Abstract
Inflammation triggers systemic growth restrictions, a process well characterised in tumour cachexia. Whether inflammatory tissue damage also induces growth restrictions, and how regenerating tissue overcome them, is less explored. Using a tissue damage model in Drosophila, we identify metabolic and signaling adaptations that both induce and bypass systemic growth restrictions. Expression of eiger, the Drosophila TNF-α homolog, in imaginal discs causes systemic insulin restriction and insulin resistance, reducing protein translation and proliferation in peripheral tissues. Regenerating cells overcome this by upregulating Pdk1, which is necessary and sufficient to promote protein translation via an Insulin/Akt-independent mechanism. JAK/STAT acts upstream to elevate Pdk1, defining a JAK/STAT-Pdk1-S6K axis essential for regenerative proliferation. Regenerating cells also upregulate amino acid transporters and rely on mTORC1. Similar signatures in RasV12, scrib tumors indicate that tumors co-opt these pathways to sustain growth under insulin restriction. This physiological program thus integrates systemic nutrient mobilization and local metabolic reprogramming, with implications for tissue repair but also pathologies, such as chronic wounds and cancer.
Similar content being viewed by others
Introduction
Tissue damage and inflammation trigger a dynamic interplay between cellular signals and cell behaviours to promote repair and regeneration. Proper orchestration of these responses is crucial, as failure can lead to chronic wound healing pathologies or diseases like cancer1,2,3,4,5. To better understand these diseases, a wide range of recent studies aim to dissect the pathological reprogramming of relevant metabolic circuits6,7,8. However, a surprising gap exists in our knowledge about the precise metabolic adaptations employed by the normal physiological programmes of tissue repair and regeneration1,9,10,11. With this study, we aim to provide insight into the local and systemic metabolic adaptations during physiological tissue repair and regeneration.
Central to both metabolism and cellular growth are the Insulin/PI3K/Akt and mTORC1 signalling pathways, which are evolutionarily conserved from invertebrates to vertebrates. Both pathways converge on their shared effector ribosomal protein S6 kinase (S6K), which drives protein synthesis and cellular growth by activating protein translation. The Insulin/PI3K/Akt signalling branch is activated by binding of Insulin to its receptor, stimulating Phosphoinositide-3-kinase (PI3K) to produce Phosphatidylinositol (3,4,5)-trisphosphate (PIP3). PIP3 recruits Phosphoinositide-dependent kinase 1 (Pdk1) and activates Akt, which inhibits nuclear translocation of the transcription factor FOXO. Pdk1 phosphorylates S6K, initiating its activation, whereas optimal S6K activity requires an additional phosphorylation by mTORC112,13,14,15,16. mTORC1 specifically responds to amino acid availability and is therefore central to anabolic growth17. While previous studies implicate a role for Insulin/PI3K/Akt and mTORC1 signalling in tissue repair processes, the precise metabolic adaptations remain to be investigated18,19,20,21.
Drosophila models have advanced our understanding of tissue repair, regeneration and metabolism22,23. Specifically, studies in developing imaginal discs or the adult gut have highlighted the role of two key signalling pathways - JNK/AP1 and JAK/STAT - in repair and proliferation. These pathways coordinate a range of responses, from senescent-like cell cycle arrest in damaged cells to compensatory proliferation in adjacent cells3,20,24,25,26,27. Importantly, arrested, JNK-signalling cells produce Unpaired (Upd) cytokines, which activate JAK/STAT signalling in surrounding cells at the site of inflammatory damage, promoting survival and rapid regenerative proliferation25,26,28,29,30,31,32,33,34. The distinct functional demands of senescent and rapidly proliferating cells raise the question about how these distinct cell populations metabolically adapt to successfully support tissue repair.
Tissue repair and tumour development share striking similarities; in fact, tumours have long been compared to non-healing wounds35. Accordingly, Drosophila tumour models activate JNK/AP1 and JAK/STAT signalling, which promote tumour progression30,36,37. To support their growth, tumours secrete signalling molecules that initiate inter-organ signalling and systemic metabolic responses38,39,40. For instance, the Insulin-like peptide 8 (Dilp8), when secreted by tumours, disrupts hormone balance by reducing Ecdysone and Insulin production through direct effects on the ring gland and Insulin-producing cells (IPCs), with the effect of halting developmental progression of the tumour-bearing host41,42. Other cytokines, such as Ecdysone-inducible gene L2 (ImpL2), or the TNFα homologue Eiger (Egr), directly or indirectly trigger lipolysis and proteolysis to promote nutrient release from muscles as well as the fat body, an adipose tissue central for nutrient storage and energy homoeostasis40,43,44,45,46,47,48,49,50. Amino acids or sugars are released in this manner and subsequently absorbed by tumours, and facilitate their anabolic growth51,52,53. This inter-organ signalling network and metabolic state resembles cachexia, a clinical syndrome characterised by weight loss, muscle atrophy and fatigue, typically observed in chronic inflammatory conditions, including cancer38,39,40,54,55. While these studies reveal oncogenic metabolic signalling networks, the metabolic signalling networks employed during physiological tissue repair and regeneration remain poorly understood. Previous studies demonstrate that systemically acting cytokines may also be released upon tissue damage in the absence of oncogenic transformation41,42,56,57,58,59, and fat body break-down changing Methionine, S-adenosylmethionine, and Kynurenine metabolism promotes imaginal disc regeneration60,61. However, the integration of local and systemic metabolic adaptations promoting physiological repair and regeneration remains poorly characterised. In our study, we combine genetic analysis, quantitative imaging, untargeted metabolomics and information from single-cell RNA sequencing data to outline the local and systemic adaptations that selectively support fast-proliferating cells during regeneration through an Insulin-independent JAK/STAT-Pdk1-S6K signalling axis.
Results
Regenerating cells maintain high levels of translation
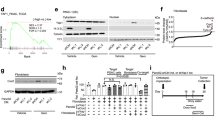
To induce regeneration, we expressed the Drosophila homologue of TNF-α, known as Eiger (Egr), for a 24 h period within the imaginal wing pouch using the rn-GAL4 driver62. As expected, egr-expression caused significant cell death, accompanied by the activation of the JNK/AP-1 activity reporter TRE-RFP (Fig. 1A–C and Supplementary Fig. S1A, B)24,26,28,63. The central disc region with high JNK signalling exhibited markers of cellular senescence, including increased senescence-associated β-galactosidase activity (Supplementary Fig. S1C, D), upregulation of the matrix metalloprotease MMP-1 and cytokines of the Upd family, as well as a JNK-signalling induced cell cycle arrest in G224,25,26,32,64,65. In contrast, cells within 40 µm surrounding this central JNK signalling domain were highly proliferative, as detected by EdU incorporation (Fig. 1A, B, D)29,62. To facilitate quantification of cell behaviours and account for disc size variation, we defined a conservative 20 µm band outside the high JNK signalling domain as the ‘proliferative domain’ (PDegr) for the remainder of this study.
A, B Control (A) and genetically ablated wing disc (B) after 24 h of egr-expression in the wing pouch (see Fig. 1C) under control of the rn-GAL4 driver (rnts and rnts > egr). TRE-RFP visualises JNK-pathway activity (magenta/grey), EdU visualises DNA replication (cyan/grey), and DAPI visualises nuclei. C Schematic of a third instar wing imaginal disc with pouch, notum, and hinge (left) and after 24 h of egr-expression in the pouch (right). Magenta identifies the JNK-signalling domain representing inflammatory damage. D Quantification of mean EdU intensity in the DAPI area contained within 10 µm bands segmented inward and outward from the edge of the high JNK domain in egr-expressing discs. Magenta = high JNK-signalling domain, cyan = domain of proliferative regeneration. Mean and 95% CI (confidence interval) shown, One-way ANOVA followed by Tukey’s post-hoc test for multiple comparisons (n = 7 discs). P-values: Band 1 vs Band 5 = 0.0001; Band 5 vs Band 8 = 0.0004; Band 1 vs Band 8 = 0.9984 (ns). E, F Protein synthesis visualised by O-proparyl-puromycin (OPP) incorporation into newly synthesised proteins in control wing disc (E) and egr-expressing disc (F). G Mean OPP intensity in three different regions of control (pouch, hinge, and notum) and egr-expressing discs (high JNK signalling domain - approximating the central pouch, the proliferating region - approximating the peripheral pouch and hinge, and the notum. Mean and 95% CI are shown. One-way ANOVA followed by Tukey’s post-hoc test for multiple comparisons (control: n = 8, egr-expressing disc: n = 8). P-values: Proliferative region vs Quiescent notum < 0.0001; Notum vs Quiescent notum < 0.0001; Pouch vs JNK domain = 0.9992 (ns). H, I OPP visualises protein synthesis in nota of control (H) and egr-expressing discs (I). J, K EdU visualise DNA replication in nota of control (J) and egr-expressing discs (K). L EdU area per DAPI area in the notum of control and egr-expressing discs, approximating percentage cells undergoing DNA replication. Mean and 95% CI, two-tailed Unpaired t test (control: n = 9, egr-expressing disc: n = 8), with a p-value = 0.0001. M Mean EdU intensity per EdU-positive area in the notum of control and egr-expressing discs, approximating relative DNA replication speed. Mean and 95% CI, two-tailed Unpaired t-test (control: n = 9, egr-expressing disc: n = 8), with a p-value = 0.0011. N Schematic of protein synthesis rates (blue shades) and mitotic activity (black dots) in control (left) and egr-expressing discs (right). Scale bars: 100 μm. and DAPI visualises nuclei. Fluorescence intensities reported as arbitrary units. Source data in graphs are provided as a Source Data file.
How do these proliferating cells meet their metabolic needs? To answer this question, we monitored protein translation using O-propargyl-puromycin (OPP)-incorporation assays66. This assay revealed uniform levels of protein synthesis throughout control wing imaginal discs. In egr-expressing discs, protein synthesis in the proliferative and the JNK signalling domains proceeded at levels similar to control discs (Fig. 1E–G). Interestingly, the notum of egr-expressing discs exhibited a significant decrease in OPP incorporation, which correlated with a marked reduction in EdU incorporation (Fig. 1G–M). The contrasting levels of protein synthesis, proliferation and signalling between different regions reveal that inflammatory damage induces at least three distinct cell populations with different cellular programmes: (1) a senescent cell population exhibiting high protein synthesis and JNK/AP-1 activity; (2) an cycling cell population exhibiting high protein synthesis and low JNK/AP-1 activation; and (3) notum cells exhibiting low protein synthesis, low JNK/AP-1 activity and slow cell cycling (Fig. 1N). While these observations mirror proliferation dynamics visualised in earlier studies33,62,67, we wanted to better understand how these differences reflect a need to integrate local and systemic metabolic demands during regeneration.
Tissue damage induces systemic insulin restriction
A reduction in protein translation rates in peripheral tissue domains like the notum suggested that insulin signalling, normally supporting protein translation through S6K activation, is reduced. Notably, cells with active JNK signalling express high levels of Dilp8, a known antagonist of insulin production by insulin-producing cells (IPCs) in the larval brain (Fig. 2A, B)42,68. Expression of eiger could thus limit anabolic growth despite nutrient intake by feeding43,44,46,59,68,69,70. To determine if egr-expressing larvae indeed restrict Insulin expression, we analysed the expression of the Drosophila Insulin-like peptides dILP2 and dILP571. We found that dILP2 and dILP5 expression was significantly reduced in egr-expressing larvae, and approached levels seen in larvae starved for 24 h (Fig. 2C), a condition known to reduce insulin expression due nutrient limitation71,72.
A, B Dilp8-GFP (cyan) in control (A) and egr-expressing discs (B). TRE-RFP visualises JNK-pathway activity (magenta). dFOXO-GFP visualised using BDSC 38644. C Relative dilp2 and dilp5 mRNA in control, egr-expressing, and starved male larvae. Egr-expression was induced for 24 h in the wing pouch using rn-GAL4, while starved larvae underwent 24 h starvation. Mean and 95% CI, Kruskal-Wallis test followed by Dunn’s multiple comparison (control larvae: n = 13, egr-expressing larvae: n = 11, starved larvae: n = 11). P-values for dilp2: Control vs egr = 0.0075; Control vs Starved < 0.0001; egr vs Starved = 0.1138 (ns). P-values for dilp5: Control vs egr = 0.0440; Control vs Starved < 0.0001; egr vs Starved = 0.0073. D, E dFOXO-GFP (cyan/grey, BDSC: 38644) in notum of control (D) and egr-expressing wing discs (E). DAPI (magenta/grey) visualises nuclei. Nuclear dFOXO indicates low insulin signalling. D’ and E’ show magnified boxed regions. F Quantification of mean nuclear dFOXO-GFP intensity in notum of control (D) and egr-expressing wing discs (E). Mean and 95% CI, two-tailed Unpaired t test, p-value = 0.0427 (control: n = 6, egr-expressing disc: n = 8). G, H OPP incorporation visualised protein synthesis in eye-antennal disc, dissected from larvae with control (G) or egr-expressing (H) wing discs using salm-GAL4 expressed in the central pouch. I, J EdU incorporation visualises DNA replication in eye-antennal discs, dissected from larvae with control (I) or egr-expressing (J) wing discs using salm-GAL4 driver. The yellow line outlines the disc. K, L OPP incorporation visualised protein synthesis in the leg disc, dissected from larvae with control (K) or egr-expressing (L) wing discs using salm-GAL4. M, N EdU incorporation visualises DNA replication in leg imaginal discs, dissected from larvae with control (M) or egr-expressing (N) wing discs using salm-GAL4. The yellow line outlines the disc. O, P OPP incorporation visualised protein synthesis in the eye disc, dissected from larvae with control (O) or egr-expressing (P) wing discs using rn-GAL4. Q Mean OPP intensity in eye discs, dissected from larvae with control or egr-expressing wing discs. Mean and 95% CI, two-tailed Unpaired t test, p-value = 0.0002 (control: n = 5, egr-expressing disc: n = 5). R, S EdU visualises DNA replication (magenta) in eye discs, dissected from larvae with control (R) or egr-expressing (S) wing discs using rn-GAL4. Photoreceptors are marked by Elav (cyan). T Mean EdU intensity per EdU-positive area (relative DNA replication speed), in eye discs dissected from larvae with control or egr-expressing wing discs. Mean and 95% CI, two-tailed Unpaired t test, p-value = 0.0015 (control: n = 6, egr-expressing disc: n = 6). U Model: dILP-8 from senescent-like cells in the high JNK domain causes downregulation of dILP-2 and dILP-5 from insulin-producing cells (IPCs) in the larval brain, reducing systemic Insulin/Akt/FOXO signalling, protein synthesis and proliferation. Scale bars: 100 μm. and DAPI visualises nuclei. Fluorescence intensities reported as arbitrary units. Source data in graphs are provided as a Source Data file. Experiments depicted in 2G-N were carried out twice with n > 6 discs for control or experiment. Illustrations were created in Biorender Classen, A. (2025) https://BioRender.com/h63vwwi.
To understand if this reduction in dILP2 and dILP5 expression correlates with reduced Insulin signalling in peripheral tissues, we examined nuclear localisation of the dFOXO, a key downstream effector of canonical Insulin signalling16,73. We observed elevated nuclear dFOXO in the notum of egr-expressing discs (Fig. 2D–F), demonstrating that reduced dILP2 and dILP5 expression correlated with systemic attenuation of insulin signalling. This attenuation would explain the low rates of protein translation observed in the notum. In support of this conclusion, we found that low rates of protein translation in nota did not correlate with activation of JNK stress signalling or apoptosis, indicating that low Insulin signalling likely causes the observed reduction in protein synthesis (Supplementary Fig. S2A, B). Importantly, this systemic reduction in protein translation was also evident in other imaginal discs, such as the leg and the eye, and this effect was robustly reproduced by eiger expressed under the more restricted spatial pattern of salm-GAL4 (Fig. 2G–T and Supplementary Fig. S2C–K). In all cases, the reduction in translational capacity correlated with decreased proliferation, with the developing eye showing reduced EdU incorporation overall and in the second mitotic wave, specifically (Supplementary Fig. S2H, I). Taken together, these findings reveal a widespread decline in cell proliferation and protein synthesis in egr-expressing larvae, which correlates with restricted Insulin production and signalling (Fig. 2U). These observations are consistent with previous reports of reduced disc sizes following other types of tissue damage and resemble systemic changes induced by inflammatory tumours40,42,44,45,47,48,53,62,67,74,75.
Nutrient importers and TORC1 support regenerative growth
How can the proliferative domain maintain high rates of protein translation and proliferation in a systemic environment that does not rely on Insulin signalling? In this environment, proliferating cells must solve two problems: (1) They must obtain and take up the right nutrients, and (2) they must drive protein translation and growth under Insulin-limiting conditions. To address the first question, we investigated if egr-driven Insulin restriction may induce nutrient release from the fat body, the largest nutrient storage organ in larvae. Previous studies have shown that fat body breakdown provides nutrients for tumour growth, thereby revealing an ancient programme of nutrient supply under inflammatory conditions44,45,47,48,52,53,76. To understand if similar catabolic changes may be induced in egr-expressing larvae, we employed Nile Red staining to visualise lipid droplets within the fat body. We observed morphological changes, such as increased droplet size and ‘roundness’ (defined as the relationship between area and length of major axis), consistent with molecular changes associated with lipid mobilisation (Fig. 3A–C and Supplementary Fig. S3.1A)44,48,50,77,78. Correspondingly, fat bodies of these larvae exhibited a significant decrease in triglyceride content, similarly to fat bodies from starved larvae (Fig. 3D, E). Moreover, we find that levels of total and activated p-Akt in fat body from egr-expressing larvae were reduced, similar to insulin restriction observed in starved larvae (Supplementary Fig. S3.1B). In addition, levels of nuclear dFOXO-GFP were elevated (Supplementary Fig. S3.1C–E), supporting the notion that egr-induced insulin restriction reduces systemic Insulin signalling and promotes a catabolic fat body state.
A, B Nile Red staining (cyan/grey) of fat body dissected from larvae with control (A) or egr-expressing (B) wing discs. DAPI (magenta) visualises nuclei. C Mean area of lipid droplets in fat body dissected from larvae with control (A) or egr-expressing (B) wing discs. Mean and 95% CI, two-tailed Unpaired t test. p-value = 0.0004. (control: n = 15, egr-expression in discs: n = 18). D, E Triacylglycerides (TAG) levels in fat bodies dissected from larvae with egr-expressing wing disc, control or starved (16 h) larvae. Mean and 95% CI, One-way ANOVA followed by Dunnett’s test for multiple comparison (control larvae: n = 9, egr-expressing larvae: n = 9 and starved larvae: n = 4). p-values: control vs egr = 0.0353; control vs starved =0.0076. F, G ImpL2-GFP in control (A) and egr-expressing discs (B). DAPI visualises nuclei. H Mean ImpL2-GFP intensity in control (F) and egr-expressing discs (G). Mean and 95% CI, two-tailed Welch’s t test, p-value = 0.0001 (control: n = 9, egr-expressing discs: n = 11). I, J Nile Red staining of fat body (cyan/grey) from larvae with control (I) or ImpL2-expressing wing discs using rn-GAL4 (J). K Lipid droplet areas in fat body from larvae with either control or ImpL2-expressing wing discs using rn-GAL4 (24 h). Mean and 95% CI, two-tailed Mann-Whitney test, p-value < 0.0001 (control: n = 23, experiment: n = 18). L Heat map showing relative changes of metabolite concentrations in the larval hemolymph from control and egr-expressing larvae. Data were quantile normalised and analysed using a two-sided, unpaired Wilcoxon rank-sum test. Metabolites with at least < 0.75 and > 1.5-fold change were selected. Metabolites were ordered by log2 fold changes shown as Z-scores. Sample size n = 3 per condition. M, N CG5535-GFP in control (cyan/grey) (M) and egr-expressing discs (N). TRE-RFP visualises JNK- activity (magenta). O Mean CG5535-GFP intensity in the pouch of control and the proliferative domain of egr-expressing discs (PDegr). Mean and 95% CI, two-tailed Unpaired t test, p-value = 0.0092 (control: n = 4, egr-expressing discs: n = 5). P, Q OPP visualises protein synthesis in egr-expressing wing discs from larvae fed on food without (P) or with rapamycin (200 μM) (Q) for 24 h during egr-expression. R. Mean OPP intensity in the pouch of control and the proliferative domain of egr-expressing discs (PDegr) from larvae fed on food without or with rapamycin. Mean and 95% CI, One-way ANOVA followed by Tukey’s post-hoc test for multiple comparisons (control: non-fed: n = 9, fed: n = 10; egr-expressing disc: non-fed: n = 10, fed: n = 10). PDegr vs PDegr + Rapamycin: p < 0.0001, Control + Rapamycin vs PDegr + Rapamycin: p = 0.0167 and Control vs Control + Rapamycin: p = 0.0017. S Model: ImpL2 from senescent-like cells in high JNK domain represses fat body anabolism via inducing insulin resistance, promoting nutrient mobilisation. This branch may reinforce the metabolic switch induced by Insulin restriction. Released nutrients (e.g., amino acids) enter hemolymph and support mTORC1/S6K activation in the proliferative domain. Scale bars: 100 μm. Fluorescence intensities are reported as arbitrary units. Source data in graphs are provided as a Source Data file. Illustrations were created in Biorender Classen, A. (2025) https://BioRender.com/h63vwwi.
Notably, JNK-signalling cells in egr-expressing discs also express ImpL2, upd2 and upd3, factors previously implicated in fat body breakdown during tumour growth in larval and adult hosts (Fig. 3F–H and Supplementary Fig. S3.1 F–H, S3.2)44,47,49,79. When we ectopically expressed ImpL2 for 24 h using the wing-pouch-specific driver rn-GAL4, we found that this was sufficient to induce lipid droplet changes consistent with fat body catabolism (Fig. 3I–K and Supplementary Fig. S3.1 I). Moreover, egr-expressing larvae display activation of JAK/STAT signalling in fat body, which has also been associated with fat body catabolism (Supplementary Fig. S3.1J–L)47,80,81. Of note, we did not observe alterations in muscle morphology or evidence of autophagy in imaginal discs, suggesting that these tissues do not serve as a primary source for nutrients after just 24 h of egr-expression, different to what is observed in the chronic presence of tumours (Supplementary Fig. S3.1M–P)44,45,47,48,52,53,76. Combined, we conclude that the combinatorial expression of systemically acting cytokines from JNK-signalling cells in egr-expressing discs drives detectable nutrient mobilisation from the fat body via multiple pathways.
To determine if the observed break-down of the fat body could also generate amino acid compounds supporting the high rates of protein translation in the proliferative domain, we performed an untargeted metabolomic analysis of the hemolymph from control and egr-expressing larvae. Our results revealed a shift in the composition of amino acids in egr-expressing larval hemolymph. We observed an enrichment of many amino acids, including Leucine, Arginine or Glutamine, which are reported to activate mTORC182,83. Surprisingly, many come enriched in form of mixed dipeptide species. Furthermore, elevated levels of dipeptide species and free amino acids associated with glutamate metabolism were observed, including increased concentrations of glutamine, glutamate, and glutamyl dipeptides (Fig. 3L). Notably, a signature of arginine metabolism emerged, characterised by increased levels of ornithine, N-acetyl-arginine, and aspartate alongside decreased levels of argininosuccinic acid, citrulline, and N-acetyl-ornithine. This altered metabolite profile in the hemolymph represents a flexible supply of building blocks not only for protein synthesis, but also for energy production, co-factor generation, and glutathione-based redox metabolism84,85,86,87.
We next examined the ability of proliferating cells to absorb these amino acids. We investigated the expression of solute carrier (SLC) transporters and found that several transcripts were expressed and elevated in egr-expressing discs. For instance, CG15279 (a putative glycine and proline transporter from the SLC6 family), path (a potential alanine and glycine transporter from the SLC36 family), mnd (an amino acid/polyamine transporter involved in leucine import), and CG5535 (a putative lysine, arginine, and ornithine transporter from the SLC7 family) were all elevated (Supplementary Fig. S3.2). Upregulation of CG5535 was confirmed by immunofluorescence, and we similarly detected increased levels of CG1139 (Arcus), another SLC36 family member that may transport alanine, glycine, and proline (Fig. 3M–O and Supplementary Fig. S3.1Q, R). In addition, a potential sugar transporter, CG3168 of the SLC22 family, and TRET-1, which is predicted to import trehalose, were strongly upregulated (Supplementary Fig. S3.1 S-U). These findings suggest that proliferating cells have an enhanced capacity for importing both amino acids and energy sources.
One regulatory branch promoting protein translation in response to amino acid availability is mediated by mTORC182,83,88,89,90. Moreover, mTORC1 activity itself is promoted by sugar import, boosting ATP production, and hence inhibiting the mTORC1 antagonist AMPK88,89,90. The observed upregulation of amino acid and sugar transporters in egr-expressing discs could therefore facilitate mTORC1 activation. To demonstrate that the mTORC1 pathway supports protein synthesis during wing disc regeneration, we inhibited mTORC1 activity by feeding egr-expressing larvae the mTOR inhibitor Rapamycin for 24 h during egr-expression15,91. This treatment resulted in a pronounced reduction in protein synthesis within the proliferative domain of egr-expressing discs, demonstrating that mTORC1 activity is essential for protein translation during regeneration, as in wild-type control discs (Fig. 3P–R and Supplementary Fig. S3.1V, W). Overall, our data support a model in which fat body catabolism, enhanced amino acid uptake and mTORC1 activation together support the rapid growth and proliferation of the regenerating domain (Supplementary Fig. 3S).
Pdk1 is upregulated in the proliferating domain
S6K, an important effector of mTORC1, directly activates protein translation15,90. S6K transcripts are upregulated in the wound-associated cell populations of egr-expressing discs (Supplementary Fig. S3.2D)25, suggesting that S6K is positively regulated during tissue repair. Importantly, staining for phosphorylated ribosomal S6 (p-S6), a direct target of S6K, revealed that the proliferating domain of egr-expressing discs exhibited strongly elevated p-S6 levels (Fig. 4A–C). However, elevated p-S6 levels cannot be explained solely by mTORC1 activation of S6K because S6K requires co-activation by the Pdk1, a kinase central to canonical Insulin/PI3K signalling14,15,16.
A, B Phosphorylated-S6 (pS6, cyan/grey) in control (A) and egr-expressing wing discs (B). TRE-RFP visualises JNK activity (magenta or grey). C Mean p-S6 intensity in the pouch of control and the proliferative domain of egr-expressing discs (PDegr). Mean and 95% CI, two-tailed Mann-Whitney test, p-value < 0.0001 (control: n = 9, egr-expressing discs: n = 9). D Schematic of canonical Insulin/Akt/FOXO signalling. Phosphatidylinositol kinase-1 (Pdk1) is a key serine/threonine kinase in this pathway. E, F dFOXO-GFP (cyan/grey, BDSC: 38644) in control (E) and egr-expressing wing discs (F). TRE-RFP visualises JNK activity (magenta/grey). Box marks the inset region. G Mean nuclear dFOXO-GFP intensity in the pouch of control or proliferative domain of egr-expressing discs (PDegr). Mean and 95% CI, two-tailed Mann-Whitney test, p-value = 0.0006 (control: n = 7, egr-expressing disc: n = 7). H, I Pdk1-GFP (cyan or grey) in control (H) and egr-expressing discs (I). TRE-RFP visualises JNK activity (magenta or grey), DAPI visualises nuclei. Box marks the inset region. J. Mean Pdk1-GFP intensity in the pouch of control or proliferative domain of egr-expressing discs (PDegr). Mean and 95% CI, Two-tailed Welch’s t test, p-value = 0.0025 (control: n = 10, egr-expressing disc: n = 10). K, L Pdk1-GFP (K) and OPP (L) in hid-expressing discs. an asterisk marks the damaged region. M, N Pdk1-GFP (cyan/grey) and p-S6 (magenta) in control (M) and egr-expressing wing discs (N). Scale bars: 100 μm. DAPI visualises nuclei. Fluorescence intensities are reported as arbitrary units. Source data in graphs are provided as a Source Data file. Experiments depicted in K-N were carried out twice with n > 6 discs for control or experiment. Illustrations were created in Biorender Classen, A. (2025) https://BioRender.com/h63vwwi.
However, how can proliferating cells promote activation of S6K by Pdk1, considering that egr-expressing larvae exhibit insulin restriction? To understand how Pdk1 may be activated, we examined if canonical Insulin/PI3K signalling was active in the proliferating domain of egr-expressing discs. We therefore visualised the nuclear localisation of dFOXO, the most downstream effector of Insulin/PI3K activity, using two independent GFP-tagged lines. We observed high nuclear levels of dFOXO in the proliferating domain of egr-expressing discs, suggesting that Insulin/PI3K signalling there is low, which is consistent with our evidence for systemic Insulin restriction (Fig. 4D–G and Supplementary Fig. S4A–C). This conclusion is further supported by unchanged InR levels and even reduced levels of Akt and phosphorylated Akt (P-Akt) in the proliferative domain (Supplementary Fig. S4D–K). In contrast, we found that Pdk1 expression is strongly upregulated in the proliferative domain (Fig. 4H–J) (see Materials & Methods, section Drosophila genetics). Similar upregulation of Pdk1 was also observed in other models of tissue damage and regeneration, including those using expression of hid or reaper to drive cell death. In these models, elevated Pdk1 levels also correlated with high protein translation, demarcating the proliferating domains (Fig. 4K, L and Supplementary Fig. S4L–Q). In fact, we found that high levels of Pdk1 correlated with high levels of p-S6 staining in egr-expressing discs (Fig. 4M, N). Collectively, these observations suggest that Pdk1 levels may play a central role in regenerative proliferation.
Pdk1 upregulation is sufficient to drive growth and is necessary for regenerative proliferation
To understand if Pdk1 upregulation is a central mechanism driving regenerative proliferation, we expressed either a wild-type or a constitutively active form of Pdk1 in the wing pouch for 24 h using rn-GAL4. Compared to control discs, both forms of Pdk1 resulted in higher levels of protein translation and larger pouch sizes. Larger pouch sizes correlated with more cell proliferation, as EdU incorporation in Pdk1 expressing domains was elevated (Fig. 5 A–F and Supplementary Fig. S5A–D). These results demonstrate that high Pdk1 levels are sufficient to support a metabolic programme characteristic of regenerative proliferation. Notably, the fact that overexpression of a wild-type Pdk1 alone enhances protein synthesis and proliferation suggests that Pdk1 activity scales with expression levels, functioning independently of other canonical upstream signalling inputs, such as Insulin/PI3K. Of note, Pdk1 can auto-activate via transphosphorylation when recruited to the plasma membrane92,93, reflecting potentially Insulin-independent means for Pdk1activation.
A, B OPP incorporation in control (A) and wing discs expressing Pdk1 for 24 h using rn-GAL4 (B). DAPI visualises nuclei. Line indicates boundary of pouch based on landmark folds, approximating the rn-GAL4 domain. C, D EdU incorporation in control (C) and discs expressing UAS-Pdk1 for 24 using rn-GAL4 (D). The line indicates the boundary of the rn-GAL4 expression domain. E. Mean OPP intensity in the pouch of control and Pdk1-expressing wing discs. Mean and 95% CI, two-tailed Unpaired t test, p-value < 0.0001 (control: n = 6, Pdk1-expressing disc: n = 6). F Mean EdU intensity per DAPI area as proxy for relative DNA replication speed, in control and Pdk1-expressing pouches. Mean and 95% CI, two-tailed Welch’s t test, p-value = 0.0078 (control: n = 8, Pdk1-expressing disc: n = 8). G–J OPP incorporation in control (G, H) and egr-expressing wing discs (I, J), either wild type (G, I) or heterozygous mutant for the Pdk15 null allele (H, J). K Mean OPP intensity in the pouch of control or proliferative domain of egr-expressing discs (PDegr), either wild type or heterozygous mutant for Pdk15 null allele. Mean and 95% CI, One-way ANOVA followed by Tukey’s post-hoc test for multiple comparisons. (control:n = 6, control, Pdk15/+: n = 6; egr:n = 8, egr, Pdk15/+: n = 7). PDegr vs PDegr: Pdk15/+: p-value = 0.0300 and Control: Pdk15/+ vs PDegr: Pdk15/+: p-value = 0.0350. L, M DUAL Control system (DCS) allows to manipulate the proliferative domain. A single heat shock activates both egr-expression in the salm-domain (tracked by pyknotic nuclei and MMP-1 upregulation) and genetic manipulation in the proliferative domain via dve-GAL4 (tracked by UAS-GFP co-expression in L). MMP1 is a target gene of JNK activated by Egr, approximating the domain of salm>egr. Dve-GAL4 expresses in the pouch and pouch fold, morphological landmarks approximating the dve-GAL4 domain driving expression of Pdk1-RNAi (M) for 24 h. Protein synthesis is visualised by OPP incorporation in control (L) and Pdk1-RNAi (M) discs. N Mean OPP intensity in the proliferative domain of control (DCSegr, ctrl) and Pdk1 knockdown (DCSegr, Pdk1 RNAi) discs with egr-expression in the salm-domain. A schematic of the egr-expressing region (cyan, trackable by cell death and MMP-1) and the dve-GAL4-expressing region (magenta, trackable by UAS-GFP co-expression) used for Pdk1 knockdown. Mean and 95% CI, two-tailed unpaired t test (p < 0.0001). (DCSegr, control: n = 10; DCSegr, Pdk1 RNAi: n = 9). O Scheme highlighting the Pdk1-branch driving S6K activation. Scale bars: 100 μm. DAPI visualises nuclei. Fluorescence intensities are reported as arbitrary units. Source data in graphs are provided as a Source Data file. Illustrations were created in Biorender Classen, A. (2025) https://BioRender.com/h63vwwi.
To provide evidence that Pdk1 function is also necessary to support proliferation in egr-expressing discs, we genetically reduced Pdk1 function by establishing heterozygosity for a null allele of Pdk194. In control discs, this heterozygosity did not affect overall protein synthesis compared to wild-type discs, indicating that under normal developmental conditions, a single copy of Pdk1 is sufficient for growth. However, protein synthesis in the proliferating domain of egr-expressing discs heterozygous for Pdk1 was significantly reduced (Fig. 5G–K). This finding demonstrates that the proliferating domain is particularly sensitive to reductions in Pdk1 levels and underscores the necessity for elevated Pdk1 to support protein synthesis during regeneration. To provide further proof, we expressed a validated RNAi against Pdk1 specifically in proliferating cells of egr-expressing discs under control of the DUAL Control genetic ablation system (Supplementary Fig. S5E–H)65. Consistent with our heterozygous mutant results, Pdk1 knockdown led to a marked decrease in protein translation in the proliferating domain, confirming a cell type specific function for Pdk1 upregulation in the proliferating domain during regeneration (Fig. 5L–N). Overall, these findings establish that Pdk1 upregulation is not only sufficient to drive protein translation and growth but is also critically required to sustain protein translation in the proliferative domain of regenerating imaginal discs under conditions of systemic insulin restriction (Fig. 5O).
Pdk1 is regulated by JAK/STAT signalling
To understand how Pdk1 levels are controlled during regeneration, we examined the role of JAK/STAT signalling, which is activated in the proliferating domain of egr-expressing discs to promote cell proliferation and survival (Fig. 6A, B)24,25,26,32,64,65. We therefore expressed STAT92E in the posterior compartment of wing imaginal discs, which elevates JAK/STAT signalling, using the en-GAL4 driver26. This manipulation caused an increase in levels of Pdk1-GFP, phospho-S6 and OPP incorporation (Fig. 6C–H and Supplementary Fig. S6C–E), demonstrating that JAK/STAT activation is sufficient to promote Pdk1 upregulation, S6 activation and protein translation. Conversely, when we expressed an RNAi construct to knock down STAT92E in the posterior compartment during normal development, we observed a decrease in both Pdk1-GFP levels and OPP incorporation, suggesting that JAK/STAT activity can limit Pdk1 levels and protein translation (Fig. 6I–L). Importantly, the JAK/STAT-Pdk1-S6 axis is also essential for regeneration: we genetically reduced STAT92E function in egr-expressing discs by establishing heterozygosity for a null allele of STAT92E and found that it reduced OPP incorporation specifically in egr-expressing discs but not in control discs (Fig. 6M–O and Supplementary Fig. S6F, G). Moreover, when we expressed an RNAi against STAT92E specifically in proliferating cells of egr-expressing discs under control of the DUAL Control genetic ablation system65, the elevated protein translation was abrogated (Fig. 6P–R and Supplementary Fig. S6H, I). Combined, these findings reveal a function for JAK/STAT signalling in tissue repair and regeneration by promoting Pdk1 upregulation, which, together with mTORC1, supports insulin-independent growth in the proliferating domain (Supplementary Fig. S6J).
A, B A control and egr-expressing wing disc expressing TRE-RFP (magenta) and Stat92E-dGFP (cyan) reporters. C, E, G Wing discs expressing STAT92E in the posterior compartment under en-GAL4, UAS-RFP. Pdk1-GFP (C), p-S6 (E), and OPP incorporation (G) shown. D, F, H Mean Pdk1-GFP (D), p-S6 (F) and OPP incorporation (H) levels, expressed as posterior-anterior (P/A) ratio in control and discs expressing STAT92E under the control en-GAL4. D two-tailed Unpaired t test, p-value < 0.0001 (control: n = 5, experiment: n = 8). F two-tailed Welch’s t test, p-value = 0.0033 (control: n = 5, experiment: n = 8). H two-tailed Unpaired t test, p-value = 0.0004 (control: n = 5, experiment: n = 5). Mean and 95% CI shown. I, J Control wing disc (I) and wing discs expressing STAT92E-RNAi (J) in the posterior compartment under control of en-GAL4, UAS-RFP. Pdk1-GFP and OPP incorporation shown. K Mean Pdk1-GFP levels expressed as posterior-anterior (P/A) ratio in control and discs expressing STAT92E-RNAi using en-GAL4. Mean and 95% CI, two-tailed Welch’s t test, p-value = 0.0004 (control: n = 8, experiment: n = 10). L Mean posterior-anterior (P/A) ratio of OPP incorporation in control and discs expressing STAT92E-RNAi in the posterior compartment using en-GAL4. Mean and 95% CI, two-tailed Unpaired t test, p-value < 0.0001 (control: n = 8, experiment: n = 10). M, N OPP incorporation in egr-expressing discs, either wild type (M) or heterozygous mutant for the Stat92E85C9 null allele (N). O Mean OPP intensity in the pouch of control and the proliferative domain of egr-expressing discs (PDegr), either wild type or heterozygous mutant for Stat92E85C9 null allele. Mean and 95% CI, one-way ANOVA followed by Tukey’s post-hoc test for multiple comparisons. (control:n = 7, control, Stat92E85C9/+: n = 7; egr:n = 7, egr, Stat92E85C9/+: n = 7). PDegr vs PDegr:Stat92E85C9/+: p-value = 0.0080 and Control: Stat92E85C9/+ vs PDegr: Stat92E85C9/+: p-value = 0.0005. P, Q DUAL Control system (DCS) activates egr-expression in the salm-domain (tracked by pyknotic nuclei and MMP-1 upregulation) and stat92E-RNAi in the proliferative domain via dve-GAL4 (tracked by UAS-GFP co-expression in P), expressing in the pouch and the proximal pouch fold (Q). Protein synthesis visualised by OPP incorporation in control (P) and Stat92E knockdown (Q) disc. R Mean OPP intensity in the proliferative domain of control (DCSegr) and STAT92E knockdown (DCSegr, Stat92E-RNAi) wing discs with egr-expression in the salm-domain. Mean and 95% CI, two-tailed Mann-Whitney’s test, p-value = 0.0006. (DCSegr, control: n = 7; DCSegr, Stat92E- RNAi: n = 8). S. Scheme highlighting JAK/STAT positively affecting Pdk1 and S6K. Scale bars: 100 μm. DAPI visualises nuclei. Fluorescence intensities are reported as arbitrary units. Source data in graphs are provided as a Source Data file. Illustrations were created in Biorender Classen, A. (2025) https://BioRender.com/h63vwwi.
JAK/STAT-Pdk-1-S6K signalling is linked to tumour growth
We wanted to understand if the JAK/STAT-Pdk1-S6 signalling axis observed during regenerative proliferation also plays a role in tumour growth, given that tumours often co-opt physiological repair processes for their pathological purposes. We thus analysed imaginal discs in which tumorigenesis was induced by both reducing the function of the tumour-suppressor gene scrib95 and ectopically expressing oncogenic RasV12 96,97. We used the rn-GAL4 driver to drive expression of scrib-RNAi and RasV12 in the entire wing pouch, which causes pronounced overproliferation26,51,53. This tumour model is characterised by the coordinated but spatially separated activation of JNK and JAK/STAT signalling networks, whose regulatory and functional characteristics resemble those active during tissue repair in egr-expressing discs (Supplementary Fig. S7A)26. In RasV12,scrib-RNAi expressing discs, we found a striking correlation between regions of JAK/STAT activation, elevated protein translation and increased Pdk1 level (Fig. 7A–F). An independent tumour model of Psc-Su(z)2 tumours also shows correlated elevation of JAK/STAT activation and protein synthesis (Supplementary Fig. S7B, C). Importantly, these tumours, like egr-expressing discs, induce all hallmarks of systemic cachexia, including reduced protein translation in the non-transformed notum (Fig. 7G–I), and must therefore overcome growth restrictions imposed on peripheral tissues40,53,76. Our observation demonstrates that tumours likely coopt JAK/STAT activation and Pdk1 elevation to support protein translation and thus tumour proliferation.
A, B Control (A) and wing disc expressing RasV12, scrib-RNAi (B) for 44 h starting at Day6 AED. OPP incorporation (cyan/grey) and JAK/STAT-reporter 10xStat92E-dGFP (magenta/grey). C Mean OPP intensity in the 10xStat92E-dGFP positive and negative region within a RasV12, scrib-RNAi-expressing wing discs. Mean and 95% CI, two-tailed Paired t test, p-value = 0.0006 (control: n = 6, experiment: n = 6). D, E Control and wing disc expressing RasV12, scrib-RNAi. Pdk1-GFP (cyan/grey) in control (D) and RasV12, scrib-RNAi-expressing wing disc (E), along with MMP1 staining to visualises JNK-activity (magenta/grey). F Mean Pdk1-GFP intensity in the control pouch and RasV12, scrib-RNAi-expressing pouch. Mean and 95% CI, two-tailed Unaired t test, p-value = 0.0004 (control: n = 10, experiment: n = 10). G, H OPP incorporation in the notum of control (G) and wing disc expressing RasV12, scrib-RNAi (H). I Mean OPP intensity in the notum of control and wing disc expressing RasV12, scrib-RNAi. Mean and 95% CI, two-tailed Welch’s t test, p-value = 0.0031 (control: n = 6, experiment: n = 6). Scale bars: 100 μm. DAPI visualises nuclei. Fluorescence intensities are reported as arbitrary units. Source data in graphs are provided as a Source Data file.
Pdk1 reduction in peripheral discs during growth restriction
Strikingly, Pdk1 levels appear to be a target of systemic regulation during Insulin restriction in imaginal discs. In egr-expressing larvae, we consistently observed that Pdk1 levels were significantly lower in peripheral imaginal disc, including the notum, leg and eye, if compared to wild type control discs (Fig. 8A–F and Supplementary Fig. S8). This observation was not limited to egr-expressing larvae; even in the peripheral imaginal discs of RasV12,scrib-RNAi tumour-bearing larvae, Pdk1 levels were markedly reduced (Fig. 8G–I). This targeted reduction of Pdk1 in peripheral imaginal discs is consistent with the overall decrease in protein synthesis and growth observed in these regions. Such downregulation suggests that systemic signals, maybe even Insulin restriction, actively suppress Pdk1 expression in non-regenerative areas. In contrast, JAK/STAT signalling in regenerative areas can counteract Pdk1 suppression and locally elevate Pdk1 levels and function. This dual regulation -upregulation in regenerative zones and downregulation in non-essential areas -highlights Pdk1’s central role in mediating the balance between local regenerative demands and systemic metabolic reprogramming. Consequently, Pdk1 emerges not only as a key driver of insulin-independent regenerative proliferation but also as an integrative node that coordinates systemic growth control during tissue repair and tumorigenesis.
A, B Pdk1-GFP in the notum of control (A) and egr-expressing discs (B). C Mean Pdk1-GFP intensity in the notum of control (A) and egr-expressing discs (B). Mean and 95% CI, two-tailed Mann-Whitney’s test, p-value = 0.0007 (control: n = 10, experiment: n = 10). D, E Pdk1-GFP in eye imaginal disc, dissected from larvae with control (D) or egr-expressing (E) wing discs. F Mean Pdk1-GFP intensity in eye discs, dissected from larvae with control (D) or egr-expressing (E) wing discs. Mean and 95% CI, two-tailed Unpaired t test, p-value < 0.0001 (control: n = 7, experiment: n = 7). G, H Pdk1-GFP in the notum of control (G) and wing disc expressing RasV12, scrib-RNAi (H). I Mean Pdk1-GFP intensity in the notum of control and wing disc expressing RasV12, scrib-RNAi. Mean and 95% CI, two-tailed Unpaired t test, p-value = 0.0022 (control: n = 5, experiment: n = 6). J Model summary illustrating growth mediated by S6K activation via canonical Insulin signalling (Insulin/AKT/FOXO and mTORC1) during normal developmental (left). Growth in the proliferative domain is maintained in an insulin-restricted environment through the JAK/STAT-Pdk1-S6K axis, supported by mTORC1 signalling. K Model summary of local and systemic metabolic adaptations supporting growth in the proliferative domain. Inter-organ signalling is initiated by senescent-like cells in the high JNK signalling domain at the centre of tissue damage via secreted Dilp8, ImpL2 and Upd1,2,3. Dilp8 reduces insulin-like peptide expression by acting on IPCs, thereby restricting systemic insulin signalling in peripheral tissues. ImpL2 acts on fat body, and combined both mechanisms facilitates nutrient mobilisation from stores. Secreted Unpaireds activate JAK/STAT in the nearby proliferative domain, upregulating Pdk1 and instructing S6K activation. Levels of Pdk1 in the proliferative and peripheral regions determine tissue growth in an insulin-restricted environment. Scale bars: 100 μm. DAPI visualises nuclei. Fluorescence intensities are reported as arbitrary units. Source data in graphs are provided as a Source Data file. Illustrations were created in Biorender Classen, A. (2025) https://BioRender.com/h63vwwi.
Discussion
Our study reveals how signalling through a JAK/STAT-Pdk1-S6K axis promotes metabolic reprogramming to drive tissue repair and regeneration in an environment of systemic growth restriction (Fig. 8J, K). We demonstrate that in response to inflammatory damage, a senescent subpopulation of cells at the centre of tissue damage induces insulin restriction via action of dILP8 on IPCs, as well as insulin resistance via action of ImpL2 on the fat body. The resulting systemic reduction of insulin signalling is evident from the decreased expression of dILP2 and dILP5 in IPCs, as well as the enhanced nuclear localisation of dFOXO, low rates of protein synthesis and proliferation in peripheral tissues, or reduced Akt signalling and nutrient mobilisation in the fat body. These findings align with previous work suggesting that systemic growth restriction, for example via repression of Ecdysone signalling, helps synchronise regeneration with developmental timing and prevents the overgrowth of undamaged tissues41,42,57,58,98.
However, slowing down peripheral growth poses a challenge to the regenerating cell population, which must continue to support high anabolic activity despite reduced canonical Insulin/PI3K/AKT signalling. The regenerating cell population circumvents this limitation by engaging a previously uncharacterised mechanism to sustain protein synthesis. Specifically, we find that Pdk1 is robustly upregulated in the proliferative domain of egr-expressing discs, and this upregulation is sufficient and necessary to drive ribosomal S6 phosphorylation and activate protein translation, even in the context of low systemic Insulin signalling. The capacity of Pdk1 to function independently of insulin positions Pdk1 as a central regulator of regenerative metabolism. Notably, our results mirror earlier observations of AKT-independent, yet Pdk1-dependent, growth during Drosophila development94,99. During regeneration, we identify JAK/STAT as an upstream regulator of Pdk1, and consequently as a regulator for phosphorylation of ribosomal S6 protein and activation of protein synthesis. Our work thus uncovers a pathway whereby JAK/STAT signalling can mediate the known proliferation-promoting function in imaginal disc development, regeneration, and tumour growth30. Of note, the increase in Pdk1 protein levels likely arises from JAK/STAT regulated post-translational mechanisms affecting Pdk1 stability, as we did not observe an increase in Pdk1 transcripts in egr-expressing discs25. Such post-translational regulation of Pdk1 may also underlie the differential regulation of Pdk1 between regenerative and peripheral tissues. While we find that the proliferative domain actively upregulates Pdk1 to overcome insulin restrictions, we also find that the peripheral imaginal discs downregulate Pdk1. Based on our experiments demonstrating that Pdk1 levels are limiting for protein translation, the peripheral reduction in Pdk1 may contribute to the reduction in protein synthesis and growth of other imaginal discs. This dual regulation emphasises the surprisingly central role of Pdk1 in integrating local demands for regeneration with systemic constraints on growth.
Furthermore, our work reveals that fat body catabolism, which mobilises nutrients necessary for regeneration, is activated in egr-expressing larvae. In addition to the remodelling of the lipid stores, proteins may also be catabolized, as suggested by an altered amino acid signature in the hemolymph. The upregulation of amino acid and carbohydrate transporters in the proliferative domain further suggests that regenerating cells actively import these mobilised nutrients, thereby fueling mTORC1 activation and subsequent protein synthesis. Earlier studies find elevated levels of ornithine, glutamate, and glutamine in rat wound fluids100, similar to the ornithine, glutamate, and glutamine signature in hemolymph from egr-expressing larvae. Our observations suggest that glutamate metabolism may be important for regeneration, consistent with its described roles in tumour growth, inflammation, and defence against oxidative stress10,84,101.
Together, our results support a model in which egr-induced inflammation induces systemic insulin restriction and insulin resistance, thereby limiting resource availability in peripheral tissues. At the same time, local activation of pathways such as JAK/STAT, Pdk1, or nutrient importers prioritises tissue repair at the damage site. A potential limitation of our study lies in the high levels of eiger-expression used in this genetic model, which may not fully reflect physiological conditions. In fact, as we previously reported, egr-expressing discs can resemble chronic wound states, where prolonged inflammatory signalling impairs effective tissue regeneration26. However, in the widely studied RasV12,scrib-RNAi tumour model, we observe that regions with high JAK/STAT activity also exhibit elevated Pdk1 expression and protein translation, suggesting that tumours which can induce cachexia also exploit this pathway to sustain growth despite low systemic insulin signalling. Given that Pdk1 can bypass the need for Insulin/AKT/FOXO signalling, it may not be surprising that Pdk1 upregulation is found in many cancers102. While oncogenic mutations may facilitate nutrient use in such a cachexic tumour environment45,52,103,104, our work specifically reveals how wild-type tissues adapt within a physiological repair programme to overcome systemic inhibition of growth.
In summary, our study uncovers a regulatory network wherein fat body catabolism, enhanced nutrient uptake, mTORC1 activation, and the instructive JAK/STAT-Pdk1 axis converge to support regenerative proliferation in a cachexic environment. Undoubtedly, this is an ancient stress response and repair programme designed to distribute nutrients from an energy store to the site of tissue damage. These findings not only advance our understanding of tissue repair mechanisms in Drosophila but may also offer insights into conserved pathways in mammals, as a domain characterised by ribosomal S6 protein phosphorylation was recently described in mouse and pig skin wounds, suggesting potential similarities in regenerative programmes across species18.
Method
Drosophila maintenance
All experiments were performed on Drosophila melanogaster. Fly strains (see Supplementary Table S1) were maintained on standard fly food (10 L water, 74,5 g agar, 243 g dry yeast, 580 g corn flour, 552 ml molasses, 20.7 g Nipagin, 35 ml propionic acid) at 18 °C – 22 °C. Larvae from experimental crosses were allowed to feed on Bloomington formulation (175.7 g Nutry-Fly,1100 ml water 20 g dry yeast, 1.45 g Nipagin in 15 ml Ethanol, 4.8 ml Propionic acid) and raised at 18 °C or 30 °C to control GAL80ts-dependent induction of GAL4/UAS. Our experimental design did not consider differences between sexes, unless for genetic crossing schemes.
Drosophila genetics
To induce expression of UAS-constructs, such as UAS-egr, under the control of rn-GAL4 in the wing pouch, experiments were carried out as described in24,28,62 with minor modifications. Briefly, larvae of genotype rn-GAL4, tub-GAL80ts and carrying the desired UAS-transgenes were staged with a 6 h egg collection and raised at 18 °C at a density of 50 larvae/vial. Overexpression of transgenes was induced by shifting the temperature to 30 °C for 24 h on the seventh day (D7) after egg deposition (AED) to relieve temperature-sensitive GAL80ts repression of GAL4. Larvae were dissected after 24 h of egr-expression. Control genotypes were generated by crossing rn-GAL4, tub-GAL80ts into a wild-type background. The DUAL Control genetic ablation system (DCS) was employed, wherein a single heat shock simultaneously activates genetic cell ablation by egr-expression within the salm domain and GAL4 expression in the dve-GAL4 domain (hsFLP; hs-p65::zip, lexAOp-egrNI/CyO; salm-zip::LexA-DBD, DVE»GAL4)65. Specifically, larvae were raised at 25 °C and subjected to a 1-hour heat shock at 37 °C on Day 4.5, followed by dissection 24 h post-heat shock. The Pdk1-GFP line (BDSC: 59836) was characterised using the following approaches: a complex expression pattern can be observed in wild type tissues in immunofluorescence, where GFP is detected in the cytoplasm but also at membranes (see for example, Fig. 6C). Tissues form larvae produce a band of the expected size on Western blot. Three independent RNAi lines (BDSC: 27725, BDSC: 34936, BDSC: 36071) targeting the 3’ region of the Pdk1 transcript downstream of the GFP-cassette insertion site can robustly knock down GFP expression (see for example, Fig. 5E). We conclude that a full-length, membrane-recruitable and genetically tractable protein is produced from the endogenous locus. Thus, the annotation of the insertion site in FlyBase is incorrectly oriented.
Immunohistochemistry of wing imaginal discs
Wing discs from third instar larvae were dissected and fixed for 15 minutes at room temperature in 4% paraformaldehyde in PBS. Washes were performed in PBS containing 0.1% TritonX-100 (PBT). The discs were then incubated with primary antibodies (listed in Supplementary Table S2) in 0.1% PBT, gently mixed overnight at 4 °C. During incubation with cross-absorbed secondary antibodies coupled to Alexa Fluorophores at room temperature for 2 h, tissues were counterstained with DAPI (0.25 ng/µL, Sigma, D9542). Tissues were mounted using SlowFade Gold Antifade (Invitrogen, S36936). To ensure comparability in staining between different genotypes, experimental and control discs were processed in the same vial and mounted on the same slides whenever possible. Images were acquired using the Leica TCS SP8 Microscope, using the same confocal settings for linked samples and processed using tools in Fiji.
Protein synthesis assays using OPP- Click-iT staining
OPP Assays were performed using Click-iT® Plus OPP Protein Synthesis Assay Kits (Invitrogen Molecular Probe) according to the manufacturer’s instructions. Briefly, larvae were dissected, and inverted cuticles were incubated with a 1:1000 dilution of Component A in Schneider’s medium for 15 min on a nutator. Larval cuticles were fixed with 4% paraformaldehyde for 15 min, rinsed twice in 0.1% PBT, and permeabilized with 0.5% PBT for 15 min. The cuticles were then stained with the Click-iT® cocktail for 30 min at room temperature, protected from light. Further immunohistochemistry analysis and sample mounting was performed as described above.
EdU labelling
EdU incorporation was performed using the Click-iT Plus EdU Alexa Fluor 647 Imaging Kit. Briefly, larval cuticles were inverted in Schneider’s medium and incubated with EdU (10 µM final concentration) at RT for 15 min. Cuticles were then fixed in 4% PFA/PBS for 15 min, washed for 30 min in PBT 0.5%. EdU-Click-iT labelling was performed according to the manufacturer’s guidelines. Further immunohistochemistry analysis and sample mounting was performed as described above.
SA-β-Gal staining
Cell senescence detection kit from Invitrogen (C10850) was used to analyse senescence-associated β-galactosidase activity. Briefly, larval cuticles were inverted in PBS, fixed with 4% PFA, washed with 1% BSA (in PBS) and then incubated in the working solution for 2 h at 37 °C, according to the manufacturer’s instructions. Washing steps were performed in PBS and PBS containing 0.1% TritonX-100 (PBT). Further immunohistochemistry analysis and sample mounting was performed as described above.
Fat body Nile Red staining
Early third instar larvae were collected in PBS and dissected, leaving the gut intact to prevent fat body loss. Inverted cuticles were transferred to an Eppendorf tube and fixed with 4% paraformaldehyde/PBS for 15 min. Samples were washed in 0.1% PBT. Cuticles were incubated with Nile Red (2 μg/mL in PBS) for 1 h, protected from light. Following incubation, samples were washed in PBS, the fat body was dissected and mounted as described above.
Hemolymph sample preparation
Fifteen larvae were collected and thoroughly washed with Milli-Q water to remove any fly food particles. Care was taken to ensure no food particles remained on the larvae surfaces, and they were dried using Kim Tech paper wipes. Each larva was punctured in the centre using forceps and transferred to a 0.5 mL microcentrifuge tube with three 1 mm holes at the bottom. The 0.5 mL microcentrifuge tube was then placed into a pre-cooled 1.5 mL microcentrifuge tube. One larva at a time was processed in this assembly and centrifuged in a microfuge for 10 seconds. After each centrifugation, the larval carcass was removed from the 0.5 mL microcentrifuge tube to prevent blockage of the holes. Hemolymph isolated from the 15 larvae was collected in the bottom 1.5 mL tube. From the total collected hemolymph, 8 µL was transferred to a fresh 1.5 mL tube, and 10 µL of ultrapure Milli-Q water, 30 µL of methanol, an internal standard, and 50 µL of MTBE were added. The solution was mixed thoroughly and centrifuged at 1000 g for 10 minutes at 4 °C. After centrifugation, the organic and polar phases were collected separately in different tubes for metabolite measurement.
Hemolymph metabolomic analyses
Non-targeted analysis of polar metabolites by LC-MS was carried out as described previously105 using an Agilent 1290 Infinity II UHPLC in line with a Bruker Impact II QTOF-MS operating in negative and positive ion mode. Scan range was from 20 to 1050 Da, and mass calibration was performed at the beginning of each run. LC separation was on a Waters Atlantis Premier BEH ZHILIC column (100 ×2.1 mm, 1.7 µm particles), buffer A was 20 mM ammonium carbonate and 5 µM medronic acid in milliQ H2O, and buffer B was 90:10 acetonitrile:buffer A and the solvent gradient was from 95% to 55% buffer B over 14 min. Flow rate was 180 µL/min, column temperature was 35 °C, autosampler temperature was 5 °C and injection volume was 3 µL. Data processing, including feature detection, feature deconvolution, and annotation of features, was performed using MetaboScape (version 2023b). Quantile normalisation was performed to minimise sample size effects, and further statistical processing were performed with R. Metabolites with missing values were eliminated from the dataset. Both the Shapiro-Wilk test and the Q-Q plot, showed that the data was not normally distributed. Therefore, a two-sided, unpaired Wilcoxon rank-sum test was performed on the quantile-normalised data. Only metabolites with at least < 0.75 and > 1.5-fold change and a p-value < 0.19 and were selected for further analysis and were displayed in a heatmap with row-wise normalization (Fig. 3L).
Thin layer chromatography (TLC)
Fat bodies were dissected in cold PBS from third instar (L3) control (rn-GAL4, tubGAL80ts) and egr-expressing (rn-GAL4, UAS-egr, tubGAL80ts)larvae, and from wild type larvae starved for 24 h. Three fat bodies were pooled per sample and immediately transferred into 100 μL chloroform: methanol (3:1) solution, followed by storage on ice. Samples were mechanically homogenised and centrifuged at 15,000 × g for 5 min at 4 °C. For semi-quantitative analysis of triglycerides, a standard curve was prepared using lard as a reference material, dissolved in chloroform: methanol (3:1) at the following concentrations: STD1 (75 μg/μL); STD2 (60 μg/μL); STD3 (50 μg/μL); STD4 (37.5 μg/μL); STD5 (25 μg/μL); STD6 (15 μg/μL); STD7 (10 μg/μL). A blank control containing only the solvent mixture (chloroform:methanol (3:1)) was included. Samples and standards were loaded onto a glass silica gel TLC plate (Millipore). The mobile phase consisted of a hexane:diethyl ether (4:1) solvent system. Chromatography was performed, and after air-drying, the TLC plate was treated with ceric ammonium heptamolybdate (CAM) staining solution and developed at 80 °C, with band development monitored at 5 min intervals. Images were captured using a gel documentation system (gelONE); image analysis and quantification were performed using ImageJ. Automatic image thresholding was applied using the Otsu method, and regions of interest (ROIs) were defined for each band. Integrated intensity analysis was performed, and triglyceride density was quantified based on the standard curve.
Real-time PCR
Third instar (L3) control (rn-GAL4, tubGAL80ts), egr expressing (rn-GAL4, UAS-egr, tubGAL80ts), and wild type larvae starved for 24 h larvae were carefully collected from the food and pooled in groups of three male larvae per sample. Samples were homogenised in 100 µL of TRIzol, and RNA was extracted using chloroform (20 µL), followed by precipitation with 50 µL of isopropanol. The purified RNA was suspended in nuclease-free water, subjected to DNase treatment and reverse transcription was performed using RevertAid Reverse Transcriptase (Thermo Fisher Scientific) according to the manufacturer’s guidelines. qPCR was performed using the Blue S’Green qPCR Kit Separate ROX (Biozym) on a LightCycler 480 (Roche). Standard dilution series (1:5) were generated for each primer pair to determine primer efficiency based on the regression curve. The following primers were used (for, rev 5’−3’): Rpl1 (TCCACCTTGAAGAAGGGCTA, TTGCGGATCTCCTCAGACTT), Ilp2 (ATCCCGTGATTCCACCACAAG, GCGGTTCCGATATCGAGTTA), Ilp5 (GCCTTGATGGACATGCTGA, AGCTATCCAAATCCGCCA)106,107. Gene expression levels were normalised to the reference gene Rpl1, and relative expression was calculated using the Pfaffl method. Statistical outliers were identified using the ROUT test (Q = 1%) and were removed to maintain data consistency. Statistical analysis was performed using the Kruskal-Wallis test.
Western blot analysis
Fat bodies were dissected in cold PBS from third-instar (L3) control (rn-GAL4, tubGAL80ts) and egr-expressing (rn-GAL4, UAS-egr, tubGAL80ts) larvae, and from wild-type larvae starved for 24 h. Five fat bodies were lysed on ice (300 mM NaCl, 50 mM Tris-HCl ph 7,5, 1% Triton X-100, 0.1 mM EDTA, 0.1% SDS, 5% Glycerol, Roche Complete protease inhibitor cocktail and Complete phosphate inhibitor cocktail). Tissues were further homogenised and centrifuged at 6000 × g for 3 min at 4 °C. Supernatants were incubated in Laemmli buffer at 85 °C for 5 min. Samples were loaded onto Mini-PROTEAN TGX gels (4–15%) corresponding to one fat body per lane. After electrophoresis, proteins were transferred onto a nitrocellulose membrane, blocked in 1 × Tris-buffered saline containing bovine serum albumin, incubated overnight with primary antibodies at 4 °C and secondary antibody for 1 h at RT. Proteins were visualised using SuperSignal West Femto Maximum Sensitivity Substrate and a Bio-Rad ChemiDoc-MP imaging system.
Image quantification and statistical analysis - General comments
For all quantifications, control and experimental samples were processed together and imaged in parallel, using the same confocal settings. Images were processed, analysed and quantified using tools in Fiji (ImageJ 2.14)108. Care was taken to apply consistent methods to control and experimental samples (i.e., number of projected sections, thresholding methods, processing) for image analysis and quantifications. See Supplementary Fig. S1 for macros used in FIJI during image segmentation and quantification. Further details on segmentation and quantification are provided below. Figure panels were assembled using Affinity Designer 2.4.0. Statistical analyses were performed in Graphpad Prism. Illustrations were created in Biorender Classen, A. (2025) https://BioRender.com/h63vwwi. To quantify fluorescence intensities inside the proliferative domain (PDegr), a mask of the high JNK-signalling domain region was generated, followed by creating a 20 μm band on the outside to mark the proliferative domain. Fluorescence intensities in proliferative domains was always compared to fluorescence intensities within the pouch domain of control discs, unless otherwise noted.
Quantification of cycling cells using EdU incorporation
-
(a)
EdU quantification in notum: An xy-section with the maximum number of notum epithelial cells was selected, excluding any myoblast cells. A mask was created from the DAPI staining to represent nuclei and saved as an ROI. A binary operation (AND) was used to compute a notum DAPI mask with the aid of a manually selected notum region (FileS1, Macro-1L(a)). This mask was then used to measure EdU intensity.
-
(b)
EdU quantification within 10μm bands: In egr-expressing discs, TRE-RFP expression was used to generate a mask of the high JNK-signalling domain (FileS1, Macro-1D(a)). The boundary of this mask defined the regions inside and outside the high JNK region. Five 10μm bands were created outside the high JNK-signalling domain, and three 10μm bands inside of the JNK-signalling domain. Also applying a DAPI mask to each band (FileS1, Macro-1D(b)), the mean EdU intensity in each of the 10μm bands was measured. Consistent with previous studies24,26, the highest EdU intensities were observed in 3 bands outside the JNK-signalling domain, which was denoted as the proliferative domain.
-
(c)
EdU quantification in eye disc: For the eye disc, an EdU mask (FileS1, Macro-2T(a)) was generated using the EdU staining, which was then applied to measure EdU intensity in the entire visible eye disc.
Quantification of Upd3-LacZ levels
A maximum projection of TRE-RFP expression was used to generate a mask of the high JNK-signalling domain in egr-expressing disc (FileS1, Macro-S3H(a)). Upd3-LacZ intensity was measured within the TRE-RFP mask of the high JNK-signalling domain in egr-expressing disc, while for the control disc, Upd3-LacZ intensity was measured in the pouch.
Quantification of ImpL2-GFP levels
A pouch mask was generated using the maximum projection of an anti-Nubbin staining (FileS1, Macro-3H(a)) for both control and egr-expressing discs. ImpL2-GFP intensities were measured within the Nubbin mask on the sum projection of the ImpL2-GFP signals within the stack (seven slices in each disc).
Quantification of OPP levels
-
(a)
in different regions of wing imaginal disc: In the control disc, mean OPP intensity was measured separately in the pouch, hinge, and notum regions. In egr-expressing discs, mean OPP intensity was measured in the high JNK signalling region (TRE-RFP mask), the proliferative region (20 µm band around the TRE-RFP mask), and the notum region. In control discs, the pouch, hinge, and notum regions were selected using wing fold landmarks. In egr-expressing discs, a max projection of TRE-RFP intensities was used to create a mask of the high JNK signalling domain (FileS1, Macro-1G(a)), which was applied to measure OPP intensity in the high JNK region. A 20μm band outside the high JNK region was generated to mark the proliferative region, and OPP intensity in this domain was measured. For the notum, a manual selection along the first notum fold and outlining the edges of the notum was carried out.
-
(b)
in the eye disc: For the eye-antennal disc, the DAPI staining was used to generate a mask of the tissue outline (FileS1, Macro-2Q(a)) and combined with a manual selection of the eye disc to generate a region of interest (ROI) within which we measured OPP intensity.
Quantification of nuclear dFOXO levels
In control discs, a slice with the maximum number of pouch cells was selected, and a DAPI mask (Supplementary File S1, Macro-4G(a)) was generated and combined with manual pouch selection to create a pouch nuclear mask. This mask was used to measure mean dFOXO intensity in the pouch nucleus. In egr-expressing discs, a single slice from the Z-stack with the maximum number of cells in the proliferative domain was selected. A max projection of TRE-RFP intensities was used to create a mask of the high JNK signalling domain, and a 20 μm band was created to locate the proliferative region. A DAPI mask (Supplementary File S1, Macro-4G(b)) was generated from the selected slice and combined with the 20 μm band to create a new mask for the proliferative cell nuclei, which was used to measure dFOXO intensity in the nucleus. For the analysis of nuclear dFOXO in the notum, a Z-slice containing the maximum number of notum cells was selected. A DAPI mask (Supplementary File S1, Macro-2F(a)) was generated and combined with manual selection of notum, guided by wing fold, to create a notum nuclear mask. Please note that discs and fat body were treated with Leptomycin B (413 nM) for 30 min before dissection to block nuclear export.
Quantification of Pdk1 GFP, p-S6 and OPP levels in the en-GAL4 domain
The slice with the maximum number of pouch cells was selected, and masks of the anterior and posterior pouch compartments were generated by segmenting the enGAL4, UAS-RFP expression domain. Levels of p-S6(Supplementary File S1, Macro 6 F(a)) and Pdk1-GFP intensities were measured within these masks, and the P/A ratio (en-GAL4 region/non-en-GAL4 region) was calculated. Mean OPP intensity in the control and experimental pouch en-GAL4 domain was measured and reported.
Quantification of 10xStat92E-GFP levels in the fat body
A single slice from the Z-stack with the maximum number of nuclei was selected. A 200 × 200 pixel square region was defined and used as a standard area for measurement of GFP intensities.
Quantification of lipid droplet size and shape
A single slice from the Z-stack through the most anterior fat body containing the maximum number of nuclei was selected. A published STAR protocol was applied to measure lipid droplet area and the ‘roundness’ of lipid droplets (defined as the relationship between area and length of major droplet axis) in both control and experimental samples109.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data, workflows and FIJI algorithms necessary to interpret the data are included within the manuscript. Because of the large data set size, microscopy images and segmentation results are available upon request. Requests should be addressed to and will be fulfilled within a week by AKC. Source data are provided in this paper.
References
Eming, S. A., Wynn, T. A. & Martin, P. Inflammation and metabolism in tissue repair and regeneration. Science 356, 1026–1030 (2017).
Huang, W., Hickson, L. J., Eirin, A., Kirkland, J. L. & Lerman, L. O. Cellular senescence: the good, the bad and the unknown. Nat. Rev. Nephrol. 18, 611–627 (2022).
Pena, O. A. & Martin, P. Cellular and molecular mechanisms of skin wound healing. Nat. Rev. Mol. Cell Biol. 25, 599–616 (2024).
MacCarthy-Morrogh, L. & Martin, P. The hallmarks of cancer are also the hallmarks of wound healing. Sci. Signal. 13, https://doi.org/10.1126/scisignal.aay8690 (2020).
Rybinski, B., Franco-Barraza, J. & Cukierman, E. The wound healing, chronic fibrosis, and cancer progression triad. Physiol. Genomics 46, 223–244 (2014).
Martinez-Reyes, I. & Chandel, N. S. Cancer metabolism: looking forward. Nat. Rev. Cancer 21, 669–680 (2021).
Stine, Z. E., Schug, Z. T., Salvino, J. M. & Dang, C. V. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 21, 141–162 (2022).
Wong, K. K. L. & Verheyen, E. M. in DMM Disease Models and Mechanisms (Company of Biologists Ltd, 2021).
Meacham, C. E., DeVilbiss, A. W. & Morrison, S. J. Metabolic regulation of somatic stem cells in vivo. Nat. Rev. Mol. Cell Biol. 23, 428–443 (2022).
Zhu, J. & Thompson, C. B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 20, 436–450 (2019).
Eming, S. A., Murray, P. J. & Pearce, E. J. Metabolic orchestration of the wound healing response. Cell Metab. 33, 1726–1743 (2021).
Manning, B. D. & Cantley, L. C. AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 (2007).
Holz, M. K., Ballif, B. A., Gygi, S. P. & Blenis, J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569–580 (2005).
Hopkins, B. D., Goncalves, M. D. & Cantley, L. C. Insulin-PI3K signalling: an evolutionarily insulated metabolic driver of cancer. Nat. Rev. Endocrinol. 16, 276–283 (2020).
Wu, X., Xie, W., Xie, W., Wei, W. & Guo, J. Beyond controlling cell size: functional analyses of S6K in tumorigenesis. Cell Death Dis. 13, 646 (2022).
Hoxhaj, G. & Manning, B. D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 20, 74–88 (2020).
Saxton, R. A. & Sabatini, D. M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976 (2017).
Ring, N. A. R. et al. The p-rpS6-zone delineates wounding responses and the healing process. Dev. Cell 58, 981–992 e986 (2023).
Kakanj, P. et al. Insulin and TOR signal in parallel through FOXO and S6K to promote epithelial wound healing. Nat. Commun. 7, 12972 (2016).
Nakamura, M. et al. Autocrine insulin pathway signaling regulates actin dynamics in cell wound repair. PLoS Genet. 16, e1009186 (2020).
Wei, X., Luo, L. & Chen, J. Roles of mTOR signaling in tissue regeneration. Cells 8, https://doi.org/10.3390/cells8091075 (2019).
Fox, D. T., Cohen, E. & Smith-Bolton, R. Model systems for regeneration: Drosophila. Development 147, https://doi.org/10.1242/dev.173781 (2020).
Worley, M. I. & Hariharan, I. K. Imaginal Disc Regeneration: Something Old, Something New. Cold Spring Harb. Perspect. Biol. 14, https://doi.org/10.1101/cshperspect.a040733 (2022).
Cosolo, A. et al. JNK-dependent cell cycle stalling in G2 promotes survival and senescence-like phenotypes in tissue stress. Elife 8, https://doi.org/10.7554/eLife.41036 (2019).
Floc’hlay, S. et al. Shared enhancer gene regulatory networks between wound and oncogenic programs. Elife 12, https://doi.org/10.7554/eLife.81173 (2023).
Jaiswal, J. et al. Mutual repression between JNK/AP-1 and JAK/STAT stratifies senescent and proliferative cell behaviors during tissue regeneration. PLoS Biol. 21, e3001665 (2023).
Stevens, L. J. & Page-McCaw, A. A secreted MMP is required for reepithelialization during wound healing. Mol. Biol. Cell 23, 1068–1079 (2012).
La Fortezza, M. et al. JAK/STAT signalling mediates cell survival in response to tissue stress. Development 143, 2907–2919 (2016).
Crucianelli, C. et al. Distinct signaling signatures drive compensatory proliferation via S-phase acceleration. PLoS Genet. 18, e1010516 (2022).
Herrera, S. C. & Bach, E. A. JAK/STAT signaling in stem cells and regeneration: from Drosophila to vertebrates. Development 146, https://doi.org/10.1242/dev.167643 (2019).
Santabarbara-Ruiz, P. et al. ROS-Induced JNK and p38 Signaling Is Required for Unpaired Cytokine Activation during Drosophila Regeneration. PLoS Genet. 11, e1005595 (2015).
Worley, M. I. et al. Ets21C sustains a pro-regenerative transcriptional program in blastema cells of Drosophila imaginal discs. Curr. Biol. 32, 3350–3364 e3356 (2022).
Sustar, A., Bonvin, M., Schubiger, M. & Schubiger, G. Drosophila twin spot clones reveal cell division dynamics in regenerating imaginal discs. Dev. Biol. 356, 576–587 (2011).
Herrera, S. C., Martin, R. & Morata, G. Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet. 9, e1003446 (2013).
Dvorak, H. F. Tumors: wounds that do not heal. N. Engl. J. Med. 315, 1650–1659 (1986).
La Marca, J. E. & Richardson, H. E. Two-faced: roles of JNK signalling during tumourigenesis in the drosophila model. Front. Cell Dev. Biol. 8, 42 (2020).
Enomoto, M. & Igaki, T. Cell-cell interactions that drive tumorigenesis in Drosophila. Fly. 16, 367–381 (2022).
Liu, Y., Saavedra, P. & Perrimon, N. Cancer cachexia: lessons from Drosophila. Dis. Model Mech. 15, https://doi.org/10.1242/dmm.049298 (2022).
Bilder, D., Ong, K., Hsi, T. C., Adiga, K. & Kim, J. Tumour-host interactions through the lens of Drosophila. Nat. Rev. Cancer 21, 687–700 (2021).
Hodgson, J. A., Parvy, J. P., Yu, Y., Vidal, M. & Cordero, J. B. Drosophila Larval Models of Invasive Tumorigenesis for In Vivo Studies on Tumour/Peripheral Host Tissue Interactions during Cancer Cachexia. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22158317 (2021).
Colombani, J., Andersen, D. S. & Leopold, P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336, 582–585 (2012).
Garelli, A., Gontijo, A. M., Miguela, V., Caparros, E. & Dominguez, M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336, 579–582 (2012).
Agrawal, N. et al. The drosophila TNF eiger is an adipokine that acts on insulin-producing cells to mediate nutrient response. Cell Metab. 23, 675–684 (2016).
Figueroa-Clarevega, A. & Bilder, D. Malignant Drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev. Cell 33, 47–55 (2015).
Song, W. et al. Tumor-derived ligands trigger tumor growth and host wasting via differential MEK activation. Dev. Cell 48, 277–286 (2019).
Rajan, A. & Perrimon, N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151, 123–137 (2012).
Ding, G. et al. Coordination of tumor growth and host wasting by tumor-derived Upd3. Cell Rep. 36, 109553 (2021).
Lodge, W. et al. Tumor-derived MMPs regulate cachexia in a Drosophila cancer model. Dev. Cell 56, 2664–2680 (2021).
Romao, D., Muzzopappa, M., Barrio, L. & Milan, M. The Upd3 cytokine couples inflammation to maturation defects in Drosophila. Curr. Biol. 31, 1780–1787 (2021).
Gutierrez, E., Wiggins, D., Fielding, B. & Gould, A. P. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature 445, 275–280 (2007).
Cong, B. et al. JNK and Yorkie drive tumor malignancy by inducing L-amino acid transporter 1 in Drosophila. PLoS Genet. 17, e1009893 (2021).
Newton, H. et al. Systemic muscle wasting and coordinated tumour response drive tumourigenesis. Nat. Commun. 11, 4653 (2020).
Khezri, R. et al. Host autophagy mediates organ wasting and nutrient mobilization for tumor growth. EMBO J. 40, e107336 (2021).
Argiles, J. M., Busquets, S., Stemmler, B. & Lopez-Soriano, F. J. Cancer cachexia: understanding the molecular basis. Nat. Rev. Cancer 14, 754–762 (2014).
Setiawan, T. et al. Cancer cachexia: molecular mechanisms and treatment strategies. J. Hematol. Oncol. 16, 54 (2023).
Sun, F. & Poss, K. D. Inter-organ communication during tissue regeneration. Development 150, https://doi.org/10.1242/dev.202166 (2023).
Halme, A., Cheng, M. & Hariharan, I. K. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr. Biol. 20, 458–463 (2010).
Hackney, J. F. & Cherbas, P. Injury response checkpoint and developmental timing in insects. Fly. 8, 226–231 (2014).
Gontijo, A. M. & Garelli, A. in Mechanisms of Development 154, 44–50 (Elsevier Ireland Ltd, 2018).
Kashio, S. et al. Tissue nonautonomous effects of fat body methionine metabolism on imaginal disc repair in Drosophila. Proc. Natl. Acad. Sci. USA 113, 1835–1840 (2016).
Kashio, S. & Miura, M. Kynurenine Metabolism in the Fat Body Non-autonomously Regulates Imaginal Disc Repair in Drosophila. iScience 23, 101738 (2020).
Smith-Bolton, R. K., Worley, M. I., Kanda, H. & Hariharan, I. K. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev. Cell 16, 797–809 (2009).
Chatterjee, N. & Bohmann, D. A. versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PLoS ONE 7, e34063 (2012).
Worley, M. I., Alexander, L. A. & Hariharan, I. K. CtBP impedes JNK- and Upd/STAT-driven cell fate misspecifications in regenerating Drosophila imaginal discs. Elife 7, https://doi.org/10.7554/eLife.30391 (2018).
Harris, R. E., Stinchfield, M. J., Nystrom, S. L., McKay, D. J. & Hariharan, I. K. Damage-responsive, maturity-silenced enhancers regulate multiple genes that direct regeneration in Drosophila. Elife 9, https://doi.org/10.7554/eLife.58305 (2020).
Aviner, R. The science of puromycin: From studies of ribosome function to applications in biotechnology. Comput. Struct. Biotechnol. J. 18, 1074–1083 (2020).
Sustar, A. & Schubiger, G. A transient cell cycle shift in Drosophila imaginal disc cells precedes multipotency. Cell 120, 383–393 (2005).
Vallejo, D. M., Juarez-Carreno, S., Bolivar, J., Morante, J. & Dominguez, M. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science 350, aac6767 (2015).
Ingaramo, M. C., Sanchez, J. A., Perrimon, N. & Dekanty, A. Fat Body p53 regulates systemic insulin signaling and autophagy under nutrient stress via drosophila Upd2 repression. Cell Rep. 33, 108321 (2020).
Wang, M. C., Bohmann, D. & Jasper, H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell 121, 115–125 (2005).
Semaniuk, U. et al. Drosophila insulin-like peptides: from expression to functions – a review. Entomol. Exp. Appl. 169, 195–208 (2020).
Sudhakar, S. R. et al. Insulin signalling elicits hunger-induced feeding in Drosophila. Dev. Biol. 459, 87–99 (2020).
Manning, B. D. & Toker, A. in Cell 169, 381–405 (Cell Press, 2017).
Boulan, L., Andersen, D., Colombani, J., Boone, E. & Leopold, P. Inter-organ growth coordination is mediated by the Xrp1-Dilp8 axis in drosophila. Dev. Cell 49, 811–818 e814 (2019).
Jiang, H., Kimura, T., Hai, H., Yamamura, R. & Sonoshita, M. Drosophila as a toolkit to tackle cancer and its metabolism. Front Oncol. 12, 982751 (2022).
Katheder, N. S. & Rusten, T. E. Microenvironment and tumors-a nurturing relationship. Autophagy 13, 1241–1243 (2017).
Beller, M. et al. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 12, 521–532 (2010).
Ugrankar, R. et al. Drosophila snazarus regulates a lipid droplet population at plasma membrane-droplet contacts in adipocytes. Dev. Cell 50, 557–572 e555 (2019).
Kwon, Y. et al. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev. Cell 33, 36–46 (2015).
Shin, M., Cha, N., Koranteng, F., Cho, B. & Shim, J. Subpopulation of Macrophage-Like Plasmatocytes Attenuates Systemic Growth via JAK/STAT in the Drosophila Fat Body. Front. Immunol. 11, 63 (2020).
Hersperger, F. et al. DNA damage signaling in Drosophila macrophages modulates systemic cytokine levels in response to oxidative stress. Elife 12, https://doi.org/10.7554/eLife.86700 (2024).
Yue, S., Li, G., He, S. & Li, T. The Central Role of mTORC1 in Amino Acid Sensing. Cancer Res. 82, 2964–2974 (2022).
Lama-Sherpa, T. D., Jeong, M. H. & Jewell, J. L. Regulation of mTORC1 by the Rag GTPases. Biochem. Soc. Trans. 51, 655–664 (2023).
Altman, B. J., Stine, Z. E. & Dang, C. V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016).
Marti, I. L. A. A. & Reith, W. Arginine-dependent immune responses. Cell Mol. Life Sci. 78, 5303–5324 (2021).
Keshet, R., Szlosarek, P., Carracedo, A. & Erez, A. Rewiring urea cycle metabolism in cancer to support anabolism. Nat. Rev. Cancer 18, 634–645 (2018).
Lieu, E. L., Nguyen, T., Rhyne, S. & Kim, J. Amino acids in cancer. Exp. Mol. Med 52, 15–30 (2020).
Jewell, J. L., Russell, R. C. & Guan, K. L. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139 (2013).
Shimobayashi, M. & Hall, M. N. in Cell Research 26, 7–20 (Nature Publishing Group, 2016).
Liu, G. Y. & Sabatini, D. M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020).
Li, J. & Kim, S. G. & Blenis. J. Cell Metab. 19, 373–379 (2014).
Levina, A., Fleming, K. D., Burke, J. E. & Leonard, T. A. Activation of the essential kinase PDK1 by phosphoinositide-driven trans-autophosphorylation. Nat. Commun. 13, 1874 (2022).
Komander, D. et al. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 23, 3918–3928 (2004).
Rintelen, F., Stocker, H., Thomas, G. & Hafen, E. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. USA 98, 15020–15025 (2001).
Bilder, D., Li, M. & Perrimon, N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113–116 (2000).
Pagliarini, R. A. & Xu, T. A genetic screen in Drosophila for metastatic behavior. Science 302, 1227–1231 (2003).
Dillard, C., Reis, J. G. T. & Rusten, T. E. Ras(V12); scrib(-/-) Tumors: a cooperative oncogenesis model fueled by tumor/host interactions. Int. J. Mol. Sci. 22, https://doi.org/10.3390/ijms22168873 (2021).
Cao, X., Rojas, M. & Pastor-Pareja, J. C. Intrinsic and damage-induced JAK/STAT signaling regulate developmental timing by the Drosophila prothoracic gland. Dis. Model Mech. 15, https://doi.org/10.1242/dmm.049160 (2022).
Radimerski, T. et al. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell Biol. 4, 251–255 (2002).
Albina, J. E., Abate, J. A. & Mastrofrancesco, B. Role of ornithine as a proline precursor in healing wounds. J. Surg. Res 55, 97–102 (1993).
Ling, Z. N. et al. Amino acid metabolism in health and disease. Signal Transduct. Target Ther. 8, 345 (2023).
Zheng, N., Wei, J., Wu, D., Xu, Y. & Guo, J. Master kinase PDK1 in tumorigenesis. Biochim. Biophys. Acta Rev. Cancer 1878, 188971 (2023).
Lee, J., Ng, K. G., Dombek, K. M., Eom, D. S. & Kwon, Y. V. Tumors overcome the action of the wasting factor ImpL2 by locally elevating Wnt/Wingless. Proc. Natl. Acad. Sci. USA 118, https://doi.org/10.1073/pnas.2020120118 (2021).
Santabarbara-Ruiz, P. & Leopold, P. An Oatp transporter-mediated steroid sink promotes tumor-induced cachexia in Drosophila. Dev. Cell 56, 2741–2751.e2747 (2021).
Edwards-Hicks, J., Mitterer, M., Pearce, E. L. & Buescher, J. M. Metabolic dynamics of in vitro CD8+ T cell activation. Metabolites 11, https://doi.org/10.3390/metabo11010012 (2020).
Kierdorf, K. et al. Muscle function and homeostasis require cytokine inhibition of AKT activity in Drosophila. Elife 9, https://doi.org/10.7554/eLife.51595 (2020).
Maniere, G., Ziegler, A. B., Geillon, F., Featherstone, D. E. & Grosjean, Y. Direct sensing of nutrients via a LAT1-like transporter in drosophila insulin-producing cells. Cell Rep. 17, 137–148 (2016).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. methods 9, 676–682 (2012).
Dark, C., Cheung, S. & Cheng, L. Y. Analyzing cachectic phenotypes in the muscle and fat body of Drosophila larvae. STAR Protoc. 3, https://doi.org/10.1016/j.xpro.2022.101230 (2022).
Acknowledgements
We thank the staff of the Life Imaging Centre (LIC) in the Hilde Mangold House (HMH) of the Albert-Ludwigs-University of Freiburg for help with their confocal microscopy resources, and the excellent support in image recording. We specifically thank the DFG for supporting our imaging work through project number 414136422. We thank the Lighthouse Core Facility and its staff, as well as Elisabeth Savkova, Janhvi Jaiswal and Carlo Crucianelli for help with our experimental work. We thank Robin Harris, Susumi Hirobayashi, Hugo Stocker, Aurelio Teleman and Hong Xu, Iswar Hariharan, Dirk Bohmann for sharing reagents and the Bloomington Drosophila Stock Centre (BDSC), the Vienna Drosophila Stock Collection (VDRC), the University of Zurich ORFeome Project (FlyORF), the Developmental Studies Hybridoma Bank (DSHB) and the Monoclonal Antibody Core Facility at the Helmholtz Zentrum Munich for providing fly stocks and antibodies. We thank the IMPRS-EBM and SGBM graduate schools for supporting our doctoral researchers. Funding for this work was provided by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany ́s Excellence Strategy (CIBSS – EXC-2189) to K.K. and A.K.C., and by the DFG Heisenberg Programme to A.K.C. (668189) and to K.K. (544402801), as well as DFG grants to AKC (667603) and to K.K. (529943809). Funding was furthermore provided by the Boehringer Ingelheim Foundation (BIF Plus3 & Rise Up) to A.K.C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
Conceptualisation A.V.M. and A.K.C.; Data curation A.V.M., L.N.e, L.N.o, L.H., and A.K.C.; Formal analysis A.V.M., L.N.e, L.H., J.B., and A.K.C.; Funding acquisition K.K. and A.K.C.; Investigation A.V.M., L.N.e, L.N.o, A.S., and A.K.C.; Methodology A.V.M., L.N.e, L.N.o, A.S., I.G., J.B., and A.K.C.; Project administration I.G. and A.K.C.; Supervision K.K. and A.K.C.; Validation A.V.M., L.N.e, L.N.o, A.S., K.K., and A.K.C.; Visualisation A.V.M., L.N.e, and L.H.; Writing – original draft A.V.M. and A.K.C.; Writing – review & editing A.V.M., K.K., and A.K.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Vijayakumar Maya, A., Neuhaus, L., Nogay, L. et al. A JAK/STAT-Pdk1-S6K axis bypasses systemic growth restrictions to promote regeneration. Nat Commun 16, 10944 (2025). https://doi.org/10.1038/s41467-025-66995-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66995-z