Abstract
Coronaviruses (CoVs) maintain large RNA genomes that frequently undergo mutations and recombination, contributing to their evolution and emergence. In this study, we find that SARS-CoV-2 has greater RNA recombination frequency than other human CoVs. In addition, CoV RNA recombination primarily occurs at uridine (U)-enriched RNA sequences. Therefore, we next evaluate the role of SARS-CoV-2 NSP15, a viral endonuclease that targets uridines (EndoU), in RNA recombination and virus infection. Using a catalytically inactivated EndoU mutant (NSP15H234A), we observe attenuated viral replication in vitro and in vivo. However, the loss of EndoU activity also dysregulates inflammation resulting in similar disease in vivo despite reduced viral loads. Next-generation sequencing (NGS) demonstrates that loss of EndoU activity disrupts SARS-CoV-2 RNA recombination by reducing viral sub-genomic mRNA but increasing recombination events that contribute to defective viral genomes (DVGs). Overall, the study demonstrates that NSP15 plays a critical role in regulating RNA recombination and SARS-CoV-2 pathogenesis.
Similar content being viewed by others
Introduction
The emergence of the SARS-CoV-2 in 2019 resulted in a global pandemic with unprecedented economic disruption and 700 million cases worldwide1,2. While initial efforts to quell the outbreak focused on vaccination3, the development of SARS-CoV-2 variants of concern demonstrated the ability of the virus to evolve and evade host immunity4,5. As a result, “herd” immunity to COVID-19 has rendered a less deadly, but a still quite infectious and transmissible SARS-CoV-2. Importantly, the continued evolution of SARS-CoV-2 suggests that most people will face multiple infections and potential long-term complications including numerous manifestations of long COVID6,7.
Genomic mutation and recombination are the main drivers of CoV evolution. While employing an error prone polymerase like other RNA viruses, CoVs have a significantly lower mutation rate governed by a proofreading viral 3′ exonuclease8,9. Yet, the sheer number of SARS-CoV-2 infections worldwide has led to accumulation of advantageous mutations and evolution of variants5. RNA recombination offers a second mechanism for adaptation, shuffling of chunks of genetic sequence within and between virus strains10. Importantly, recombination is required for the CoV lifecycle including its generation of sub-genomic messenger RNA (sgmRNA) from discontinuous genome segments11. In addition, genetic and experimental analyzes reveal extensive recombination between virus strains of the CoV families12,13. Giving rise to hybrid and novel strains, these recombinant viruses may be the key to viral emergence and immune evasion. Finally, RNA recombination gives rise to defective viral genomes (DVGs) which play a complex and still unresolved role in engaging host immunity following infection14. These activities highlight the importance of RNA recombination to CoV infection and identify the need to better understand its underpinnings.
Despite playing a critical role, CoV RNA recombination is still poorly understood. Prior to the development of next-generation sequencing (NGS), analysis of recombination was severely limited and difficult to study15. Even early NGS approaches have complicated analysis with PCR duplication, error rate, and other sequencing artifacts reducing confidence in potential findings. However, as the SARS-CoV-2 pandemic continued, novel techniques and approaches have allowed further insights into CoV recombination. Initial work by our group found that SARS-CoV-2 is more recombinogenic than MERS-CoV15. The work has also implicated viral exonuclease NSP14 in playing a role in promoting recombination in mouse hepatitis virus. Similarly, recombination has been reported to induce significant DVG production following SARS-CoV-2 infection driving host immune responses14. Notably, SARS-CoV-2 was also shown to primarily recombine at uridine rich sequences15; these U-rich tracts are potential targets for CoV NSP15, a highly conserved viral endonuclease targeting uridines (EndoU)16,17,18,19,20. Prior work has shown CoV EndoU plays a critical role in preventing host sensor recognition by cleaving viral RNA and preventing interferon responses17,18,21,22,23. Given that RNA recombination junctions primarily occur at uridine-rich tracts, NSP15 may contribute to CoV RNA recombination.
In this study, we explore RNA recombination in the context of SARS-CoV-2 and other human CoVs. Using a refined analysis pipeline, we demonstrate that SARS-CoV-2 RNA is more recombinogenic than other human CoVs. We also show that RNA recombination occurs primarily at uridine-enriched tracts across each of the human coronaviruses (HCoVs). Mechanistically, the uridine-rich sequence at the RNA recombination junctions suggests a role for CoV endoribonuclease NSP15. Therefore, we generate a catalytically inactive NSP15 mutant (NSP15H234A). NSP15H234A shows attenuated viral replication in vitro and in vivo, but similar in vivo pathogenesis as wild-type (WT) infection, which is driven by augmented host responses characterized by both antiviral activity and inflammation mediated tissue damage. Surprisingly, loss of NSP15 activity increases recombination events in vitro including deletions and micro-deletions; yet, NSP15H234A also has reduced viral subgenomic mRNA formation. In vivo, NSP15H234A continues to show reduced viral subgenomic mRNA formation. In addition, NSP15H234A infected animals contain a viral population with reduced diversity but strong selection of particular DVG populations. Overall, our results highlight a critical role for NSP15 in modulating differential RNA recombination (facilitating sgmRNA formation but antagonizing DVGs), which may contribute to the development of viral infection, pathogenesis, and host immune responses.
Results
Increased RNA recombination in SARS-CoV-2 compared to other human coronaviruses
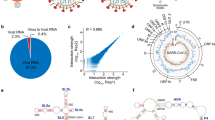
Our prior studies suggested that SARS-CoV-2 RNA was more recombinogenic than MERS-CoV RNA15. To determine if SARS-CoV-2 RNA produced greater recombination frequency than other HCoVs, we conducted additional experiments with SARS-CoV-2 and two common cold HCoVs strains, HCoV-OC43 and HCoV-229E. Briefly, we chose appropriate cell lines to infect with SARS-CoV-2 (Vero E6), HCoV-229E (HUH7), or HCoV-OC43 (HCT8) at low MOI. These cell lines were used to maximize infection of each CoV, and we harvested after significant cytopathic effect (>40%) was observed to maximize viral RNA yield. Total cellular RNA was collected, and NGS libraries were constructed using the random-primed ClickSeq approach24. NGS reads were processed and aligned to corresponding virus genomes, and analysis was conducted with bioinformatic pipeline “ViReMa (Virus Recombination Mapper)” to map the distribution of RNA recombination events (Fig. 1a)25,26. In addition, our previously published MERS-CoV data15 were reanalyzed using the same bioinformatic pipeline to facilitate comparisons.
a Schematic of RNA recombination reads that consist of gaps in linear genome, and the nucleotide positions flanking the recombination start (donor) and stop (acceptor) sites. b Cell culture infected with human coronaviruses and sequenced with random primers. SARS-CoV-2 RNA showed higher recombination tendency than other human coronaviruses. “JFreq” (junction frequency) measures the number of recombination events per 104 sequenced virus reads. c The RNA recombination of human coronaviruses utilizes U-rich sequences flanking the recombination junctions. b, c N = 3 biological replicates for SARS-CoV-2 and MERS-CoV; N = 2 biological replicates for hCoV-229E and hCoV-OC43. Statistics are conducted with one-way ANOVA, α = 0.05. Data are presented as: mean ± standard deviation.
Analyzing junction frequency (JFreq: the number of ViReMa-detected recombination junctions per 104 mapped viral reads15), revealed a significantly higher frequency of recombination for SARS-CoV-2 than MERS-CoV, HCoV-229E, and HCoV-OC43 (Fig. 1b). While the other HCoVs had comparable recombination events that hovered at JFreq of 70-76, SARS-CoV-2 had a ~1.8 fold greater JFreq of ~125. These results demonstrate that SARS-CoV-2 RNA is more recombinogenic than HCoVs 229E and HCoV-OC43, which is consistent with our earlier studies with MERS-CoV15. It remains unclear if the higher recombination rate is unique to SARS-CoV-2, but not to other HCoVs. While representative alpha (HcoV-229E) and beta (MERS-CoV, HCoV-OC43) CoVs lack the higher recombination rate, further study with other CoVs is required to determine conservation of the phenotype.
Human coronavirus RNA recombination occurs most frequently at U-rich tracts
Our previous studies found that SARS-CoV-2 RNA recombination was enriched at uridine-rich tracks flanking the “start” and “stop” sites of recombination junctions, both in cell culture and from human clinical specimens15,27. Here, we evaluated whether the U-favored RNA recombination applies to other HCoVs. The uridine and other nucleotide percentages were calculated at each upstream (−25 to −1) or downstream (+1 to +25) positions of the recombination junction, as well as the linear genome positions (Fig. 1a). Each recombination event was also weighted by abundance to provide a representative understanding of the nucleotide frequency at each position. However, contrasting the prior approach15, sub-genomic mRNA (sgmRNA) events were excluded due to their predominance and putatively different recombination mechanisms. We observed distinct peaks of uridine percentage (U%) flanking recombination junctions in all 4 HCoV infections (Fig. 1c), while no robust trends were observed in the other nucleotides (Supplementary Fig. 1). Our results indicate that the RNA recombination of all four HCoVs are most frequent at uridine(U)-rich sequences near both start (donor) sites and stop (acceptor) sites of RNA recombination and that the U-enriched RNA recombination is not unique to SARS-CoV-2 but applies to the other HCoVs tested.
Loss of EndoU activity attenuates SARS-CoV-2 replication in vitro
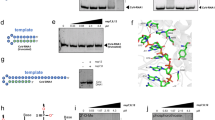
Given the propensity for CoV recombination to occur in uridine-enriched RNA tracts, we next focused on CoV non-structural protein 15 (NSP15), an endoribonuclease that cleaves RNA at uridine rich sites (EndoU)17. NSP15 is known to play a key role in evading type I interferon (IFN) by targeting viral RNA for cleavage and disrupting recognition by host sensors17,21. Importantly, NSP15 activity is maintained across the entire CoV family, and the active site residues are conserved (Fig. 2a, b)28. While established to play a role in CoV infection and immune evasion, we sought to determine if NSP15 EndoU activity impacts SARS-CoV-2 viral RNA recombination.
a Sequence alignment of Nsp15 endoribonuclease domain from different coronaviruses. b SARS-CoV-2 Nsp15 hexamer (grey) with catalytic amino acid residues labeled (blue). The histidine-to-alanine mutation at amino acid position 234 is in red. c Schematic of the Nsp15 structure showing the N-terminal domain (ND), middle domain (MD), and endoribonuclease domain (endoU). Nucleotides in red represent the 2-bp substitution in the H234A mutant. d Viral replication in Vero E6 cells infected with WT (black) or H234A (red) at MOI = 0.01. e Viral replication of Calu-3 2B4 cells infected with WT (black) or H234A (red) at MOI = 0.01. f Vero E6 cells were treated with control (solid) or 100 U type I interferon (IFN) (hashed) 16 h prior to infection with WT (black) or H234A (red) at MOI = 0.01. Viral replication was measured at 48 h post infection. The fold change relative to control is shown in brackets for each virus. d–f Data are presented as mean ± SD. Statistics are conducted with two-tailed Student’s t-test, α = 0.05, N = 6 from two experiments with three biological replicates each.
While NSP15 deletion mutants are not viable, catalytically inactive mutants have been recovered and characterized in several CoVs including mouse-hepatitis virus (MHV) and MERS-CoV21,29. Several studies have already established NSP15H234A and other mutations result in loss of NSP15 activity in biochemistry assays23,30,31,33. In this study, we took a similar approach by targeting amino acid H234 to ablate catalytic activity as previously described23 (Fig. 2a–c). Using our established SARS-CoV-2 reverse genetic system33,34, we generated a NSP15 mutant (NSP15H234A) capable of robust replication without significant changes in plaque morphology. Following inoculation of Vero E6 cells at MOI 0.01, the NSP15H234A exhibited modest attenuation (~0.5log) in viral replication at both 24- and 48-h post infection (HPI) relative to the WT SARS-CoV-2 virus (Fig. 2d). These results suggest that the NSP15H234A had a small impact on the viral replication capacity. We then examined viral replication in Calu-3 2B4 cells, an interferon (IFN)-responsive human respiratory cell line. We observed a more robust reduction (~1log) in viral replication of the NSP15H234A at both 24 and 48 HPI (Fig. 2e). Taken together, our data demonstrate that the disruption of the catalytic endoU domain in NSP15 attenuates viral replication in vitro.
NSP15H234A has increased sensitivity to type I interferon
Prior studies demonstrate the importance of CoV NSP15 in controlling the type I IFN response following infection17,21. To examine IFN sensitivity of NSP15H234A, we pretreated Vero E6 cells with 100 units of universal type I IFN and infected at MOI 0.01. Compared to untreated cells, WT SARS-CoV-2 had a modest reduction in viral replication (~6 fold) following type I IFN pretreatment, consistent with previous findings (Fig. 2f)34. In contrast, the NSP15 mutant virus had a 27-fold reduction in viral titers (Fig. 2f). These results indicate that the NSP15H234A is more sensitive to type I IFN responses than WT SARS-CoV-2. These results are consistent with findings from MHV and MERS-CoV21,29.
NSP15H234A attenuates viral replication, but not disease in vivo
Having established attenuation in vitro, we next evaluated the NSP15H234A in vivo using the Golden Syrian Hamster model of infection35,36. Briefly, 3-to-4-week-old golden Syrian hamsters were challenged with either WT SARS-CoV-2 or NSP15H234A at 105 focus forming units (FFU) and monitored for weight loss and disease over a 7-day time course (Fig. 3a). At 2, 4, and 7 days post infection, cohorts of animals were nasal washed under anesthesia, subsequently euthanized, and lung tissues were collected for further analyzes of viral titers and histopathology. Surprisingly, hamsters infected with NSP15H234A exhibited similar weight loss and disease as WT-infected animals (Fig. 3b). These results contrast in vitro findings and indicate that NSP15H234A maintains the capacity to cause significant disease in vivo.
a Schematic of golden Syrian hamster infection with WT or H234A SARS-CoV-2. Three-to-four-week-old golden Syrian hamsters were intranasally infected with 105 plaque forming units (PFU) of WT or H234A and monitored for weight changes and signs of disease over a 7-day time course. Hamsters were nasal washed, and lung lobes were collected at days 2, 4, and 7 post infection for viral titers and histopathology analyses. Created in BioRender. Menacherylab, T. (2025) https://BioRender.com/6ula8ve. b Percent body weight change from starting weight for WT or H234A infected groups. Viral titers were measured for nasal wash (c) and lung (d) for animals infected with WT or H234A at day 2, 4, and 7 post infection. b–d Data are presented as mean ± SD. Statistical analysis was measured using a two-tailed Student’s t-test. Statistics are conducted with two-tailed Student’s t-test, α = 0.05, N = 5 biological replicates. ND: not detected.
Examining viral load in the nasal wash and lung, we observed that both WT and NSP15H234A infected animals had similar viral titers at day 2 post infection (Fig. 3c, d). However, by day 4 post infection, we observed significant reductions in viral titers in the nasal wash (~2 log). While significantly reduced in the lung, the attenuation (~0.5 log) in NSP15H234A was small relative to WT-infected animals (Fig. 3c, d). By day 7, WT and NSP15H234A were both cleared from hamsters (Fig. 3c, d). Together, the data argued that the NSP15H234A mutant had modest attenuation in the lung versus a more robust attenuation in the upper airways.
Similarly, viral antigen staining in the lung also showed reduced infection in the NSP15H234A infected animals as compared to WT controls (Fig. 4). Airway, parenchyma, and overall lung antigen scoring showed a significant deficit in the NSP15H234A compared to WT at day 4 with similar trends at day 2 (Fig. 4a–c). While antigen distribution between the airway and parenchyma was similar, overall staining intensity and area were diminished in the mutant relative to WT at days 2 and 4 (Fig. 4d, e). Similar to viral titer (Fig. 3c, d), day 7 viral antigen staining was absent in both infection groups which is consistent with viral clearance (Fig. 4f, g). Together, the viral titer data and antigen staining demonstrate modest attenuation of viral replication in NSP15H234A despite significant weight loss following infection.
Scores of nucleocapsid antigen staining in (a) parenchyma, (b) airway, and (c) total from hamster left lung infected with WT or H234A. a–c Data are presented as mean ± SD. Statistics are conducted with two-tailed Student’s t-test, α = 0.05, N = 5 biological replicates. Representative images of viral antigen staining (nucleocapsid) of hamster lung sections at (d) day 2, (e) day 4, (f) day 7, and (g) mock.
Significant disease and damage observed following NSP15H234A infection
Having established reduced viral loads and antigen staining in the lung, we further evaluated disease and damage in the lung of NSP15H234A infected hamsters. Utilizing H&E staining, a certified pathologist examined lung sections from days 2, 4, and 7 following WT and NSP15H234A infection (Supplementary Fig. 2a). Despite differences in viral antigen staining, both WT and NSP15H234A infected hamsters had significant immune infiltration and damage relative to mock (Supplementary Fig. 2b–e). At day 2, little pathology or immune infiltration were observed, consistent with previous studies of SARS-CoV-2 in hamsters36,37,38 (Supplementary Fig. 2c). However, both WT and NSP15H234A viruses had significant disease at day 4 characterized by bronchiolitis, interstitial pneumonia, vasculitis, and alveolar cytopathology (Supplementary Fig. 2d). At day 7, we observed severe disease in both WT and NSP15H234A infected hamsters (Supplementary Fig. 2e); while the damage score from virus associated lesions were slightly reduced, the NSP15H234A mutant still produced significant disease despite reduced viral loads in the lung (Fig. 4). Together, the inflammation and immune infiltration likely lead to lung damage resulting in the similar weight loss and disease observed between mutant and WT virus infected animals.
NSP15H234A mutant induced augmented host immune responses in vivo
Coronavirus NSP15 has been shown to play an important role in cleaving viral RNA and preventing recognition by host innate immune sensors16,17,32,39,40. To evaluate changes in host responses, we profiled the transcriptomes of WT and NSP15H234A mutant at days 2 and 4 post infection. Total cellular RNA from hamster lung tissues was sequenced with Poly(A)-ClickSeq as previously described41,42. By mapping reads to the Mesocricetus auratus (Golden Hamster) reference genome, we obtained gene counts across 15606 annotated and unknown genes. Our results show divergent transcriptomic profiles between WT and NSP15H234A, especially at day 2 (Fig. 5a, red box). By day 4, this divergence was mostly diminished with the NSP15H234A-infected lungs having a similar gene expression profile to WT infected lungs. Principal component assay (PCA) confirms that the host responses against NSP15H234A diverged from WT at day 2, while day 4 infections and PBS mock controls clustered respectively (Fig. 5b). Gene expression analyzes (Supplementary Fig. 3a) revealed 1266 differentially expressed genes between WT and H234A at day 2 (Padj < 0.1, |fold change| > 1.5); 864 with increased expression in H234A and 402 with decreased expression (Fig. 5c). In contrast, only 57 differentially expressed genes were observed at day 4: 42 with increased expression in H234A and 15 with decreased expression (Supplementary Fig. 3a). The results indicate that differential host responses occur between NSP15H234A and WT infection at early times post infection.
a Hierarchical gene clusters based on average read count of each gene. H234A and WT showed distinctive gene expression profiles with particular clusters of genes (red box) at day 2 (D2) but not at day 4 (D4). b Principal component assay (PCA) showed that host responses against H234A progressed faster than WT at D2 but comparable at D4. c Up- and down-regulated genes between H234A and WT at D2 (Padj < 0.1, |log2FC| > 0.585, N = 3 biological replicates). d Top 10 gene ontology (GO) terms discovered from significantly (Padj < 0.1, |log2FC| > 1, N = 3 biological replicates) up- or down- regulated genes (WT vs H234A) at D2. e At D2, WT and H234A showed both correlated and anti-corelated gene expression profiles in comparison to mock. Red box: genes that are up-regulated for both WT and H234A; blue box: genes that are down-regulated by WT but up-regulated by H234A. f RNA fold expression (Log2 transformed) of individual innate immune genes in infected WT (black) and H234A (red) relative to time-matched mock hamster lungs. Data are presented as mean ± SD. Statistics are conducted with two-tailed Student’s t-test, α = 0.05, N = 3 biological replicates.
We further scrutinized how NSP15H234A induced different host responses than WT at day 2 (Fig. 5c). Among the upregulated genes, we identified enrichment of immune modulatory genes (e.g., Ifnb1, Ifnl3, Cxcl10) as well as several genes known for their function in dsRNA-binding and RNA cleaving/modification (e.g., Zbtb32, Rpp25, Aicda, Upp1, Ddx60, and Dhx58). Our results suggest that the catalytically active NSP15 endonuclease activity suppresses the activation of these host immune factors. We subsequently conducted DAVID43 gene ontology (GO) analysis using a curated list of 444 differentially upregulated genes and 171 differentially downregulated genes of H234A(D2) (Padj < 0.1, |log2FC| > 1, relative to WT(D2)). The top 10 (ranked by p-value) GO terms in biological process, cellular component, and molecular function are presented (Fig. 5d, and Supplementary Fig. 3b). Examining the GO biological processes, several are associated with either innate immunity or response to viral infection (innate immune response, defense response to virus, cellular response to lipopolysaccharide (LPS), response to virus, immune response), suggesting loss of NSP15 activity is driving increased host sensing and type I interferon responses. However, several GO processes augmented in the NSP15H234A-infected animals are associated with inflammation, overactive immune responses, and apoptosis (inflammatory response, positive regulation of NIK/NF-kB signaling, positive regulation of inflammatory response, apoptotic process). Together, the results indicate that NSP15H234A induces a robust antiviral response controlling virus infection coupled with excessive inflammation driving immune-mediated pathology.
To further understand the host responses, we compared the transcriptomics of WT and NSP15H234A vs. PBS mock control at day 2 and day 4 (Fig. 5e, and Supplementary Fig. 4a). At day 2, most differentially expressed genes (Padj < 0.1) between WT and NSP15H234A showed a clear correlation, indicating that host responses to NSP15H234A follow the same trajectory as WT, but elicit greater gene expression relative to WT (Fig. 5e). Examination of genes upregulated in both infections (Fig. 5e, red box) finds increased antiviral factors in the NSP15H234A-infected animals including interferon genes (e.g., Ifnb1, Ifnl3) and interferon stimulated genes (e.g., Rsad2, Ifit3, Ifit2, Mx1, Isg15, Bst2). These results are consistent with the finding that the loss of NSP15 activity increased host sensing of viral RNA and augmented innate immune responses. At the same time, the cluster of genes upregulated in both infection groups also has several inflammatory markers (e.g., Tnfaip6, Il1rn, Socs1, Tnfsf13b) and cytokines (e.g., Cxcl10, Ccl5, Cxcl9, Ccl19). This demonstrates that the lack of NSP15 activity also elicits stronger inflammatory responses. In addition, the anti-correlatively expressed genes between WT and NSP15H234A (suppressed in WT but induced in NSP15H234A, Fig. 5e, blue box) also have several factors associated with inflammation (e.g., Csf3, Pla2g2a, Ptgds, IL1b, MMp7), suggesting that NSP15H234A induces the expression of inflammatory genes which are typically suppressed in WT infection. By day 4, NSP15H234A gene expression is largely comparable to that of WT, with only 42 upregulated genes in the mutant (Supplementary Fig. 5a); these genes show a similar trajectory but with increased magnitude in the NSP15H234A (Supplementary Fig. 5b).
To further examine early innate host responses, we profiled individual gene expressions in the hamster lung at both day 2 and 4 post infection. Looking at type I (Ifnb1) and type III IFN (Ifnl3), we found that NSP15H234A had a significant increase in expression as compared to WT at day 2 (Fig. 5f). Similarly, interferon stimulated genes (Rsad1, Mx1, Isg15, Ifit3, Bst2) also had increased expression in the NSP15H234A compared to WT at day 2 (Fig. 5f, and Supplementary Fig. 4c). By day 4, gene expression in WT infected animals had recovered to similar levels as the mutant. While higher IFN responses correspond to diminished viral load in the NSP15H234A, we also noted increases in cytokines and inflammatory genes. NSP15H234A infected hamster lungs have augmented cytokine expression at day 2 including Cxcl10, Ccl5, Cxcl11, and Cxcl9 (Fig. 5f, and Supplementary Fig. 4d). In addition, inflammatory genes including Tnfaip6, Tnfsf13b, Pla2g2a and Il1rn were also increased at day 2 in NSP15H234A as compared to WT control (Fig. 5f, and Supplementary Fig. 4e). We also conducted qRT-PCR to verify that Ifnb1, Ifnl3 and Il1rn are significantly induced in NSP15H234A-infected lungs at D2 but not D4 (Supplementary Fig 4f). Combined with the broad network analysis, our results demonstrate that NSP15H234A drives an augmented immune response, pairing antiviral activity with excessive inflammation and damage during early infection.
NSP15H234A increased RNA recombination but reduced sgmRNAs in vitro
Having established viral replication attenuation and altered host responses, we next evaluated changes in viral RNA recombination of WT and NSP15H234A. Total cellular RNA was collected from Vero E6 cells infected with WT or NSP15H234A (Fig. 6a). NGS libraries were made using Tiled-ClickSeq (TCS)27, an approach that uses >300 primers targeting the entire SARS-CoV-2 genome to provide sensitive detection and even coverage across the virus genome. TCS also allows for improved RNA recombination resolution27 and better effectiveness than a random priming approach for sequencing SARS-CoV-2 genome from cell lysate (Supplementary Fig. 5a). We subsequently analyzed SARS-CoV-2 sequencing data and RNA recombination events with “ViReMa” as described above25,26.
a Schematic of RNA sequenced from monolayered cells infected by viruses. Created in BioRender. Zhou, T. (2025) https://BioRender.com/b8kgz7t. b 2D maps of recombination events and their frequencies from cell monolayer-infected by WT (black) or H234A (red). Each event represents a recombination sequence mapped to a start position (Y-axis, donor site) and a stop position (X-axis, acceptor site). c Across the entire genome, H234A showed significantly higher recombination (JFreq: number of recombination reads per 104 viral genome reads) than WT. d H234A showed altered utilization frequencies of uridines flanking the recombination junctions. e H234A showed increased JFreq of deletion, micro-deletion (MicroDel), but reduced intracellular sgmRNA. f The most abundant 9 canonical sgmRNAs, their mean recombination rates and the percentage change of sgmRNA between WT and H234A; g 1D map of deletion events (>25nts.) showed that H234A in general contained more genomic deletions than that of WT in cell lysate. h 1D map of micro-deletions events (3−25nts.) showed H234A contained more genomic micro-deletions than WT in cell lysate. A substantial micro-deletion in NSP6 was shared for both WT and H234A. Another predominant microdeletion in spike protein is unique to H234A but not WT. c, e, f Data are presented as mean ± SD. Statistics are conducted with two-tailed Student’s t-test, α = 0.05, N = 3 biological replicates. b, d, g, h Data are presented as mean (N = 3 biological replicates).
Using two-dimensional scatter plots, we visualized the frequency and location of RNA recombination events relative to their start and stop position and normalized the frequency of each recombination event to sequencing depth at the junction (Fig. 6b). This depicts several types of recombination events including: (1) canonical and non-canonical sub-genomic mRNA, bound on the X axis and capturing events between the Transcription Regulatory Sequence (TRS)-leader and the rest of genome (Fig. 6b, blue box); (2) micro-deletions (MicroDel) of <25 nucleotides along the X = Y axis (Fig. 6b, magenta box); (3) deletions (>25nts.) dispersed below the X = Y axis (Fig. 6b, green triangle). In addition, we also detected other RNA recombination events such as end fusion (recombination between 3′-end of genome to 5′-start of genome) and insertion events (long and micro-insertions). However, no statistical differences were found between WT and H234A for these events, and they are therefore depicted separately for clarity (Supplementary Fig. 5b).
While RNA recombination was observed in both infection groups, NSP15H234A infected cells produced significantly more RNA recombination than WT (Fig. 6b). Specifically, the NSP15H234A showed a ~16% increase in JFreq (recombination junctions per 104 mapped viral reads) compared to WT (Fig. 6c). Having established that the lack of NSP15 activity increases RNA recombination, we next evaluated if the loss of NSP15 activity altered the uridine-enriched sequences flanking SARS-CoV-2 RNA recombination junctions (Fig. 6d). Following NSP15H234A infection, we observed modest increase of uridine frequencies near RNA recombination start/stop sites mainly upstream of the junction, but no substantial differences in other nucleotides (Supplementary Fig. 6). These results suggest that endoU activity of NSP15 is not required for the uridine-favored RNA recombination in SARS-CoV-2.
Examination of the types of recombination indicated significant shifts between the WT and NSP15H234A (Fig. 6e). For example, despite more overall abundant recombination events, NSP15H234A infection produces less sub-genomic mRNAs (sgmRNA) as compared to WT (~27% reduction). From the nine canonical sgmRNAs, we found that six had significant reduction compared to WT (Fig. 6f). We also use the linear viral sequencing across the intact TRS-L as a proxy for viral genomic RNA and evaluate the ratio of sgmRNA to viral genomic RNA (Supplementary Fig. 5d). While the frequency of intact TRS-L is similar between infections, NSP15H234A showed a decreased TRS-L:TRS-B junction frequency that gives rise to sgmRNAs (Supplementary Fig. 5e). These changes result in a lower subgenomic/genomic RNA ratio and confirm reduced sgmRNA formation in NSP15H234A compared to WT.
In contrast to the sgmRNA, the frequency of deletion (>25 nts.) and micro-deletions (<25nts.) was significantly higher in the NSP15H234A as compared to WT (Fig. 6e), which are major contributors to DVGs14. For deletions, the NSP15H234A had a ~77% increase in JFreq relative to WT. Examination of the deletion distribution found them spread throughout the genome for both infections, but more abundant in NSP15H234A (Fig. 6g). Notably, despite more overall deletions in NSP15H234A, some sites were more abundant in WT cell lysate (e.g., start positions at nts.1854, 1988, and 6949). For micro-deletions, NSP15H234A also showed an increase in JFreq (~48%) compared to WT (Fig. 6e). Similar to deletions, NSP15H234A micro-deletions showed increased frequencies and distribution, especially towards the 3′end of the viral genome (Fig. 6h). Together, this demonstrates that NSP15 plays a role in divergent regulation of RNA recombination events: limiting deletions and micro-deletions but also facilitating the formation of sgmRNAs.
Notably, while micro-deletion events were mainly with low frequency (<1.5%), two high-frequency events were observed in our analysis. The first event, a high-frequency micro-deletion between nts.11078-11080 (NSP6, ~4.2–4.3%), was shared by both WT and NSP15H234A. This micro-deletion recombination results in a single nucleotide change (nt. 11079) and a frame shift in the open reading frame. In contrast, the second high-frequency micro-deletion recombination between nts. 23583 and 23599 was found to be ~30-fold more frequent in NSP15H234A than WT. This site corresponds to a deletion of the QTQTN motif found in the furin-cleavage loop of SARS-CoV-2 spike, a common attenuating mutation observed following passage in Vero cells36. We further scrutinized the accumulation of Δ11078-11080 (NSP6) and Δ23583-23599 (QTQTN) by sequencing the parental virus strains P1 stock (Supplementary Fig. 7). We found that both parental WT and NSP15H234A had the Δ11078-11080 (NSP6) with comparable frequencies, which may be the result of T7/RDRP processibility in adjacent regions (11075-(U)8-11082) or a consistent sequencing artifact at this poly U region. In contrast, neither parental virus showed elevated frequency of Δ23583-23599 (QTQTN). Together, the loss of NSP15 activity led to the rapid accumulation of this micro-deletion conferring a fitness advantage in Vero cells36.
NSP15H234A increased accumulation of defective viral genomes in virions
Cell associated RNA represents viral replication occurring in a complex intracellular environment under pressure by host anti-viral and immune processes. In contrast, purified virions provide a controlled microenvironment to investigate if cellular RNA recombination events carry forward during infection, such as viral particles composed of DVGs44,45,46. To this end, we collected the supernatant from Vero E6 cells infected with WT SARS-CoV-2 or NSP15H234A, pelleted virus particles using sucrose cushion (Fig. 7a) and conducted NGS and bioinformatic analyzes as described above.
a Schematics of RNA sequenced from purified virions. Created in BioRender. Zhou, T. (2025) https://BioRender.com/nduh00b. b 2D maps of recombination events and their frequencies from virions of infected Vero E6. c The purified virions recapitulated the increased JFreq of H234A than that of WT. d H234A mutant virus particles recapitulated the altered recombination U frequencies similar to that of cell monolayer. e While little sgmRNA was encapsidated by virus particles, H234A particles contained more DVGs such as deletions and micro-deletions. f The most abundant 9 canonical sgmRNAs found in supernatant fraction. g 1D map of deletion events (>25nts.) showed that H234A in general contained more genomic deletions than that of WT in supernatant. h 1D map of micro-deletions events (3-25nts.) showed H234A contained more genomic micro-deletions than that of WT in supernatant. Error bar: standard deviation. c, e, f Data are presented as mean ± SD. Statistics are conducted with two-tailed Student’s t-test, α = 0.05, N = 3 biological replicates. b, d, g, h Data are presented as mean (N = 3 biological replicates).
From purified virions, we found that viral RNA recombination trends recapitulated the events found in cell lysates (Fig. 7b). While the frequency of recombination events in virions decreased compared to cellular lysates, the NSP15H234Avirions showed significantly increased (~25%) JFreq compared to WT (Fig. 7c). Similarly, U-rich tracts were still the primary site for recombination in the virions with or without NSP15/endoU activity (Fig. 7d, and Supplementary Fig. 8). These results confirm that the NSP15H234A increased recombination without substantially compromising U-enriched tracts adjacent to recombination events in purified virions. Consistent with cellular data, no statistical differences were found between WT and NSP15H234A for end-fusion or insertion events (Supplementary Fig. 5c).
Examination of the types of recombination revealed similarities and differences with cellular RNA analysis. Only trace amounts of sgmRNAs were detected in purified WT and NSP15H234A virions (Fig. 7e, f, and Supplementary Fig. 5f). This is consistent with sgmRNAs not being packaged into virion and reflects the relative purity of the virion preparation. Similar to viral cellular RNA, both deletions (>25nts.) and micro-deletions (<25nts.) were found to be significantly increased in the NSP15H234A mutant relative to WT (~200% and ~58% increase of JFreq, respectively). Examining their distribution, the deletions had greater frequency and distribution across the genome in NSP15H234A compared to the WT (Fig. 7g). Similarly, the micro-deletion rates were also more abundant and diverse in NSP15H234A compared to WT (Fig. 7h). Notably, both highly abundant micro-deletions (Δ11078-11080 (NSP6) & Δ23583-23599 (QTQTN)) from cellular lysates were retained in the virions. In addition, the relative abundance was similar to cellular RNA findings with Δ11078-11080 at equivalent levels and Δ23583-23599 showed a ~29-fold increase over WT in NSP15H234A. Importantly, the results demonstrate that the increased deletion and micro-deletions produced by NSP15H234A infection are recapitulated in virus packaging and carried forward as DVGs. Supporting this idea, we found that the NSP15H234A mutant has threefold higher RNA/PFU ratio in supernatant relative to WT following infection of Vero cells (Supplementary Fig. 5g). These results are consistent with reduced titer of mutant virus (Fig. 2d), and indicate that the dysregulated recombination in NSP15H234A virus contributes to increases in defective virus particles. In addition, these DVGs may contribute to augmented immune responses during NSP15 mutant virus infections14.
NSP15 H234A altered RNA recombination landscape and sgmRNA formation in vivo
Having demonstrated that the NSP15H234A can significantly increase RNA recombination in vitro, we evaluated viral recombination events in vivo. Using total RNA from homogenized hamster lung tissue (Fig. 8a), we utilized tiled-clickseq and ViReMa to evaluate recombination events following WT and NSP15H234A infection. For both WT and NSP15H234A infected hamster lungs, we observed substantially fewer recombination events (Fig. 8b, c) as compared to cell lysate or virions (Figs. 6 and 7). This ~2–3 fold reduction in recombination JFreq indicates that the in vivo environment restricts accumulation of recombination events. Importantly, reduced recombination frequency in hamsters is consistent with findings from human autopsy samples14 and suggest more restrictive selection pressure impacts the accumulation of recombination events.
a Schematic of RNA sequenced from infected hamster lung. Created in BioRender. Zhou, T. (2025) https://BioRender.com/qad6dhx. b 2D maps showing the coordinates of recombination events in hamster lung and the average frequency of each event at day 2 (D2) and day 4 (D4). c Across the entire genome, H234A showed slightly reduced recombination rate than correspondent WT control, at both D2 and D4; d H234A continued to significantly restrict the formation of sgmRNA in hamster lungs. H234A showed a trend of decreased rate of deletions and micro-deletions. e In hamster lungs, WT and H234A showed different patterns of micro-deletions at D4: WT accumulated a myriad of near-background micro-deletions, while H234A contained fewer events but with higher frequencies. f Deletions at D4 exhibit similar patterns as micro-deletions. g Stacked circle plot highlights the divergent evolution of deletions in hamster lung at D4. Each circle represents a deletion event, and area of the circle represents the frequency. WT has more events, but lower frequency of each event. H234A has less events and therefore slightly reduced JFreq, but more frequency of each event. c, d Data are presented as mean ± SD. Statistics are conducted with two-tailed Student’s t-test, α = 0.05, N = 4 biological replicates. b, e–g Data are presented as mean (N = 4 biological replicates).
In both day 2 and day 4 lung samples, the NSP15H234A showed slightly reduced recombination events than WT, contrasting cellular and virion results. In the lung tissues, NSP15H234A showed a modest increased in U% flanking RNA recombination start and stop sites relative to WT (Supplementary Fig. 8b). This result is consistent with similar findings in vitro (Supplementary Fig. 6) and suggest that the endoU activity of NSP15 is not required for U-favored recombination of SARS-CoV-2.
Also consistent with in vitro findings, NSP15H234A maintained significant reduction in sgmRNA (Fig. 8d, and Supplementary Fig. 9c). At both day 2 and 4, NSP15H234A-infected hamsters had reduced sgmRNA JFreq (30% and 52%, respectively) relative to WT infected animals. Similarly, we observed a lowered ratio of sgmRNA to genomic RNA (Supplementary Fig. 9d, e) from the lungs of animals infected with NSP15H234A relative to infection with WT. These results demonstrate that sgmRNA formation is reduced in the absence of NSP15 activity in vivo, similar to in vitro results.
Examining recombination types also found varying trends between in vitro and in vivo infection with WT and NSP15H234A. Both WT and NSP15H234A had no significant differences between end-fusion, insertion and micro-insertion (Supplementary Fig. 9a), similar to cellular and virion analyzes. However, both WT and NSP15H234A demonstrate complexity in the context of deletion and micro-deletions. The NSP15H234A-infected hamster lungs exhibited a trend towards modest reductions in their total deletion and micro-deletions frequencies relative to WT (Fig. 8d). Examining further, the diversity and frequency of the recombination events highlight differences between WT and NSP15H234A. For example, at day 4, WT-infected hamster lungs accumulated one dominant (Δ11078-11080) and abundant low frequency micro-deletions (Fig. 8e). In contrast, the NSP15H234A -infected lungs produced micro-deletions with less diversity, but with higher frequencies at specific sites (Fig. 8e). The high frequency, low diversity micro-deletions indicate strong selection as evidenced by the absence of Δ23583-23599 (ΔQTQTN) event in NSP15H234A -infected lungs (Δ%WT-H234A = −0.1% on D2, −0.02% on D4). This ΔQTQTN mutation, highly penetrant in Vero cells, has been shown to be highly attenuated in vivo36. Similar to micro-deletions, the long deletion events in hamster lungs followed the same trend (Fig. 8f, and Supplementary Fig. 9 g), with NSP15H234A produced less deletion events but with higher frequencies at certain sites, especially at D4. We further demonstrate that NSP15H234A infected animals have higher frequencies of individual deletion/micro-deletion, in spite of reduced diversity of events (Supplementary Fig. 10). At D4, NSP15H234A -infected lungs had lower number of deletion/micro-deletion events (Supplementary Fig. 10a), but individual events are of higher frequency of recombination (Supplementary Fig. 10b). H234A-infected lungs also contained more high-frequency deletions/micro-deletions than WT (Supplementary Fig. 10c). To visualize these data, we created stacked circle plots (Fig. 8g, and Supplementary Fig. 10d), with each circle representing a recombination event whereas the size of the circle represents the frequency of the event. At D2, WT and H234A show similar population of recombination events. At D4, WT generates more events, but the frequency of each event remains comparable. NSP15H234A generates fewer events, its recombination events accumulated to higher frequencies. D4 marks the peak of anti-viral responses during hamster lung infection, as RNA expression changes (Fig. 5) precede protein secretions. This suggests that the selective pressure shapes the landscape of RNA recombination in NSP15H234A-infected lung that produces reduced number of events, but higher frequency in defective genome species. Notably, the outcome of selection pressure varies across individual animals (Supplementary Fig. 9h). Among four individual hamsters, NSP15H234A-infected lungs gave rise to several high-frequency micro-deletions that only occurred in a single animal. In contrast, WT-infected lungs contained background micro-deletions with low frequency in each individual, except for the common Δ11078-11080 (NSP6) event found in all animals from both groups at days 2 and 4. The increased frequency of deletion and micro-deletion events in NSP15H234A-infected at specific genomic positions in lungs (Fig. 8e, f) is consistent with in vitro findings, that the lack of NSP15 activity can still drive the accumulation of DVGs in vivo. On the other hand, the reduced distribution of these events demonstrate that these DVGs are shaped by a strong, individually divergent selective pressure in vivo.
Discussions
While severity and lethality of COVID-19 have largely declined, SARS-CoV-2 remains a global health problem due to its ability to evade host immunity through evolution. Viral RNA recombination plays a role in this process, and we show here that SARS-CoV-2 RNA recombines at a higher rate than other human CoVs (Fig. 1b). In addition, we confirm that hotspots for RNA recombination occur at uridine-rich sites across all CoVs tested (Fig. 1c). Importantly, we also demonstrate a role for viral endonuclease NSP15 in balancing SARS-CoV-2 RNA recombination. In the absence of NSP15 catalytic activity (NSP15H234A), infected cells and purified virions accumulated more genomic deletions and micro-deletions. (Figs. 6 and 7). In contrast, the sgmRNA recombination is reduced in NSP15H234A compared to WT. The in vivo model represents a stronger selective pressure and hence, lowered overall recombination rate (Fig. 8). In vivo, we recapitulate the reduced sgmRNA formation in NSP15H234A compared to WT. In addition, the absence of NSP15 activity suppresses the diversity of recombination deletions but increases the frequency of a subset of events (Fig. 8). Combined with transcriptomic analyzes, loss of NSP15 activity renders a stronger, earlier interferon and inflammatory responses in hamster lungs (Fig. 5). The induced antiviral state reduces viral replication, but also promotes immune-mediated damage, resulting in similar disease in hamsters infected with either mutant or WT virus (Figs. 2 and 3). Overall, our studies demonstrate that NSP15 catalytic activity plays a critical role in controlling host responses, facilitating sgmRNA formation, and limiting DVG accumulation during SARS-CoV-2 infection.
NSP15 endonuclease activity has previously been found to be a critical factor in controlling the host innate immune response17,18,20,21,22,23. Showing a preference for cleaving uridines and poly U tracts28,47,48,49, NSP15 endonuclease activity suppresses production of pathogen-associated molecular patterns and impairs immune sensing. The loss of NSP15 activity has also been associated with increased sensitivity to type I interferon treatment and attenuation of CoV replication17,18,20,21,22,23. Our study largely confirms these findings with enhanced sensitivity to type I IFN pretreatment (Fig. 2) and amplification of antiviral gene expression (Fig. 5) following NSP15H234A infection. However, we also observed an increased expression of genes associated with inflammation and cytokines as well as significant lung pathology following NSP15H234A infection. Together, our results show that the loss of NSP15 activity promotes an amplified immune response resulting in both viral suppression and immune-mediated damage.
Prior studies demonstrate that loss of NSP15 in CoVs is related to the increased dsRNA levels which leads to enhanced activation of immune sensors23,32,51. Our work adds50 additional details related to CoV NSP15 activity and host responses. Prior studies have shown that dysregulated viral RNA recombination drives virus-host interactions14,45,46,51, as well as cytokine production in lung tissues20. Recent studies from COVID-19 positive patients also reveal the critical connection between RNA recombination and host IFN responses14. Here, our work demonstrated loss of NSP15 results in dysregulated RNA recombination (Figs. 6–8) and coincides with the elevated cytokine responses in hamster lungs (Fig. 5). Thus, we contend that the elevated interferon and inflammatory responses in NSP15 mutant may be attributed to dysregulated RNA recombination. The excess recombination in the NSP15 mutant would provide a mechanism for the reported increased in dsRNA formation as well as increased DVGs (Fig. 7) which both induce strong host innate immune responses.
In addition to host immune induction, our studies demonstrate the impact of selection pressure on viral recombination outcomes. Here, we utilize two distinct systems to explore recombination: in vitro and in vivo. Our in vitro model uses Vero E6 cells which are a highly permissive model lacking type I IFN responses; in this system, the majority of cells are infected, and viral RNA can evolve without notable selective pressure from intrinsic or bystander cell host immunity. In contrast, the in vivo hamster model is a restrictive model that controls and clears viral infection over time. The combination of few cells infected, robust bystander responses with interferon capacity, and infiltration of host immune cells renders significant selection pressure absent in the Vero E6 model.
A more restrictive system explains the different recombination landscape between WT and NSP15H234A in vivo (Fig. 8), as compared to in vitro (Figs. 6 and 7). Importantly, it also provides context for the divergent results of overall recombination (JFreq) between in vitro and in vivo (Figs. 6b and Fig. 8b). We postulate that the differences are a product of the more restrictive in vivo environment. The elevated host anti-viral immunity during NSP15H234A infection (Fig. 5) actively clears viral RNA with disadvantageous recombination events (e.g., Δ23583-23599(QTQTN)), resulting in reduced JFreq in vivo. Supporting this idea, the deletions and micro-deletions found in NSP15H234A infected animals start to differentiate towards high individual frequency but low overall abundance at D4 (Supplementary Fig. 10d), after eliciting mRNA expressions of anti-viral genes (Fig. 5). Loss-of-NSP15 induced high recombination frequency and is consistent in our in vitro (Fig. 6g, h) and in vivo (Fig. 8e, f) data. We think that the high frequency, low abundance deletions/ microdels in NSP15H234A infected animals are a combination of induced RNA recombination rate (high frequency) and elevated host anti-viral clearance (low abundance). Together, these factors drive viral genomic evolution in hamster lungs. In comparison, from D2 to D4, WT infected animals only show increased overall abundance (more recombination events) without increased frequency (Supplementary Fig. 10d), suggesting minimal evolutionary pressure. Notably, these recombination sites in NSP15H234A are not conserved across multiple animals; this suggests that they are not a product of selection, but rather from more divergent recombination in the mutant (Fig. 8g). We predict at early times post in vivo infection (D0-D1), NSP15H234A may produce increased overall recombination rate relative to WT; however, as host immunity is activated and suppresses NSP15H234A infection earlier, recombination events are reduced due to increased selection at the later time points captured in our analysis. Further examination of recombination kinetics is needed to clarify these questions.
Our manuscript also provides critical insights into key elements of CoV recombination and evolution. While structurally and functionally conserved, the catalytic activity of NSP15 endonuclease varies among coronavirus species49 and variants52. Recent studies showed that many of the prevalent SARS-CoV-2 variants convey decreased nuclease activity of NSP15. Here, we showed that the loss of NSP15 endoU activity leads to higher RNA recombination. This suggests a potential link between reduced endoU activity and increased recombination which may explain differences between SARS-CoV-2 and other CoVs. Consistent with this idea, a change in endoU activity may alter virus evolution and contribute to rapid development of novel variants. While more frequent RNA recombination may be unique to SARS-CoV-2, it is unclear if this trait is conserved in other sarbecoviruses and requires further study. Notably, for all CoVs tested, uridine rich tracts serve as the primary site for recombination (Fig. 1c) and the process operates independently of NSP15 activity. While we had initially postulated that NSP15 endoU serves to provide cleaved template RNA to facilitate recombination, we instead found accelerated recombination in its absence suggesting a key role in regulation of overall recombination.
Our results also provide experimental insights to connect NSP15 endoU activity to the correct formation of SARS-CoV-2 canonical sgmRNAs. The coronavirus TRS is a conserved RNA motif that resides at stem loop 3 of virus 5’UTR53. The production of subgenomic mRNA (sgmRNA) relies on the correct recognition of the complementarity between leader TRS (TRS-L) and the recombination to body TRS (TRS-B)54,55. The conserved Sarbecoviruses “AACGAAC” TRS-L motif56 is A-rich in positive sense and U-rich in negative sense viral RNAs, which is also flanked by A/U-rich sequences both up- and down-stream. It is conceivable that the endoU activity of NSP15 may play a role in mediating the template-switching between +gRNA and -sgmRNA to give rise to +sgmRNA. Indeed, previous studies speculated that NSP15 cleavage of TRS is required to form sgmRNA57,58. Our data demonstrated that the loss of endoU activity significantly downregulated the formation of TRS-L and TRS-B recombination both in vitro (Figs. 6 and 7) and in vivo (Fig. 8). This provides experimental evidence that NSP15 plays a critical role to “proofread” the correct recombination between TRS-L and TRS-B.
Our results also provide mechanistic insight into the role of NSP15 beyond viral IFN antagonist. Our data show augmented host immune responses in the NSP15H234A, but also a shift in DVG formation. The dual impact of limiting innate immune sensing and production of immunogenic DVGs highlight the crucial role for NSP15 during SARS-CoV-2 infection. It is also possible that NSP15 activity varies across the CoV family, modulating the levels of recombination and possibly explaining the increased recombination observed in SARS-CoV-2. Notably, it also questions the safety of targeting NSP15 for drug and therapeutic treatment. While targeting NSP15 activity may attenuate viral replication, it may also promote immune-mediated damage as a byproduct of treatment. In addition, NSP15 targeting may increase viral recombination permitting more rapid formation of resistance to this and other treatments. Given the mutagenesis concerns associated with molnupiravir treatment, similar safety challenges might be associated with NSP15-targeted treatments.
Overall, this research provides a detailed exploration of SARS-CoV-2 recombination in vitro and in vivo. Our results confirm higher recombination in SARS-CoV-2 primarily at uridine-rich tracts. Importantly, we show that SARS-CoV-2 NSP15 endonuclease activity is key to balancing recombination in cells, in virions, and in vivo. Together, the work highlights novel elements of CoV recombination and novel mechanistic insights into how NSP15 modulates host immunity and DVG production.
Methods
All studies were conducted under protocols approved by the institutional biosafety committee at the University of Texas Medical Branch. Similarly, animal studies were conducted under protocols approved by the UTMB Institutional Animal Care and Use Committee and complied with USDA guidelines in a laboratory accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. UTMB PHS Animal Welfare Assurance Number: A3314-01
Cell culture
Vero E6, Huh7, and HCT-8 cells were grown in high glucose Dulbecco modified Eagle medium (DMEM; Gibco #11965–092) supplemented with 10% fetal bovine serum (FBS) (HyClone #SH30071.03) and 1% antibiotic-antimycotic (Gibco #5240062). Calu3 2B4 cells were cultured in high glucose DMEM supplemented with 10% defined FBS, 1 mM sodium pyruvate (Gibco), and 1x antibiotic-antimycotic. Cells were maintained at 37 °C in a humidified incubator with 5% CO2.
Viruses
The recombinant and mutant SARS-CoV-2 viruses were generated based on the USA-WA1/2020 sequence provided by the World Reference Center for Emerging Viruses and Arboviruses, which was originally obtained by the Center for Disease Control and Prevention59. The Nsp15 mutant was constructed with restriction enzyme-based cloning techniques and our reverse genetic system as previously described32. Virus stocks were amplified using Vero E6 cells. Viral RNA was extracted from recovered viruses, and the mutation was verified using NGS as previously described27.
In vitro infection
In vitro infection of Vero E6 and Calu3 2B4 cells was performed as previously described36. Briefly, Vero E6 or Calu3 2B4 cells were seeded in a 6-well plates or T-175 flasks. HUH7 (HCoV-229E) and HCT8 (HCoV-OC43) were seeded in T-175 flasks. For experiments involving IFN-I pre-treatment, Vero E6 cells were treated with 100 units of Universal Type I IFN for 16 h prior to infection. Cell growth media was removed and infected with either WT (SARS-CoV-2, HCoV-229E, HCoV-OC43) or mutant SARS-CoV-2 virus (NSP15H234A) at an MOI of 0.01 for 45 min at 37 °C. Following adsorption, cells were washed three times with phosphate-buffered saline, and fresh growth medium was added. Three or more biological replicates were collected at each time point. Viral titers of the samples were subsequently determined using focus-forming assay as previously described35,37. Following in vitro infection, culture supernatant was harvested, clarified, and virus particles were pelleted by ultracentrifugation with previously established methods36.
In vivo infection
Hamster infection studies
Three-to-four-week-old male golden Syrian hamsters were purchased from Envigo. Procedures involving infectious SARS-CoV-2 were performed in the Galveston National Laboratory ABSL3 facility. Animals were housed in ventilated cages prior to the study. Animals were intranasally infected with 105 FFU of WT or H234A in 100-μl inoculum or mock-infected with PBS. Animals were monitored daily for weight loss and signs of clinical disease for up to 7 days post infection (DPI). On days 2, 4, and 7, five animals from each group were anesthetized with isoflurane (Henry Schein Animal Health) and nasal washed with PBS and subsequently euthanized with CO2 for organ collection. Lung lobes were collected in either PBS for viral titers, RNAlater (Invitrogen #AM7021) for NGS/gene expression, or 10% phosphate-buffered formalin (Fisher #SF100) for histopathology.
Histology
Left lung lobes were collected and fixed in 10% buffered formalin for at least 7 days. Fixed tissues were paraffin-embedded, sections cut into 5-µM thickness and stained with hematoxylin and eosin on a SAKURA VIP 6 tissue processor at the University of Texas Medical Branch Surgical Pathology Laboratory. For viral antigen staining, tissue sections were deparaffinized and reacted with SARS-CoV-2 N-specific primary antibody and incubated with a secondary horseradish peroxidase (HRP)-conjugated anti-rabbit antibody as previously described37. Viral antigen was visualized and scored blinded on a scale of 0 (none) to 3 (most) in 0.25 increments with scores averaged from at least two sections from each hamster. Scores based on N staining in the parenchyma, airways, and overall lung section and scored by trained investigators. Hemoxylin and eosin (H&E) staining was performed by the University of Texas Medical Branch Histology Laboratory and then analyzed and scored by a blinded board-certified pathologist. Pathology scored based on combination of lung damage, immune infiltration, and overall involvement across multiple lung sections per animal on scale of 0–100.
Virus quantitation
For in vitro samples, viral titers were measured using focus forming assay as previously described37. Briefly, hamster lung lobes were homogenized with zirconia beads in a MagNA Lyser instrument (Roche Life Science) and clarified with low-speed centrifugation. Vero E6 cells were seeded in 96-well plates to achieve 100% confluency at the time of titration. A tenfold serial dilution was performed for virus-containing supernatant, and 20 μL of the dilutions were transferred to Vero E6 cells after the culture medium was removed. Cells were incubated for 45 min at 37 °C with 5% CO2 to allow adsorption before 0.85% methylcellulose overlay was added. After removing the overlay, cells were washed 3 times with PBS before fixation in 10% buffered formalin for 30 min at room temperature. Cells were permeabilized and incubated with SARS-CoV-2 nucleocapsid antibody (Cell Signaling) followed by Alexa FluorTM 555-conjugated α-mouse secondary antibody (Invitrogen). Fluorescent foci images were captured on Cytation 7 imaging multi-mode reader (BioTek) and foci were counted with ImageJ.
Next generation sequencing (NGS) libraries
For NGS analyzes, RNA template was extracted from infected cells, supernatant or animal tissue with Direct-zol RNA miniprep kits (Zymo Research).
To sequence different HCoVs, a random hexamer (N6) primer was used with standard “ClickSeq”24. In brief, viral RNAs were extracted from cell lysate and reverse transcribed with N6 and 1:35 azido-ddNTP:dNTP ratio. The azido-ddNTP-terminated cDNAs were “click-ligated” with a 3′-alkyne modified adapter. The ligated cDNAs then underwent final PCR to fulfill illumina library structure. Gel selected libraries were single-end sequenced with illumina NextSeq 2K.
To further investigate the recombination rate of SARS-CoV-2, we used “Tiled-ClickSeq”27 approach, which uses >300 primers specific to SARS-CoV-2 genome and ClickSeq components24,60 to achieve sensitive detection and even coverage of coronavirus genome. The extracted RNAs from cell or animal tissue were used as template for Tiled-ClickSeq libraries with standard protocol27. Gel selected libraries were pair-end sequenced with illumina NextSeq 2K and the R2 reads were used to map primer sequences.
To understand the transcriptomic changes of infected animals, the extracted total cellular RNA from hamster lungs were subjected to PolyA-ClickSeq41, which utilizes a semi-anchored oligo(dT) primer (5′- (T)21-3′) to specifically target polyA tails of cellular mRNA and a 1:5 azido-ddNTP:dNTP ratio to ensure sufficient termination of cDNA. The PolyA-ClickSeq library was constructed with previously established protocol61, and gel selected libraries were single-end sequenced with ElementBio Aviti.
Bioinformatics and data analysis
Raw reads sequenced from Tiled-ClickSeq libraries are processed and analyzed with previously established bioinformatic pipelines TCS (https://github.com/andrewrouth/TCS) with parameters “-p PMV”. In brief, after initial quality filter and trimming of illumina adapter, the detected primer sequences from R2 reads will subsequently be trimmed from respective R1 reads (therefore excluding potential primer-genome recombination). The processed R1 reads are then mapped to SARS-CoV-2 reference genome (NCBI reference: NC_045512.2). The same processed R1 reads are also analyzed with ViReMa (version 0.28) (https://sourceforge.net/projects/virema/) with parameters “--ErrorDensity 2,30 --Seed 25 --Defuzz --X 3”. The ViReMa-output BED files are further analyzed to categorize different recombination events with custom python script “plot_cs_freq.py” (included in the TCS package), which defines the length of microindel to be within 25nts. 2D-maps of ViReMa-mapped recombination events are plotted with ViReMaShiny62 (https://routhlab.shinyapps.io/ViReMaShiny/) with modifications for cosmetic and style.
Raw reads sequenced from random primed libraries are processed with fastp63 for adapter removal and quality control. To remove potential artifacts of random hexamer, all R1 reads were trimmed by 8 bases from 3′-direction (“-t 8”). For each virus, the corresponding virus genome was first polished with Pilon64 with sequenced reads to improve mapping efficiency. This is followed by ViReMa mapping of recombination reads to the Pilon64-polished viral genomes (NCBI reference number SARS-CoV-2: NC_045512.2; MERS-CoV: NC_019843.3; hCoV-229E: NC_002645.1; hCoV-OC43: ATCC VR-1558) with the same parameters stated above.
For recombination events detected by ViReMa, “JFreq” (junction frequency) is the metric to reflect overall number of recombination events of the sample. JFreq is normalized by measuring (the total number of recombination events)/(104 sequenced virus reads of individual sample). Recombination frequency (in percentage) is the metric to reflect individual recombination event frequency at the junction, which is normalized by (the number of individual event)/(sequencing coverage at the junction).
Raw reads sequenced from PolyA-ClickSeq libraries are processed and analyzed with the previously established DPAC42 pipeline (https://github.com/andrewrouth/DPAC/) with parameters “-p PMBCD”, which detects and processes the polyA-containing reads and maps the upstream sequence of polyA tail to host reference. In this study, a PolyA-site clustering database was curated based on the published reference genome and annotation of M. auratus (Genbank accession number: GCA_000349665.1). Due to the incompleteness of the reference genome, the genome mapping criterion was slightly loosened in hisat265 stage with parameter “--score-min L,0,-0.3”. Differential gene expression profiles of mock (1X PBS), WT, and H234A infected hamster lung tissues were then analyzed with DESeq266 that has been integrated in the DPAC pipeline (D stage) to reveal the normalized read count of each annotated gene.
For differential gene expression analyzes, hierarchical gene clustering was conducted with Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/). This is followed by TreeView (http://jtreeview.sourceforge.net/) to visualize the heat map and gene clusters. A gene is considered differentially expressed with Benjamini and Hochberg Padj < 0.1 and |log2FC| > 0.585. Gene ontology assay was conducted with DAVID43 and differentially expressed genes (Padj < 0.1, |log2FC| > 1), to highlight the most direct GO terms in biological process, cellular component, and molecular function.
Stacked circle plots are made with Hiplot server (https://hiplot.org). All other statistics are conducted on GraphPad Prism (v.10.6.1).
qRT-PCR
For viral RNA, qRT-PCR is conducted with previously established method67, with QuantStudio5 qPCR system and RdRp-specific forward primer (5′-GTGARATGGTCATGTGTGGCGG) and reverse primer (5′-CARATGTTAAASACACTATTAGCATA). For hamster lung host mRNA, qRT-PCR is conducted with NEB Luna Universal qPCR system with the following primer sets: Ifnb1 (F: 5′- AATCATTAGAAGGCTCACTTG; R: 5′- CTTTGACCTTTCAGATGCT), Ifnl3 (F: 5′- TGCAGGAGTTCTGCAGTGT; R: 5′- CATTCTGTGGGGTCTGAGTC), Il1rn (F: 5′- TGTGAAGTGATGGTACAC; R: 5′- CAGAGGGATTCAGACTATTG). GAPDH (F:5′-CCTGGATCATCCATCCAC; R: 5′-GAAACTGTGAGGGAGATG) is used as a reference gene to normalize gene expression levels which are represented as fold change over mock samples, as previously described67.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequencing data of this study are available in the NCBI sequence read archive (SRA) with accession number: PRJNA1131338, PRJNA1154272. MERS-CoV data are reanalyzed from existing SRA project: PRJNA62301615. Supplementary Figs. 1-10 are provided as Supplementary Figs. A list of sorted recombination deletion and micro-deletion frequencies from in vitro and in vivo samples are provided in Supplementary data 1. Raw reads count and mapping rate are listed in Supplementary data 2. A description of all supplementary files is included. Source data are provided in Source Data File. Source data are provided with this paper.
References
WHO COVID-19 dashboard, https://data.who.int/dashboards/covid19/cases (2024).
Gralinski, L. E. & Menachery, V. D. Return of the Coronavirus: 2019-nCoV. Viruses 12, https://doi.org/10.3390/v12020135 (2020).
Amanat, F. & Krammer, F. SARS-CoV-2 vaccines: status report. Immunity 52, 583–589 (2020).
DeGrace, M. M. et al. Defining the risk of SARS-CoV-2 variants on immune protection. Nature 605, 640–652 (2022).
Plante, J. A. et al. The variant gambit: COVID-19’s next move. Cell Host Microbe 29, 508–515 (2021).
Davis, H. E., McCorkell, L., Vogel, J. M. & Topol, E. J. Author correction: long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21, 408 (2023).
Bowe, B., Xie, Y. & Al-Aly, Z. Acute and postacute sequelae associated with SARS-CoV-2 reinfection. Nat. Med. 28, 2398–2405 (2022).
Fehr, A. R. & Perlman, S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23 (2015).
Minskaia, E. et al. Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 103, 5108–5113 (2006).
Wells, H. L. et al. The coronavirus recombination pathway. Cell Host Microbe 31, 874–889 (2023).
Sola, I., Almazan, F., Zuniga, S. & Enjuanes, L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2, 265–288 (2015).
Keck, J. G. et al. In vivo RNA-RNA recombination of coronavirus in mouse brain. J. Virol. 62, 1810–1813 (1988).
Makino, S., Keck, J. G., Stohlman, S. A. & Lai, M. M. High-frequency RNA recombination of murine coronaviruses. J. Virol. 57, 729–737 (1986).
Zhou, T. et al. Generation and functional analysis of defective viral genomes during SARS-CoV-2 infection. mBio 14, e0025023 (2023).
Gribble, J. et al. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 17, e1009226 (2021).
Deng, X. et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. USA 114, E4251–E4260 (2017).
Hackbart, M., Deng, X. & Baker, S. C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. USA 117, 8094–8103 (2020).
Deng, X. et al. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 93, https://doi.org/10.1128/JVI.02000-18 (2019).
Otter, C. J. et al. SARS-CoV-2 nsp15 endoribonuclease antagonizes dsRNA-induced antiviral signaling. Proc Natl Acad Sci USA. 121, e2320194121 (2024).
Caobi, A. et al. SARS-CoV-2 nsp15 enhances viral virulence by subverting host antiviral defenses. Proc. Natl. Acad. Sci. USA 122, e2426528122 (2025).
Kindler, E. et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 13, e1006195 (2017).
Volk, A. et al. Coronavirus endoribonuclease and deubiquitinating interferon antagonists differentially modulate the host response during replication in macrophages. J. Virol. 94, https://doi.org/10.1128/JVI.00178-20 (2020).
Otter, C. J. et al. SARS-CoV-2 nsp15 endoribonuclease antagonizes dsRNA-induced antiviral signaling. Proc. Natl. Acad. Sci. USA 121, e2320194121 (2024).
Routh, A., Head, S. R., Ordoukhanian, P. & Johnson, J. E. ClickSeq: fragmentation-free next-generation sequencing via click ligation of adaptors to stochastically terminated 3’-Azido cDNAs. J. Mol. Biol. 427, 2610–2616 (2015).
Sotcheff, S. et al. ViReMa: a virus recombination mapper of next-generation sequencing data characterizes diverse recombinant viral nucleic acids. Gigascience 12, https://doi.org/10.1093/gigascience/giad009 (2023).
Routh, A. & Johnson, J. E. Discovery of functional genomic motifs in viruses with ViReMa-a Virus Recombination Mapper-for analysis of next-generation sequencing data. Nucleic Acids Res. 42, e11 (2014).
Jaworski, E. et al. Tiled-ClickSeq for targeted sequencing of complete coronavirus genomes with simultaneous capture of RNA recombination and minority variants. eLife 10, e68479 (2021).
Pillon, M. C. et al. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat. Commun. 12, 636 (2021).
Comar, C. E. et al. MERS-CoV endoribonuclease and accessory proteins jointly evade host innate immunity during infection of lung and nasal epithelial cells. Proc. Natl. Acad. Sci. USA 119, e2123208119 (2022).
Bhardwaj, K., Sun, J., Holzenburg, A., Guarino, L. A. & Kao, C. C. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 361, 243–256 (2006).
Kang, H. et al. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J. Virol. 81, 13587–13597 (2007).
Xie, X. et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc. 16, 1761–1784 (2021).
Xie, X. et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 27, 841–848 e843 (2020).
Lokugamage, K. G. et al. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 94, https://doi.org/10.1128/JVI.01410-20 (2020).
Johnson, B. A. et al. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog. 18, e1010627 (2022).
Vu, M. N. et al. QTQTN motif upstream of the furin-cleavage site plays a key role in SARS-CoV-2 infection and pathogenesis. Proc. Natl. Acad. Sci. USA 119, e2205690119 (2022).
Garvanska, D. H. et al. The NSP3 protein of SARS-CoV-2 binds fragile X mental retardation proteins to disrupt UBAP2L interactions. EMBO Rep. 25, 902–926 (2024).
Schindewolf, C. et al. SARS-CoV-2 uses nonstructural protein 16 to evade restriction by IFIT1 and IFIT3. J. Virol. 97, e0153222 (2023).
Deng, X. et al. Inactivating three interferon antagonists attenuates pathogenesis of an enteric coronavirus. J. Virol. 94, https://doi.org/10.1128/JVI.00565-20 (2020).
Deng, X. & Baker, S. C. An “Old” protein with a new story: Coronavirus endoribonuclease is important for evading host antiviral defenses. Virology 517, 157–163 (2018).
Routh, A. et al. Poly(A)-ClickSeq: click-chemistry for next-generation 3-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res. 45, e112 (2017).
Routh, A. DPAC: a tool for differential poly(A)-cluster usage from poly(A)-targeted RNAseq data. G3 9, 1825–1830 (2019).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Zhou, Y. & Routh, A. Mapping RNA-capsid interactions and RNA secondary structure within virus particles using next-generation sequencing. Nucleic Acids Res. 48, e12 (2020).
Vignuzzi, M. & López, C. B. Defective viral genomes are key drivers of the virus–host interaction. Nat. Microbiol. 4, 1075–1087 (2019).
Lopez, C. B. Defective viral genomes: critical danger signals of viral infections. J. Virol. 88, 8720–8723 (2014).
Frazier, M. N. et al. Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease. Nucleic Acids Res. 50, 8290–8301 (2022).
Frazier, M. N. et al. Characterization of SARS2 Nsp15 nuclease activity reveals it’s mad about U. Nucleic Acids Res. 49, 10136–10149 (2021).
Bhardwaj, K., Guarino, L. & Kao, C. C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78, 12218–12224 (2004).
Wang, X. & Zhu, B. SARS-CoV-2 nsp15 preferentially degrades AU-rich dsRNA via its dsRNA nickase activity. Nucleic Acids Res. 52, 5257–5272 (2024).
Manzoni, T. B. & Lopez, C. B. Defective (interfering) viral genomes re-explored: impact on antiviral immunity and virus persistence. Future Virol. 13, 493–503 (2018).
Wilson, I. M., Frazier, M. N., Li, J. L., Randall, T. A. & Stanley, R. E. Biochemical characterization of emerging SARS-CoV-2 Nsp15 endoribonuclease variants. J. Mol. Biol. 434, 167796 (2022).
Miao, Z., Tidu, A., Eriani, G. & Martin, F. Secondary structure of the SARS-CoV-2 5’-UTR. RNA Biol. 18, 447–456 (2021).
Malone, B., Urakova, N., Snijder, E. J. & Campbell, E. A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 23, 21–39 (2022).
Hartenian, E. et al. The molecular virology of coronaviruses. J. Biol. Chem. 295, 12910–12934 (2020).
Yang, Y., Yan, W., Hall, A. B. & Jiang, X. Characterizing transcriptional regulatory sequences in coronaviruses and their role in recombination. Mol. Biol. Evol. 38, 1241–1248 (2021).
Liang, J. et al. How the replication and transcription complex functions in jumping transcription of SARS-CoV-2. Front Genet 13, 904513 (2022).
Li, X. et al. A negative feedback model to explain regulation of SARS-CoV-2 replication and transcription. Front Genet 12, 641445 (2021).
Harcourt, J. et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 26, 1266 (2020).
Jaworski, E. & Routh, A. in Next Generation Sequencing: Methods and Protocols (eds Steven R. Head, Phillip Ordoukhanian, & Daniel R. Salomon) 71–85 (Springer New York, 2018).
Elrod, N. D., Jaworski, E. A., Ji, P., Wagner, E. J. & Routh, A. Development of Poly(A)-ClickSeq as a tool enabling simultaneous genome-wide poly(A)-site identification and differential expression analysis. Methods 155, 20–29 (2019).
Yeung, J. & Routh, A. L. ViReMaShiny: an interactive application for analysis of viral recombination data. Bioinformatics 38, 4420–4422 (2022).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Kar, M. et al. CD4+ and CD8+ T cells are required to prevent SARS-CoV-2 persistence in the nasal compartment. Sci. Adv. 10, eadp2636 (2024).
Acknowledgements
Special thanks to UTMB next generation sequencing core staff (Haiping Hao, Jill K. Thompson) for sequencing support. Research was supported by grants from NIAID of the NIH to (AI168232, AI153602, and U19AI171413) to VDM. ZY and ALR was supported by an Institute of Human Infection and Immunity at UTMB COVID-19 Research Fund. Research was also supported by Burroughs Wellcome Fund (Investigators in Pathogenesis of Infectious Disease) to VDM and Chan Zuckerberg Initiative (Single-Cell Analysis Inflammation Grant) to VDM & ALR. Trainee funding provided by NIAID of the NIH to REA (T32AI007526-24).
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.Z., Y.P.A., V.D.M., A.L.R. Formal analysis: Y.Z., Y.P.A., A.L.M., K.G.L., R.E.A., D.H.W., B.A.J., A.L.R., V.D.M. Funding acquisition: A.L.R., V.D.M. Investigation: Y.Z., Y.P.A., K.G.L., R.E.A., J.R.R., L.K.E., W.M.M., A.M.M., A.L.M., J.T.M., J.R.R., DHW, B.A.J. Methodology: Y.Z., Y.P.A., K.G.L., R.E.A., M.K., M.S.S., D.H.W., A.L.R., V.D.M. Project Administration: A.L.R., V.D.M. Supervision: M.S.S., B.A.J., A.L.R., V.D.M. Visualization: Y.Z., Y.P.A., A.L.R., V.D.M. Writing – original draft: Y.Z., Y.P.A., V.D.M. Writing – review and editing: Y.Z., Y.P.A., K.G.L., R.E.A., D.H.W., B.A.J., A.L.R., V.D.M.
Corresponding authors
Ethics declarations
Competing interests
A.L.R. is a co-founder and co-owner of “ClickSeq Technologies LLC.”, a next-generation sequencing service provider of ClickSeq protocols and downstream analyses such as those presented in this manuscript. VDM has filed a patent on the reverse genetic system for SARS-CoV-2. All other authors declare no conflicts of interest.
Peer review
Peer review information
Nature Communications thanks Yan Sun, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, Y., Ahearn, Y.P., Lokugamage, K.G. et al. SARS-CoV-2 EndoU-ribonuclease regulates RNA recombination and impacts viral fitness. Nat Commun 17, 280 (2026). https://doi.org/10.1038/s41467-025-67001-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67001-2