of particular base pairs. In the future, developing new probes for the specific detection of Z-form structures will be essential.
The inhibition of ADAR1 triggers PANoptosis in cells, which activates an immune response that helps kill cancer cells41,78,79. As a result, there is significant research dedicated to developing ADAR1 inhibitors. Shortly, we can expect a variety of these inhibitors to be available. In this context, our work would provide new insight to explore additional potential applications for regulating gene expression and to conduct a comprehensive evaluation of the functions of these inhibitors.
Collectively, our findings highlight Z-DNA as a pivotal factor in transcriptional regulation, orchestrating the interplay between chromatin remodeling and RNAP II dynamics. This work not only advances our understanding of Z-DNA biology but also emphasizes its potential as a therapeutic target in MYC-driven cancers, where aberrant transcriptional activity and chromatin dysregulation are hallmarks.
Methods
Cell culture and cell lines
U2OS, HeLa, Astrocyte, and HEK293T cells were cultured in DMEM (Thermo Fisher Scientific, Cat. #C11965500BT) supplemented with 1% GlutaMax, 10% FBS, and 1% antibiotic solution (penicillin/streptomycin). HGC27 cells were cultured in RPMI 1640 (Thermo Fisher Scientific, Cat. #C11875500BT) supplemented with 1% GlutaMax, 20% FBS, and 1% antibiotic solution (penicillin/streptomycin). Human small cell lung cancer lines (H526, H82 and H211) and mouse SCLC lines (RPS1, RPP-mTmG and RPP-GSDME-KO) were cultured in RPMI 1640 (Thermo Fisher Scientific, Cat. #C11875500BT) supplemented with 1% GlutaMax, 10% FBS, and 1% antibiotic solution (penicillin/streptomycin). All mouse SCLC lines were derived from a p53, Rb, and Pten triple-knockout background, in which “RPP” denotes Rb, p53, and Pten. All cells were cultured at 37 °C with 5% CO2.
Knockdown cell lines were generated by shRNAs, which were constructed into pLKO.1 vector and listed in Supplementary Table 1.
Immunofluorescence for adherent cells (U2OS, HeLa, Astrocyte, HEK293T and HGC27)
Immunofluorescence was performed as described previously80. Cells were seeded onto coverslips in a 24-well plate. After 24 h, cells were washed with 1 ml PBS twice and then fixed with 500 μL of 4% PFA for 10 mins. After being washed with 1 ml PBS twice, cells were permeabilized with 0.5% Triton X-100 solution (20 mM HEPES pH 7.5, 50 mM NaCl, 3 mM MgCl2, and 0.3 M sucrose) with 5% BSA (Sangon Biotech, Cat. #A602440) for 1 h at room temperature. Cells were washed with PBS twice. Primary antibodies were diluted at 1:100 in PBS-T (1×PBS with 0.1% Tween 20) to be incubated with cells overnight at 4 °C. After being washed with 1 ml PBS-T three times, cells were incubated with Alexa Fluor 488/594/647-conjugated antibodies (1:1000 dilution in PBS-T) for 1 h in the dark. Cells were then washed with 1 ml PBS-T 3 times in the dark. DAPI (Invitrogen, Cat. #D1306) was diluted at 1:1000 in PBS and added to cells for 5 min in the dark. After being washed with PBS twice in the dark, slides with antifade reagents (Biosharp, Cat. #BL701A) were sealed with mounting medium and observed under a fluorescence microscope (Zeiss, LSM 880/LSM 900).
Immunofluorescence for suspension cells (SCLC lines)
A total of 2 × 105 suspension cells were resuspended in 50 μL PBS and carefully applied onto glass coverslips pre-coated with 100 μg/mL poly-L-lysine (incubated for 10 min at room temperature, rinsed with sterile water, and air-dried) to promote adherence. After allowing cells to attach for 20–30 min at 37 °C, coverslips were gently washed with PBS. Cells were then fixed with 4% PFA for 30 min at room temperature. After being washed with 1 mL PBS twice, cells were permeabilized with 0.5% Triton X-100 solution (20 mM HEPES pH 7.5, 50 mM NaCl, 3 mM MgCl2, and 0.3 M sucrose) with 5% BSA (Sangon Biotech, Cat. #A602440) for 1 h at room temperature. Cells were washed with PBS twice. Primary antibodies were diluted at 1:100 in PBS-T (1×PBS with 0.1% Tween 20) to be incubated with cells overnight at 4 °C. After being washed with 1 mL PBS-T three times, cells were incubated with Alexa Fluor 488/594-conjugated antibodies (1:1000 dilution in PBS-T) for 1 h in the dark. Cells were then washed with 1 mL PBS-T three times in the dark. DAPI (Invitrogen, Cat. #D1306) was diluted at 1:1000 in PBS and added to cells for 5 min in the dark. After being washed with PBS twice in the dark, slides with antifade reagents (Biosharp, Cat. #BL701A) were sealed with mounting medium and observed under a fluorescence microscope (Zeiss, LSM 900).
Nucleases treatment
U2OS cells expressing HA-tagged MYC, GFP-tagged SSRP1, and SPT16 for 3 days were fixed with 500 μL of 4% PFA for 10 min. After being washed with 1 ml PBS twice, cells were permeabilized with 0.5% Triton X-100 solution (20 mM HEPES pH 7.5, 50 mM NaCl, 3 mM MgCl2, and 0.3 M sucrose) for 1 h at room temperature. Cells were washed with PBS twice and then treated with 0.2 U/μL DNase I, 1 mg/mL RNase A, and 0.2 U/μL RNase H, respectively, at 37 °C for 3 h. After two washes with PBS, cells were blocked with PBST containing 5% BSA for 1 h, washed twice again with PBS, and then incubated with primary antibodies to continue immunofluorescence analysis. Cells were stained with Acridine Orange (AO, 5 µg/mL) (Absin, Cat. #abs9735) for 10 min at room temperature after DAPI staining.
FRAP analysis
FRAP experiments were carried out with a Zeiss LSM 880 microscope. U2OS cells were seeded into a 4-chamber 35 mm glass-bottom dish (Cellvis, Cat. #D35C4-20-1-N). After 24 h, the droplets of SSRP1 or SPT16 protein were photobleached by the 100% laser intensity of 488 nm (for GFP) or 594 nm (for mCherry). The recovery process was recorded at the indicated time point.
Live cell imaging
Live cell images were acquired continuously on an Olympus IX83 spinning disk confocal microscope. U2OS cells were transfected with SSRP1-GFP or SPT16-GFP, and 2 days later, they were seeded into a 4-chamber 35 mm glass-bottom dish (Cellvis, Cat. #D35C4-20-1-N). The cells were observed 1 day after seeding. The droplets of SSRP1 or SPT16 proteins were imaged using a 2% laser intensity at 488 nm, and captured for 24 h with 1-min intervals at 37 °C in a 5% CO2 atmosphere.
In vivo immunoprecipitation
Immunoprecipitation was performed as described previously81. HEK293T cells were lysed in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, and protease inhibitor). The lysates were dounced with 50 passages followed by rotation at 4 °C for 0.5 h. Samples were centrifuged at 12,000 × g for 15 min at 4 °C. 5% of the supernatants were taken as input and boiled with SDS loading buffer.
For pull-down of overexpressed FLAG/HA-tagged proteins, pre-washed FLAG-Seflnose Resin beads (Sigma, Cat. #M8823)/HA-Seflnose Resin beads (Sigma, Cat. # SAE0197) were added to the supernatants and rotated at 4 °C overnight; For pull-down of endogenous MYC protein, rabbit monoclonal anti-MYC antibody was incubated with the supernatants at 4 °C for 3 h, followed by the addition of pre-washed protein A beads and overnight rotation at 4 °C.
The samples were then washed with washing buffer (50 mM HEPES pH 7.4, 100 mM NaCl, 0.01% NP-40, 10% glycerol, and 1 mM EDTA) 5 times before being boiled for Western blotting.
Genetic encoding of phosphoserine into SSRP1
The site-specific incorporation of phosphoserine into SSRP1 was achieved using the genetic code expansion system. Specifically, the SSRP1-S510 codon (AGT) was mutated to an amber stop codon (TAG). The resulting SSRP1-S510TAG sequence was cloned into a pET28a vector containing a C-terminal His-tag. This construct was then co-transformed with the pEVOL-SEP plasmid, which carries the SEP tRNA and SEP synthetase sequences, into the engineered E. coli ΔC321.ΔSerB strain. This strain, generously provided by Dr. Shixian Lin, was previously modified by knocking out the SerB gene from the C321.ΔA.exp strain (Addgene #49018). When cultured to an optical density at 600 nm (OD600) of 0.6, arabinose (final concentration of 0.2%) and glucose (final concentration of 0.08%) were added with the corresponding pSer. Cells were induced for protein expression by adding 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 22 °C. After 16 h, cells were lysed and sonicated in 20 ml lysis buffer (50 mM Tris–HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole pH 8.0, 5% glycerol, and 0.5 mM NaF). The lysates were bound to NI-NTA Seflnose Resin beads (Sangon Biotech, Cat. #C600033) at 4 °C for 4 h. After extensive washing, the beads-bound proteins were eluted by 250 mM imidazole and dialyzed to storage buffer (50 mM Tris pH 7.5 and 150 mM NaCl) before usage.
FACT purification
The wild-type and mutant SSRP1 and SPT16 were cloned into the His-tagged pFast-Bac vector. The plasmids were packaged into viruses and transfected into SF9 cells together. After 60 h, 300 mL of cells were collected and lysed by lysis buffer (50 mM Tris-HCl pH 7.5, 500 mM NaCl, and 10% glycerol) and sonication. The cell lysates were bound with NI-NTA Seflnose Resin beads (Sangon Biotech, Cat. #C600033) at 4 °C for 4 h. The beads were washed extensively and further eluted with 250 mM imidazole. The purified proteins were dialyzed into storage buffer (25 mM HEPES pH 7.5 and 150 mM NaCl) before use.
Calf intestinal phosphatase (CIP) treatment
In vitro purified and in vivo immunoprecipitated (IPed) FACT complex was treated with 250 U/mL of CIP or denatured CIP at a temperature of 25 °C for a duration of 30 min. The denatured CIP was produced by incubating CIP at 85 °C for 10 min.
Reconstitution of the octamer and nucleosome
Histone extraction was performed as previously described82. pRSFDuet1-hH3.3/H4 and pRSFDuet1-hH2A/H2B, which were kind gifts from Dr. Ruiming Xu, were used. For the nucleosome assembly, the ‘601’ 167 bp DNA and histone were titrated at a molar ratio of 0.9:1, and mixed in initiation buffer (2 M NaCl, 10 mM Tris pH 7.5, 1 mM EDTA, and 1 mM DTT). The mixture was stepwise dialyzed to the final buffer (0.2 M NaCl, 10 mM Tris, pH 7.5, 1 mM EDTA, and 1 mM DTT) at 4 °C for 36 h. The final mixture included nucleosomes and a small proportion of free DNA. The fractions containing mononucleosomes were collected by gel filtration and further loaded into centrifugal filters for purification and concentration.
In vitro pull-down assay
To identify the binding affinity of FACT and nucleosome, FACT, FACT-S510A, or FACT treated with CIP/denatured CIP was mixed with nucleosome or H3/H4 wrapped with DNA, and incubated at 4 °C for 3 h in a rotisserie-style tube rotator. SSRP1 antibody was then added to the mixture, followed by incubation at 4 °C overnight. Next, protein G beads were added to the binding system and incubated by rotating at 4 °C for 1 h. Finally, the beads were extensively washed with TBS and boiled with SDS loading buffer before analysis.
To analyze the binding affinity of circle DNA and RNAP II, the biotin-modified CC or CL Z-SCPs were mixed into fresh cell lysis from HEK293T cells and incubated at 4 °C overnight. The cell lysis was obtained by RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, and protease inhibitor) and 50 passages of douncing. Pre-washed streptavidin beads (Smart-lifesciences, Cat. #SM007002) were then added to the mixture, followed by incubation at 4 °C for 1 h. The beads were then extensively washed with TBS three times. The purified DNA was extracted by boiling beads with 95% formamide, and the purified protein was extracted by boiling beads with SDS loading buffer.
EMSA (electrophoretic mobility shift assay)
EMSA was performed as previously described83. In brief, to induce the Z-form of nucleosome by FACT, FACT-S510A or FACT treated with CIP/denatured CIP was mixed with 1 μM nucleosome at a molar ratio of 3:1 (nucleosome:FACT) in 20 μL of reaction buffer (10 mM HEPES pH 7.5 and 10 mM MgCl2). After incubation on ice for 1 h, the sample was incubated with Z22 antibody at a molar ratio of 3:1 (nucleosome:Z22) in 30 ul of reaction buffer at room temperature for 2.5 h. Ten μL of each mixture was run on a 6% Native-PAGE gel and the gel was stained with GelRed to indicate the presents of DNA. The interaction of H3/H4 wrapped with DNA was done with the same process.
To determine the DNA structure of CC/CL Z-SCPs, CC/CL Z-SCPs were mixed with Z22 antibody at a molar ratio of 3:1 (DNA:Z22) in a total volume of 20 μL, followed by incubation at room temperature for 2.5 h. 10 μL of each mixture was run on a 6% Native-PAGE gel and stained with GelRed to indicate the presence of DNA.
Nucleocytoplasmic fractionation
HEK293T cells were lysed in Hypo buffer (20 mM HEPES pH 8.0, 5 mM KCl, 1.5 mM MgCl2, 0.1 mM DTT, and proteinase inhibitor) with RNase A and incubated at 37 °C for 1 h. The lysates were then centrifuged at 12,000 × g for 30 min at 4 °C. The supernatants were collected as cytoplasmic fractions and boiled with SDS loading buffer for Western blotting. The pellets were washed with hypo buffer once and boiled with SDS loading buffer as the nuclear fraction.
Quantitative reverse transcription PCR
Total RNA was extracted with TRIzol reagent (Sangon Biotech, Cat. #B610409) and reversely transcribed into cDNA by HiScript II Q RT SuperMix Kit according to the manufacturer’s instructions (Vazyme, Cat. #R222-01). The cDNA was proceeded for quantitative PCR with SYBR Green (Yeasen, Cat. #11201ES03) by using Quantagene q225 qPCR system (Kubo Technology, Beijing). β-ACTIN was used as a control to normalize the expression of target genes. The 2-(Δ-Δcontrol) method was used to represent the relative gene expression. Primers used for qPCR were listed in Supplementary Table 1.
Synthesis of single-stranded DNA circles
The single-stranded Z-SCP DNA sequences, either modified with biotin or not, were synthesized by Sangon Biotech. Sequences were listed in Supplementary Table 2. The DNA circle synthesis was performed as previously described59. In brief, a phosphate group was enzymatically introduced to the 5’-position of single-stranded DNA substrate with the T4 polynucleotide kinase (Thermo Scientific, Cat. #EK0031). The phosphorylation system (50 μL), containing DNA substrate (20 μM), ATP (1.6 mM), and T4 polynucleotide kinase (30 U) in 1 × Reaction Buffer A, was incubated at 37 °C for 8 h. The reaction was terminated by incubating the mixture at 75 °C for 10 min. Then the cyclization of ssDNA was carried out using the T4 DNA Ligase (Thermo Scientific, Cat. #EL0011). The cyclization system contained the splint DNA (8 μM) and T4 DNA ligase (75 U) in 120 μL of 0.2 × T4 DNA ligase buffer. Phosphorylated single-stranded linear DNA substrate (120 μL, 4 μM in 0.1 × T4 DNA ligase buffer) was divided into 10 portions and added into the cyclization system every 20 mins at 25 °C in steps, followed by incubation at 25 °C for 12 h. The reaction was terminated by another 10 min incubation at 65 °C. Exonuclease I (New England Biolabs, Cat. # M0293V) was used to remove the remaining substrate DNA and linear byproducts at 37 °C for 2 h. The final DNA circle samples were purified with 10% denaturing polyacrylamide gel containing 8 M urea (1×TBE buffer containing 89 mM Tris-borate and 2 mM EDTA) dissolved in the buffer of 0.1 × TE (1 mM Tris-HCl and 0.1 mM EDTA pH 8.0).
Preparation of CC Z-SCP and CL Z-SCP
The synthesis was performed as previously described59. In short, for CC Z-SCP, the single-stranded DNA circle (C-F) and its complementary circle (C-R) were mixed to a final concentration of 4 μM in 10 mM HEPES pH 7.5 and 10 mM MgCl2 buffer, and annealed. The annealing steps were as follows: 90 °C for 5 min, cooling to 60 °C at a rate of 0.1 °C s−1, 60 °C for 10 min, cooling to 20 °C with a rate of 0.1 °C s−1, 20 °C for 10 min.
For CL Z-SCP, the circle C-F (4 μM) and its complementary linear L-R (4 μM) in 1 × T4 DNA ligase buffer were firstly annealed from 90 °C to 20 °C as described above. The ligation was achieved by incubation at 25 °C for 12 h with T4 DNA ligase followed by termination at 65 °C for 10 min. The final products were treated with Exonuclease I at 37 °C for 2 h followed by phenol/chloroform/isoamyl alcohol (25:24:1 (v/v/v), pH 7.8) purification. Samples were dissolved in 10 mM HEPES pH 7.5 and 10 mM MgCl2 before usage.
Circular dichroism (CD) spectroscopy
The formation of stable Z-DNA in the chimera hybrids was confirmed by CD spectroscopy (Applied Photophysics Ltd, LHA19100029). The spectra were recorded from 215 nm to 320 nm.
Digestion of the CC Z-SCP chimera hybrids
Nuclease S1 has been reported to recognize the junction between Z-DNA and contiguous B-DNA84,85. The digestion was performed as previously described59. The CC chimera hybrid (0.2 μM) was treated with nuclease S1 (9 U) (Thermo Scientific, Cat. # EN0321) at 37 °C for 35 min, which was terminated by adding 1 μL EDTA (0.5 M, pH 8.0) and incubating at 75 °C for 10 min. The products were analyzed with 8% Native polyacrylamide gel. To confirm the locations of junctions between B-DNA and Z-DNA, the linear DNA products were then digested by BsaAI and Hpy188I and separated by a 20% Native polyacrylamide gel.
ChIP-seq
ChIP-seq was performed as previously described82. In brief, U2OS cells for ChIP-seq were crosslinked with 1% paraformaldehyde for 10 min and then quenched in 125 mM glycine for 5 min at room temperature with rotation. After being washed with cold PBS, cells were lysed in lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, and 0.5% NP-40) on ice for 10 min. The nuclei were spun down and then resuspended in 500 µL of 1× ChIP-seq buffer (50 mM Tris-HCl pH 8.1, 10 mM EDTA, 100 mM NaCl, 1% Triton X-100, and 0.1% sodium deoxycholate). Sonication was performed for 5 cycles of 30 s on and 30 s off in the Bioruptor. After centrifugation at 15,000 × g at 4 °C for 10 min, the supernatant was used for chromatin immunoprecipitation. The same amounts of Drosophila S2 cell chromatin were spiked in for normalization. Ten µg of Z22 antibodies were added to each sample and rocked at 4 °C overnight. Protein A beads were added and incubated for 3 h. After the beads were extensively washed, the bound DNA was eluted with elution buffer (10 mM Tris-HCl pH 8.0, 10 mM EDTA, 150 mM NaCl, 5 mM DTT, and 1% SDS). Reverse crosslinking was performed at 65 °C overnight. The eluted DNA was treated with proteinase K and RNase A, and then purified with the Min-Elute PCR purification kit (Qiagen, Cat. #28006). Samples extracted were subjected to library preparation with VAHTS Universal DNA Library Prep Kit for Illumina V3 (Vazyme, Cat. #ND607-1) following the manufacturer’s instructions.
MNase-seq
MNase-seq was performed as previously described86. In brief, 4 × 106 U2OS cells were collected and lysed in lysis buffer (10 mM Tris-HCl pH 7.5, 10 mM NaCl, and 0.5% NP-40) on ice for 10 min. The nuclei were spun down and then digested in 500 µL MNase digestion buffer (20 mM Tris-HCl pH 7.5, 15 mM NaCl, 60 mM KCl, and 1 mM CaCl2) with 1000 units of MNase (NEB, Cat. #M0247S) by incubating at 37 °C for 20 min. The reaction was stopped by adding 2X STOP buffer (100 mM Tris-HCl pH 8.1, 20 mM EDTA, 200 mM NaCl, 2% Triton X-100, and 0.2% sodium deoxycholate). After being centrifuged at 15,000 × g at 4 °C for 10 min, the supernatant was collected and added with the same amounts of Drosophila S2 cell chromatin as spike-in for normalization. The sample was then treated with proteinase K and RNase A, and purified with the Min-Elute PCR purification kit (Qiagen, Cat. #28006). Extracted DNA was subjected to library preparation with VAHTS Universal DNA Library Prep Kit for Illumina V3 (Vazyme, Cat. #ND607-1) following the manufacturer’s instructions.
CUT&Tag
CUT&Tag was performed as previously described80. In brief, 105 U2OS cells were harvested in NE buffer (20 mM HEPES pH 7.5, 0.5 mM spermidine, 10 mM KCl, 0.1% TritonX-100, 10% Glycerol, and 1 mM PMSF) and incubated on ice for 10 mins. ConA beads were pre-washed and resuspended in binding buffer (20 mM HEPES pH 7.5, 10 mM KCl, 1 mM CaCl2, and 1 mM MnCl2). Ten µL of beads were added to each sample and incubated at room temperature for 10 min. The beads were washed with washing buffer (20 mM HEPES pH 7.5, 0.5 mM spermidine, 150 mM NaCl, and 0.1% BSA) and resuspended in blocking buffer (20 mM HEPES pH 7.5, 0.5 mM spermidine, 150 mM NaCl, 0.1% BSA, and 2 mM EDTA) at room temperature for 5 min. Primary MYC, RNAP II, and SSRP1 antibodies were added at a 1:100 dilution and incubated at room temperature for 2 h, respectively. After being washed with washing buffer, secondary antibodies were added at a 1:100 dilution and incubated at room temperature for 30 min. A total of 1.2 µL of PA-Tn5 transposons was added to each sample and incubated at room temperature for 30 min. Beads were resuspended in 30 µL of washing buffer with 10 mM MgCl2 and incubated at 37 °C for 1 h. Reactions were stopped by adding 5.5 µL of stop buffer (2.25 µL of 0.5 M EDTA, 2.75 µL of 10% SDS, and 0.5 µL of 20 mg/ml Proteinase K) and incubated at 55 °C for 30 min, followed by 70 °C for 20 min to inactivate Proteinase K. 0.9× of VAHTS DNA Clean Beads (VAHTS, Cat. #N411-03) were added to each sample to extract the tagmentated DNA.
RNA-seq
For MYC and ADAR1 overexpression RNA-seq, total RNA was extracted with TRIzol reagent from U2OS cells (Sangon Biotech, Cat. #B610409). A total of 500 ng RNA was reverse transcribed into cDNA by Maxima H Minus (Thermo Fisher, Cat. #EP0751). cDNA was digested by 2 µL of Tn5 transposons in 20 µL of 1×DMF buffer at 37 °C for 5 min. Reactions were stopped by adding 5.5 µL of stop buffer (2.25 µL of 0.5 M EDTA, 2.75 µL of 10% SDS, and 0.5 µL of 20 mg/ml Proteinase K) and incubated at 55 °C for 30 min, followed by 70 °C for 20 min to inactivate Proteinase K. 0.9× of VAHTS DNA clean beads (VAHTS, Cat. #N411-03) were added to each sample to extract the tagmentated DNA.
For RNA-seq of circular DNA-transfected U2OS cells, total RNA was extracted with TRIzol reagent (Sangon Biotech, Cat. #B610409) and subjected to library preparation with RNA-seq library prep kit (Abclonal Cat. # RK20306) following the manufacturer’s instructions.
TT-seq (transient transcriptome sequencing)
TT-seq was performed as previously described87. In brief, 15-cm-dish U2OS cells were cultured in medium containing 1 mM 4-SU (Aladdin, Cat. #T122953) for 15 min. Cells were collected for RNA extraction by TRIzol reagent (Sangon Biotech, Cat. #B610409). A total of 100 µg of 4-SU labeled RNA and 1 µg of Drosophila S2 4-SU labeled RNA were mixed in a total volume of 100 µL RNase-free water. A total of 20 µL of 1 M NaOH was added to the mixture and incubated on ice for 20 min followed by RNA purification using Phenol/Chloroform pH 6.7. Then, 3 μL of biotin buffer (1 M Tris-HCl pH 7.4 and 0.5 M EDTA) and 50 μL of 0.1 mg/ml Biotin MTSEA (Biotium, Cat. #90064-90066-1) were added to 100 µL of purified RNA. After another Phenol/Chloroform pH 6.7 purification, RNA was reconstituted in 50 μL of RNase-free water and incubated at 65 °C for 10 min followed by rapid cooling on ice for 5 min. Pre-washed streptavidin beads (Smart-lifesciences, Cat. #SM007002) were added to the supernatants and rotated at room temperature for 1 h in the dark. The samples were then washed with high salt washing buffer (100 mM Tris HCl pH 7.4, 1 M NaCl, 0.1% Tween 20, and 10 mM EDTA) 5 times before being eluted twice with 100 μL of 100 mM DTT (Sangon Biotech, Cat. #A620058). The eluted RNA was purified by EtOH precipitation and subjected to library preparation with the RNA-seq library prep kit (Abclonal Cat. #RK20306) following the manufacturer’s instructions.
Antibodies
Rabbit polyclonal anti-Histone H3 (Abcam, Cat. #ab1791); Rabbit polyclonal anti-FLAG Tag (Sangon Biotech, Cat. #D110005); Mouse monoclonal anti-β-Tubulin (Cell Signaling Technology, Cat. #86298); Mouse monoclonal anti-HA Tag (Abcam, Cat. #ab18181); Rabbit polyclonal anti-c-MYC (Abcam, Cat. #ab32072); Mouse monoclonal anti-POLR2 (EMD Millipore, Cat. #05623); Rabbit monoclonal anti-SPT16 (Cell Signaling Technology, Cat. #12191); Rabbit monoclonal anti-SSRP1 (Abclonal, Cat. #13636); Mouse monoclonal anti-SSRP1 (Santa Cruz, Cat. #sc-56781); Rabbit monoclonal anti-MAX (Cell Signaling Technology, Cat. # 17471); Mouse monoclonal anti-ADAR1 (Santa Cruz, Cat. #sc-271854); Mouse monoclonal anti-ZBP1 (Santa Cruz, Cat. #sc-271483); Rabbit polyclonal anti-Histone H2B (Abcam, Cat. #ab177430); Rabbit monoclonal anti-PP2AC (Cell Signaling Technology, Cat. #2259); Mouse monoclonal anti-S9.6 antibody (Kerafast, Cat. #ENH001); Rabbit monoclonal anti-γ-Histone H2A.X antibody (Huabio, Cat. #ET1602-2); Rabbit monoclonal anti-RIPK3 antibody (Abcam, Cat. #ab195117). Rabbit monoclonal anti-RIPK3-P antibody (Sigma, Cat. # HPA055087). Rabbit monoclonal anti-top1 antibody (Cell Signaling Technology, Cat. # 79971). Peroxidase AffiniPure Goat anti-Rabbit IgG (H + L) (Peroxidase AffiniPure Goat anti-Rabbit IgG (H + L), Cat. #111-035-003); Peroxidase AffiniPure Goat anti-mouse IgG (H + L) (Peroxidase AffiniPure Goat anti-Rabbit IgG (H + L), Cat. #115-035-003); Alexa Fluor® 647 AffiniPure™ Rabbit Anti-Goat IgG (H + L) (Jackson ImmunoResearch Laboratories, Cat. #305-605-003); Alexa Fluor® 488 AffiniPure™ Rabbit Anti-Goat IgG (H + L) (Jackson ImmunoResearch Laboratories, Cat. #305-545-003); Alexa Fluor® 594 AffiniPure™ Mouse Anti-Goat IgG (H + L) (Jackson ImmunoResearch Laboratories, Cat. #205-585-108). Alexa Fluor® 647 AffiniPure™ Goat Anti-Human IgG (H + L) (Jackson ImmunoResearch Laboratories, Cat. #109-605-088). Alexa Fluor® 594 AffiniPure™ Goat Anti-Human IgG (H + L) (Jackson ImmunoResearch Laboratories, Cat. #109-585-088). Alexa Fluor® 488 AffiniPure™ Goat Anti-Human IgG (H + L) (Jackson ImmunoResearch Laboratories, Cat. #109-545-003).
CUT&Tag analysis
Raw reads were trimmed to remove adapters and low-quality sequences using Trim Galore (version 0.6.6) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with the parameters ‘-q 20 --paired’. Then trimmed reads were uniquely mapped to the human reference genome (hg19) merged with D. melanogaster genome (dm6) using Bowtie288 (version 2.4.2). The output SAM files were converted to sorted BAM files using SAMtools89 (version 1.11). Genome coverage bigwig files were generated by BEDtools90 (version 2.29.2) genomecov and bedGraphToBigWig (version 4) (https://www.encodeproject.org/software/bedgraphtobigwig/) with the parameter ‘-scale 1,000,000/ (the number of reads mapped to dm6 genome)’. Heatmaps were generated by deepTools91 (version 3.5.0) computeMatrix and plotHeatmap. A 1 Kb sliding window across the whole genome was used to calculate the Pearson product moment correlation (Supplementary Table 3).
ChIP-seq analysis
Raw reads were trimmed to remove adapters and low-quality sequences using Trim Galore with the parameters ‘-q 20 --paired’. Then trimmed reads were uniquely mapped to the human reference genome (hg19) merged with D. melanogaster genome (dm6) using Bowtie2. The output SAM files were converted to sorted BAM files using SAMtools. PCR duplicates were removed using MarkDuplicates tool of Picard (version 2.23.3) (https://broadinstitute.github.io/picard/) with the parameter ‘--REMOVE DUPLICATES = true’. Genome coverage bigwig files were generated by BEDtools genomecov and bedGraphToBigWig with the parameter ‘-scale 1,000,000/ (the number of reads mapped to dm6 genome)’. Heatmaps were generated by deepTools computeMatrix and plotHeatmap. A 1 Kb sliding window across the whole genome was used to calculate the Pearson product moment correlation (Supplementary Table 3).
RNA-seq analysis
Raw reads were trimmed to remove adapters and low-quality sequences using Trim Galore with the parameters ‘-q 20 --paired’. Then trimmed reads were uniquely mapped to the human reference genome (hg19) merged with D. melanogaster genome (dm6) using STAR mapper92 (version 2.7.7a). Tag directories were generated using HOMER93 (version 4.11.1) makeTagDirectory, and expression matrices were counted by analyzeRepeats.pl with the parameter ‘-raw’, and further scaled by the reads of dm6.
RNA synthesis rates
RNA synthesis rates were estimated based on a method described in a previous study94. In this approach, a statistical model was applied to the read counts (kij) of gene i in sample j, where the sample could be TT-Seq or RNA-Seq (total or fragmented). The model considers both labeled (αi) and unlabeled (βi) RNA levels for each gene, alongside a gene length parameter (Li). To adjust for variations in sequencing depth, scaling factors (σj) were incorporated, and the cross-contamination rate (ϵj) represents the proportion of unlabeled reads in the TT-Seq sample. The expected value for kij was modeled as follows:
For RNA samples from total cellular material, ϵj was set to 1. Estimations of sequencing depth (σj) and cross-contamination rate (ϵj) were derived from spike-in data, where unlabeled spike-ins had αi = 0 and βi = 1, and labeled spike-ins had αi = 1 and βi = 0. Maximum likelihood estimation was used to fit the model, assuming a negative binomial distribution, with dispersion parameters calculated by DESeq295. Once the sequencing depth (σj) and cross-contamination rate (ϵj) were estimated, the model was applied to all Transcription Units to obtain the labeled and unlabeled RNA amounts (αi and βi). These quantities were then converted into synthesis and degradation rates (μi, λi) using first-order kinetics, as outlined, with the following equations96:
Where t = 15 mins. And therefore:
MNase-seq analysis
Raw reads were trimmed to remove adapters and low-quality sequences using Trim Galore with the parameters ‘-q 20 --paired’. Then, trimmed reads were uniquely mapped to the human reference genome (hg19) using the Bowtie2 mapper. The output SAM files were converted to sorted BAM files using SAMtools. PCR duplicates were removed using the MarkDuplicates tool of Picard with the parameter ‘--REMOVE DUPLICATES = true’. Mononucleosomal DNA fragments (141–180 nt) were extracted for further analysis. Genome coverage bigwig files were generated by deepTools bamCoverage with the parameter ‘--MNase --binSize 1 –smoothLength 40 --scaleFactor 10,000,000/ (the number of reads within ±1 kb of the TSS)’. Aggregation plots were generated by deepTools computeMatrix and plotProfile. A 1 Kb sliding window across the whole genome was used to calculate the Pearson product moment correlation (Supplementary Table 3).
TT-seq analysis
Raw reads were trimmed to remove adapters and low-quality sequences using Trim Galore with the parameters ‘-q 20 --paired’. Then trimmed reads were uniquely mapped to the human reference genome (hg19) merged with D. melanogaster genome (dm6) using STAR mapper. Sense reads were obtained using the parameters ‘-f 128 -F 16’ and ‘-f 64 -f 16’ with samtools, while antisense reads were obtained using the parameters ‘-f 128 -f 16’ and ‘-f 64 -F 16’. Genome coverage bigwig files were generated by BEDtools genomecov and bedGraphToBigWig with the parameter ‘-split -scale 1,000,000/ (the number of reads mapped to dm6 genome)’. To assess transcripts around the transcription start site (TSS), tag directories were generated using HOMER makeTagDirectory with the parameter ‘-sspe’, and the profiles were created using annotatePeaks.pl tss with the parameter ‘-raw’, and further scaled by the reads of dm6. A 1 Kb sliding window across the whole genome was used to calculate the Pearson product moment correlation (Supplementary Table 3).
Z-SCP RNA-seq analysis
Raw reads were trimmed to remove adapters and low-quality sequences using Trim Galore with the parameters ‘-q 20 --paired’. Then trimmed reads were uniquely mapped to the human reference genome (hg19) merged with duplicated Z-SCPs sequences and D. melanogaster genome (dm6) using STAR mapper. Genome coverage bigwig files were generated by BEDtools genomecov and bedGraphToBigWig with the parameter ‘-split -scale 1,000,000/ (the number of reads mapped to dm6 genome)’. The signals on duplicated Z-SCP sequences were merged to a uniform Z-SCP length.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The reference genome used in this study is available in the UCSC Genome Browser Database (http://genome.ucsc.edu) under human reference genome hg19. The raw and processed sequencing data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database. MNase-seq datasets are available through GSE290655. TT-seq datasets are available through GSE290656. CUT&Tag datasets are available through GSE290657. Z-SCP RNA-seq datasets are available through GSE290659. RNA-seq datasets are available through GSE290661. ChIP-seq datasets are available through GSE290662. Source data are provided with this paper.
References
Hubner, M. R. & Spector, D. L. Chromatin dynamics. Annu. Rev. Biophys. 39, 471–489 (2010).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Makova, K. D. & Weissensteiner, M. H. Noncanonical DNA structures are drivers of genome evolution. Trends Genet. 39, 109–124 (2023).
Obara, P., Wolski, P. & Panczyk, T. Insights into the molecular structure, stability, and biological significance of non-canonical DNA forms, with a focus on G-Quadruplexes and i-Motifs. Molecules 29, 4683 (2024).
Madhanagopal, B. R. et al. The unusual structural properties and potential biological relevance of switchback DNA. Nat. Commun. 15, 6636 (2024).
Wang, A. H. et al. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680–686 (1979).
Liu, H., Mulholland, N., Fu, H. & Zhao, K. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol. Cell Biol. 26, 2550–2559 (2006).
Shin, S. I. et al. Z-DNA-forming sites identified by ChIP-Seq are associated with actively transcribed regions in the human genome. DNA Res. 23, 477–486 (2016).
Roy, R., Chakraborty, P., Chatterjee, A. & Sarkar, J. Comparative review on left-handed Z-DNA. Front. Biosci. 26, 29–35 (2021).
Drew, H. R., Dickerson, R. E. & Itakura, K. A salt-induced conformational change in crystals of the synthetic DNA tetramer d(CpGpCpG). J. Mol. Biol. 125, 535–543 (1978).
Herbert, A. Z-DNA and Z-RNA in human disease. Commun. Biol. 2, 7 (2019).
Schwartz, T., Rould, M. A., Lowenhaupt, K., Herbert, A. & Rich, A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284, 1841–1845 (1999).
Schwartz, T., Behlke, J., Lowenhaupt, K., Heinemann, U. & Rich, A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 8, 761–765 (2001).
Ha, S. C., Lowenhaupt, K., Rich, A., Kim, Y. G. & Kim, K. K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature 437, 1183–1186 (2005).
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007).
Zhang, T. et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180, 1115–1129.e1113 (2020).
Zhang, T. et al. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature 606, 594–602 (2022).
Karki, R. & Kanneganti, T. D. ADAR1 and ZBP1 in innate immunity, cell death, and disease. Trends Immunol. 44, 201–216 (2023).
Wang, G. & Vasquez, K. M. Dynamic alternative DNA structures in biology and disease. Nat. Rev. Genet. 24, 211–234 (2023).
Evan, G. I. et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69, 119–128 (1992).
Wittig, B., Wolfl, S., Dorbic, T., Vahrson, W. & Rich, A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 11, 4653–4663 (1992).
Eberhardy, S. R. & Farnham, P. J. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276, 48562–48571 (2001).
Baluapuri, A., Wolf, E. & Eilers, M. Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol. 21, 255–267 (2020).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Conacci-Sorrell, M., McFerrin, L. & Eisenman, R. N. An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 4, a014357 (2014).
Patange, S. et al. MYC amplifies gene expression through global changes in transcription factor dynamics. Cell Rep. 38, 110292 (2022).
Das, S. K., Lewis, B. A. & Levens, D. MYC: a complex problem. Trends Cell Biol. 33, 235–246 (2023).
Endres, T. et al. Ubiquitylation of MYC couples transcription elongation with double-strand break repair at active promoters. Mol. Cell 81, 830–844.e813 (2021).
Herold, S. et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature 567, 545–549 (2019).
Guccione, E. et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 8, 764–770 (2006).
Kieffer-Kwon, K. R. et al. Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol. Cell 67, 566–578.e510 (2017).
Rahl, P. B. et al. c-Myc regulates transcriptional pause release. Cell 141, 432–445 (2010).
Lin, C. Y. et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012).
Kalkat, M. et al. MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol. Cell 72, 836–848.e837 (2018).
See, Y. X., Chen, K. & Fullwood, M. J. MYC overexpression leads to increased chromatin interactions at super-enhancers and MYC binding sites. Genome Res. 32, 629–642 (2022).
Nie, Z. et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151, 68–79 (2012).
Walz, S. et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014).
Carter, D. R. et al. Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci. Transl. Med. 7, 312ra176 (2015).
Solvie, D. et al. MYC multimers shield stalled replication forks from RNA polymerase. Nature 612, 148–155 (2022).
Brigido, M. M. & Stollar, B. D. Two induced anti-Z-DNA monoclonal antibodies use VH gene segments related to those of anti-DNA autoantibodies. J. Immunol. 146, 2005–2009 (1991).
Fang, Y., Bansal, K., Mostafavi, S., Benoist, C. & Mathis, D. AIRE relies on Z-DNA to flag gene targets for thymic T cell tolerization. Nature 628, 400–407 (2024).
Baranello, L., Levens, D., Gupta, A. & Kouzine, F. The importance of being supercoiled: how DNA mechanics regulate dynamic processes. Biochim. Biophys. Acta 1819, 632–638 (2012).
Patel, H. P. et al. DNA supercoiling restricts the transcriptional bursting of neighboring eukaryotic genes. Mol. Cell 83, 1573–1587.e1578 (2023).
Clapier, C. R. & Cairns, B. R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304 (2009).
Liu, Y. et al. FACT caught in the act of manipulating the nucleosome. Nature 577, 426–431 (2020).
Prendergast, L. et al. The CENP-T/-W complex is a binding partner of the histone chaperone FACT. Genes Dev. 30, 1313–1326 (2016).
Safina, A. et al. Complex mutual regulation of facilitates chromatin transcription (FACT) subunits on both mRNA and protein levels in human cells. Cell Cycle 12, 2423–2434 (2013).
Winkler, D. D. & Luger, K. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286, 18369–18374 (2011).
Belotserkovskaya, R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003).
Orphanides, G., Wu, W.-H., Lane, W. S., Hampsey, M. & Reinberg, D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288 (1999).
Kemble, D. J., McCullough, L. L., Whitby, F. G., Formosa, T. & Hill, C. P. FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell 60, 294–306 (2015).
Safina, A. et al. FACT is a sensor of DNA torsional stress in eukaryotic cells. Nucleic Acids Res. 45, 1925–1945 (2017).
Ma, J. et al. Phosphorylation of H3.3 at Serine 31 acts as a switch of nucleosome dynamics for transcription. Nucleic Acids Res. 53, gkaf891 (2025).
Nishikawa, J. -I. & Ohyama, T. Selective association between nucleosomes with identical DNA sequences. Nucleic Acids Res. 41, 1544–1554 (2012).
Heo, K. et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell 30, 86–97 (2008).
Tsunaka, Y. et al. Phosphorylated intrinsically disordered region of FACT masks its nucleosomal DNA binding elements. J. Biol. Chem. 284, 24610–24621 (2009).
Han, J. et al. Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev. 24, 1485–1490 (2010).
Juven-Gershon, T., Cheng, S. & Kadonaga, J. T. Rational design of a super core promoter that enhances gene expression. Nat. Methods 3, 917–922 (2006).
Zhang, Y. et al. Topologically constrained formation of stable Z-DNA from normal sequence under physiological conditions. J. Am. Chem. Soc. 141, 7758–7764 (2019).
An, R. et al. Highly efficient preparation of single-stranded DNA rings by T4 ligase at abnormally low Mg(II) concentration. Nucleic Acids Res. 45, e139 (2017).
Kouzine, F. et al. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 4, 344–356.e347 (2017).
Zeller, K. I. et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. USA 103, 17834–17839 (2006).
Leenen, F. A. et al. Where does transcription start? 5’-RACE adapted to next-generation sequencing. Nucleic Acids Res. 44, 2628–2645 (2016).
Formosa, T. & Winston, F. The role of FACT in managing chromatin: disruption, assembly, or repair? Nucleic Acids Res. 48, 11929–11941 (2020).
Hsieh, F. K. et al. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. USA 110, 7654–7659 (2013).
Chang, H. W. et al. Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins. Sci. Adv. 4, eaav2131 (2018).
Mason, P. B. & Struhl, K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell Biol. 23, 8323–8333 (2003).
Zumer, K. et al. FACT maintains chromatin architecture and thereby stimulates RNA polymerase II pausing during transcription in vivo. Mol. Cell 84, 2053–2069.e2059 (2024).
Tsunaka, Y., Fujiwara, Y., Oyama, T., Hirose, S. & Morikawa, K. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686 (2016).
Ohyama, T. New aspects of magnesium function: a key regulator in nucleosome self-assembly, chromatin folding and phase separation. Int. J. Mol. Sci. 20, 4232 (2019).
Xin, H. et al. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 35, 365–376 (2009).
Formosa, T. FACT and the reorganized nucleosome. Mol. Biosyst. 4, 1085–1093 (2008).
Johnson, H., Narayan, S. & Sharma, A. K. Altering phosphorylation in cancer through PP2A modifiers. Cancer Cell Int. 24, 11 (2024).
Zheng, H. et al. Identification of integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science 370, eabb5872 (2020).
Li, H. et al. Human genomic Z-DNA segments probed by the Z alpha domain of ADAR1. Nucleic Acids Res. 37, 2737–2746 (2009).
Nichols, P. J. et al. Recognition of non-CpG repeats in Alu and ribosomal RNAs by the Z-RNA binding domain of ADAR1 induces A-Z junctions. Nat. Commun. 12, 793 (2021).
Nie, Y., Zhao, Q., Su, Y. & Yang, J.-H. Subcellular distribution of ADAR1 isoforms is synergistically determined by three nuclear discrimination signals and a regulatory motif. J. Biol. Chem. 279, 13249–13255 (2004).
Kuriakose, T. & Kanneganti, T. D. ZBP1: innate sensor regulating cell death and inflammation. Trends Immunol. 39, 123–134 (2018).
Herbert, A. To “Z” or not to “Z”: Z-RNA, self-recognition, and the MDA5 helicase. PLoS Genet. 17, e1009513 (2021).
Jia, J. et al. FUS reads histone H3K36me3 to regulate alternative polyadenylation. Nucleic Acids Res. 52, 5549–5571 (2024).
Wan, X. et al. MAT2B regulates the protein level of MAT2A to preserve RNA N6-methyladenosine. Cell Death Dis. 15, 714 (2024).
Zhang, Y. et al. SMYD5 catalyzes histone H3 lysine 36 trimethylation at promoters. Nat. Commun. 13, 3190 (2022).
Liu, Y. et al. Cryo-EM structure of SETD2/Set2 methyltransferase bound to a nucleosome containing oncohistone mutations. Cell Discov. 7, 32 (2021).
Singleton, C. K., Kilpatrick, M. W. & Wells, R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J. Biol. Chem. 259, 1963–1967 (1984).
Singleton, C. K., Klysik, J. & Wells, R. D. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc. Natl. Acad. Sci. USA 80, 2447–2451 (1983).
Carone, B. R. MNase-seq to identify genome-wide DNA-protein interactions. Methods Mol. Biol. 2866, 99–109 (2025).
Gregersen, L. H., Mitter, R. & Svejstrup, J. Q. Using TT(chem)-seq for profiling nascent transcription and measuring transcript elongation. Nat. Protoc. 15, 604–627 (2020).
Langdon, W. B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 8, 1 (2015).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Quinlan, A. R. BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinforma. 47, 11 12 11–11 12 34 (2014).
Ramirez, F., Dundar, F., Diehl, S., Gruning, B. A. & Manke, T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Schwalb, B. et al. TT-seq maps the human transient transcriptome. Science 352, 1225–1228 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Miller, C. et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol. Syst. Biol. 7, 458 (2011).
Acknowledgements
We thank Hangjun Wu in the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University, for his technical assistance with the computing node setup. We are grateful to our colleagues at the core facility of the Life Sciences Institute for their assistance with computing node setup. We thank Dr. Jun Huang at the Life Sciences Institute mentor program for the discussion in preparing this manuscript. We thank Dr. Xinhua Feng for sharing the plasmids of phosphatases. We thank Dr. Deli Huang for preparing the Z22 antibodies. We thank Dr. Jing Huang for sharing the FACT proteins. This research was partly supported by grants from the National Natural Science Foundation of China to D.F. (Grant Nos. 32222017, 32361133547, 32570644, and 32370613), the National Key R&D Program of China to D.F. (No. 2022YFA1302800), and the Zhejiang Provincial Natural Science Foundation of China under Grant No. LZ24C060001 to D.F.
Author information
Authors and Affiliations
Contributions
X.W. and D.F. conceived the project. X.W., Z.L., Y.D., W.Z., Z.S., J.S., J.J., and Y.Z. performed the experiments. H.F. and Y.L. performed the bioinformatics analysis. X.W. and D.F. analyzed the data. X.W., H.F., Z.L., Y.D., and D.F. prepared the first draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Katerina Gurova, Ken Itoh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wan, X., Fan, H., Li, Z. et al. MYC drives left-handed Z-DNA formation to shape gene expression. Nat Commun 17, 199 (2026). https://doi.org/10.1038/s41467-025-66886-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66886-3