Abstract
The bidirectional transport of Ca2+ into and out of mitochondria regulates metabolism, signaling, and cell fate. While influx is mediated by the Mitochondrial Calcium Uniporter (MCU) complex, efflux mechanisms are more diversified, involving Na⁺ or H⁺ exchange pathways. We here demonstrate that TMEM65 is a fundamental component of the Ca2+ efflux machinery of mitochondria. Its overexpression specifically enhances Na⁺- and Li⁺-dependent mitochondrial Ca²⁺ extrusion. This effect is inhibited by CGP-37157 and does not depends on NCLX, currently considered the bona fide mitochondrial Na+/Ca2+ exchanger. Its downregulation chronically elevates basal [Ca²⁺]mt and impairs efflux upon stimulation. In Caenorhabditis elegans, deletion of TMEM65 homologs compromises embryonic development under mild thermal stress, causing necrotic lesions that are suppressed by genetic inhibition of MCU-1. These findings highlight a molecular component that may be relevant in pathological settings in which excessive mitochondrial Ca2+ accumulation critically contribute to degenerative pathways.
Similar content being viewed by others
Introduction
Calcium (Ca2+) acts as a universal and ubiquitous intracellular messenger, exerting control over every facet of cellular pathophysiology1. Such a remarkable versatility relies on the one hand on a large array of dedicated Ca2+ binding and/or transporting proteins, and on the other hand on the strictly controlled compartmentalization of [Ca2+] changes. In such a sophisticated framework, mitochondria significantly shape intracellular Ca2+ dynamics and downstream Ca2+-dependent pathways. In healthy cells at rest, intramitochondrial [Ca2+] ([Ca2+]mt) matches cytoplasmic [Ca2+] ([Ca2+]cyt) at approximately 100 nM. Upon cellular stimulation, mitochondria undergo a rapid transient rise in [Ca2+]mt that largely exceed bulk [Ca2+]cyt, up to 1 mM in a few cases2,3. This marked accumulation of Ca2+ is in part due to the highly negative organelle membrane potential (−180 mV), but also to the strategic positioning of mitochondria near Ca2+-releasing channels of the endoplasmic reticulum or the plasma membrane, where local micro/nano-domains of high [Ca2+] are generated4. Inside mitochondria, Ca2+ acts as a positive modulator of three dehydrogenases (PDH, IDH3 and OGDH), which stimulating oxidative metabolism and eventually ATP synthesis5. In pathological conditions, excessive Ca2+ accumulation within the organelle matrix (commonly referred to as Ca2+ overload) can potentially trigger permeability transition, ultimately leading to membrane permeabilization that may promote the release of pro-death factors6,7,8. Functioning as auxiliary cellular Ca2+ reservoirs, mitochondria contribute to buffering intracellular [Ca2+] transients, thereby influencing the spatio-temporal patterns of [Ca2+] oscillations, either within micro/nano-domains or on a bulk scale (e.g., throughout the entire cell)9. Consequently, impairment of mitochondrial Ca2+ homeostasis has been implicated in the pathogenesis of a variety of human diseases, ranging from neuromuscular diseases to brain injuries10,11.
From a biophysical perspective, [Ca2+]mt is dynamically determined by the rates of Ca2+ influx versus efflux across the IMM. Pioneering investigations conducted in the 1960s and 1970s elucidated the mechanisms underlying this regulation: Ca2+ entry into mitochondria is facilitated by an electrogenic uniporter, while Ca2+ extrusion occurs through two distinct mechanisms - one dependent on sodium ions (i.e. mediated by a Na+/Ca2+ exchanger) and the other Na+-independent (i.e. mediated by a H+/Ca2+ exchanger)12,13. At the molecular level, mitochondrial Ca2+ influx is orchestrated by the MCU complex, whose precise composition varies across tissues and may encompass components such as MCU, MCUb, EMRE, MICU1, MICU2, and MICU314. As to the efflux, the protein NCLX, encoded by the SLC8B1 gene, has been identified as the mediator of Na+-dependent Ca2+ extrusion15, whereas two distinct proteins, LETM116,17 and TMBIM518,19, have been proposed to function as mitochondrial H+/Ca2+ exchangers. Given the breakthrough discoveries of the last years, it is not possible to rule out that other molecular components may contribute to mitochondrial Ca2+ transport. Here, we identify TMEM65 as an unexpected, broadly conserved protein that enables Na+-dependent mitochondrial Ca2+ efflux. We found that genetic manipulation of TMEM65 expression causes striking effects on mitochondrial Ca2+ homeostasis in cultured cells. Specifically, TMEM65 overexpression effectively suppresses mitochondrial [Ca²⁺] transients independently of NCLX, while its silencing prevents the decay of the organellar Ca2+ rises, resulting in markedly elevated resting [Ca2+]mt levels. Consistent with the prominent contribution of TMEM65 to organelle Ca2+ clearance, we show that genetic deletion of TMEM65 homologues in Caenorhabditis elegans causes an overt necrotic phenotype that could be prevented by inhibiting mitochondrial Ca2+ influx, thereby pointing out that mitochondrial Ca2+ overload is a crucial determinant of cell demise. Since a human subject harboring a homozygous TMEM65 loss-of-function mutation exhibits a neuromuscular phenotype reminiscent of mitochondrial disease20, it is reasonable to consider MCU inhibition as a potential therapeutic strategy for conditions characterized by excessive mitochondrial Ca2+ accumulation.
Results and discussion
TMEM65 is a conserved protein of the IMM
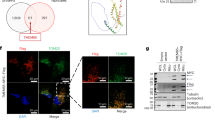
We previously performed a bioinformatic analysis and identified candidate mitochondrial proteins of the inner membrane with a putative function in Ca2+ signaling21. Besides MCU, the list contains the recently described mitochondrial H+/Ca2+ exchanger TMBIM5 (also known as GHITM/MICS1). Thus, we sought to further inspect the list of presumed Ca2+ transporters using an evolutionary analysis to look for proteins with a conservation profile similar to MCU, and focused on a protein named TMEM65 (Fig. 1a). Among these candidates, TMEM65 is a 25 kDa protein with 3 transmembrane domains and a glycine zipper motif that are well conserved in kinetoplastids, green algae (but not in other plants) and metazoan (Fig. 1b and Supplementary Fig 1a-b). Mammalian TMEM65 also shows an apparent poly-acidic tail near the C terminus. Although two works proposed TMEM65 as a protein positioned to cardiac sarcolemma22,23, multiple lines of evidence indicate that TMEM65 is a bona fide mitochondrial protein. In this regard, i) TMEM65 expression is not limited to the heart but rather is ubiquitous (highest in the cerebellum, lowest in the liver, Supplementary Fig 1c), ii) sequence analysis of TMEM65 reveals a conserved, prototypical mitochondrial targeting signal at the N-terminal (Fig. 1b, predicted MPP-cleaved peptide indicated in cyan), and iii) TMEM65 is present in all high quality mitochondrial datasets based on robust proteomic evidence, including MitoCarta24, MitoCOP25 and MitoPhos26. To confirm these predictions and assess TMEM65 localization, we cloned, tagged (3xFlag) and expressed human TMEM65 in HeLa cells. Immunoblot analyses show a major band running with high specificity at 20 kDa, in line with the predicted molecular weight of the mature protein after processing by the Mitochondrial Processing Peptidase (MPP). The unprocessed 25 kDa precursor is also detectable upon overexpression (Fig. 1c). Immunofluorescence analyses confirm co-localization with the mitochondrial marker SDHA (Fig. 1d). To test protein topology across the IMM, we performed Proteinase K (PK) protection assay in isolated mitoplasts. Digestion of intact (but not permeabilized) mitoplasts causes the loss of full-length TMEM65 and the appearance of a smaller fragment, indicating that a significant portion of TMEM65 resides within the matrix (Fig. 1e). Of note, the Flag peptide fused at the C-terminus of TMEM65 is lost after PK treatment (Fig. 1e), thereby demonstrating that TMEM65 is inserted in the IMM as depicted in Fig. 1b. This membrane orientation is also supported by other studies27, including an independent spatially-resolved proteomic screening that mapped Tyrosine 77 within the matrix (Y77, highlighted in grey in Fig. 1b)28.
a Cladogram showing the phylogenetic distribution of MCU (red) and TMEM65 (green) homologs across 1088 reference proteomes. b Schematic representation of TMEM65 membrane topology. Topological structure was generated using Protter40. c Western blot showing expression of TMEM65 and TME65-3xFlag in HeLa cells. Comparable results were obtained in at least three independent transfections. d Immunolocalization of TMEM65-3xFlag (cyan) and the mitochondrial marker SDHA (magenta) in HeLa cells, scale bar is 10 μm. Comparable staining was obtained in at least three independent transfections and with other mitochondrial markers. e Proteinase K protection assay performed in mitoplasts isolated from TMEM65-3xFlag-expressing HeLa cells. Comparable results were obtained in at least three independent experiments.
TMEM65 expression modulates mitochondrial Ca2+ homeostasis
We next tested the role of TMEM65 on mitochondrial Ca2+ handling by monitoring [Ca2+]mt in living intact HeLa cells using mitochondrial-targeted genetically-encoded Ca2+ indicators (GECI) based on the Ca2+-sensitive bioluminescent protein Aequorin29. Control and TMEM65-overexpressing cells were continuously perfused with modified Krebs-Ringer buffer and challenged with 100 μM histamine, an inositol 1,4,5-trisphosphate (IP3)-generating agonist that elicits Ca2+ release from the endoplasmic reticulum, leading to a transient increase of [Ca2+] in both cytoplasm and mitochondria. Interestingly, TMEM65 overexpression robustly inhibits (approximately 80%) the peak of [Ca2+]mt upon cellular stimulation (Fig. 2a). Importantly, this effect on mitochondrial Ca2+ handling is specific, since no major changes were detected in terms of cytosolic Ca2+ dynamics (Supplementary Fig 2a-b) and mitochondrial membrane potential (Supplementary Fig 2c), hence ruling out possible confounding factors. To obtain additional insights into the structure-to-function relationship of TMEM65, we generated a set of mutants by introducing individual substitutions in the most conserved amino acids or by disrupting relevant motifs (i.e. glycine zipper and poly-acidic tail, Supplementary Fig 2d). All TMEM65 variants are expressed at a levels similar or greater compared to wild-type, with the exception of the mutant lacking the transmembrane glycine zipper motif (GZ dead), that completely fail to express at protein level, probably due to an impairment in membrane packing (Supplementary Fig 2e-f)30. We evaluated histamine-induced [Ca2+]mt peaks in cells expressing wild-type (wt) or mutant TMEM65. Our data indicate that D132, D167, G170 and E178 are required for TMEM65 activity, while G140L and Q195E mutants are very similar to wild type (Supplementary Fig 2g-h). The TMEM65 mutant lacking the poly-acidic tail still retain some effect, although to a lower extent when compared to wt. Surprisingly, the E144Q mutant shows even stronger effect than the wt. None of these mutants affects the expression of the primary mitochondrial Ca²⁺ influx and efflux transporters (Supplementary Fig 2f). Overall, these data suggest a clear, specific involvement of TMEM65 in the regulation of mitochondrial Ca2+ handling.
a, b [Ca2+]mt measurements in HeLa cells expressing a mitochondrially targeted aequorin probe. a Representative [Ca2+]mt traces of control, TMEM65 overexpressing and silenced cells. b Histograms (mean ± s.d.) of histamine-induced [Ca2+]mt peak values. c, d [Ca2+]mt measurements in HeLa cells expressing the mitoGCaMP6 probe. c Representative traces of control, TMEM65 overexpressing and silenced cells. d Histograms (mean ± s.d.) of mitochondrial Ca2+ levels at the indicate time points. Data were analyzed by one-way ANOVA followed by Holm–Sidak’s multiple comparisons test. * indicates p < 0.01. Individual data points, descriptive statistics and exact p values are included in Source Data Table.
We next evaluated the effect of TMEM65 downregulation on organelle Ca2+ dynamics by testing different synthetic unmodified siRNAs. When agonist-induced peak [Ca2+]mt values were compared, we obtained inconsistent results among siRNA sequences (Supplementary Fig 3a-b) despite comparable efficiency in reducing TMEM65 protein level (Supplementary Fig 3e), an outcome likely due to off-target effects. To enhance specificity, we used endoribonuclease-prepared siRNAs (esiRNA)31. This approach reveals increased [Ca2+]mt peak upon specific downregulation of TMEM65 (Fig. 2a, b). More importantly, in addition to the effect on maximal [Ca2+]mt values, TMEM65 silencing alters the kinetic profile of [Ca2+]mt transients. In particular, a delayed Ca2+ clearance after the peak is noticeable, an indication of impaired mitochondrial Ca2+ extrusion. To better appreciate this effect, we re-analyzed all Ca2+ traces by normalizing [Ca2+]mt on peak [Ca2+] (Supplementary Fig 3c). The slope of the resulting trace is thus proportional to mitochondrial Ca2+ efflux rate (Supplementary Fig 3d). This analysis clearly reveals that TMEM65 overexpression enhances mitochondrial Ca2+ extrusion (notwithstanding lower [Ca2+]mt peak), while its silencing elicits the opposite effect. Remarkably, all tested siRNAs consistently decreased mitochondrial Ca2+ efflux, irrespective of their wavering effect on peak [Ca2+]mt (Supplementary Fig 3a-d). Furthermore, expression analyses of the two main mitochondrial Ca²⁺ transporters (MCU for the influx, NCLX for the efflux) revealed no significant changes (Supplementary Fig 3f). Due to the lack of specificity of the commercial antibodies tested, NCLX protein levels could not be reliably assessed (Supplementary Fig 4). Altogether, these data suggest an involvement of TMEM65 in mitochondrial Ca2+ efflux rather than in influx. To further substantiate our data, we evaluated mitochondrial Ca2+ dynamics using a GFP-based GECI, the mitoGCaMP6 probe, that also allows the more precise quantification of baseline [Ca2+]mt and extrusion kinetics32. Compared to controls, both overexpression and downregulation of TMEM65 alter mitochondrial Ca2+ dynamics, further supporting that TMEM65 is a key regulator of mitochondrial Ca2+ efflux (Fig. 2c). Specifically, TMEM65 overexpression causes a negligible decrease of resting Ca2+, a histamine induced upstroke similar to control, at least in the initial phase, but most evidently, a steep return to baseline after the peak (Fig. 2c). Conversely, TMEM65-silenced cells show chronically elevated resting matrix Ca2+ levels, and histamine-induced [Ca2+] elevations that last over several minutes (Fig. 2c, d). Altogether, our detailed analyses of mitochondrial Ca2+ dynamics uncover a prominent role of TMEM65 in controlling mitochondrial Ca2+ efflux.
TMEM65 mediates Na+-dependent mitochondrial Ca2+ efflux
We next explore the mechanism underlying TMEM65 effect on mitochondrial Ca2+. It is known that experiments carried out in intact cells have the benefit of examining mitochondria in their native environment. However, these experimental settings suffer from two main drawbacks. Firstly, mitochondrial Ca2+ influx and efflux constantly coexist, thus preventing the genuine segregation of the two phenomena. Secondly, efflux relies on two concurrent but separate mechanisms, one Na+-dependent and the other Na+-independent. To overcome these limitations, we performed [Ca2+]mt measurements in digitonin-permeabilized cells, where mitochondria can be exposed to buffers with defined ionic composition. In this experimental setup, mitochondrial Ca2+ uptake was initiated by perfusing mitochondria of plasma-membrane permeabilized cells with an intracellular-like buffer containing fixed [Ca2+], until a plateau [Ca2+]mt is reached (i.e. when influx rate equals efflux rate). Then, MCU inhibitor Ruthenium Red (RuR) was added to block the influx, resulting in a progressive decrease of [Ca2+]mt. The rate of this decrease represents a reliable measurement of mitochondrial Ca2+ efflux. Perfusion with Na+-free buffer allows the evaluation of organelle Na+-independent efflux, while incorporation of 10 mM NaCl in the RuR-containing buffer enhances mitochondrial Ca2+ extrusion, thereby enabling the quantification of Na+-dependent efflux. Li+ can also be used as Na+ surrogate, although less effective33. Using this approach, we found that both TMEM65 overexpression and downregulation showed no effects on mitochondrial Ca2+ influx or Na+-independent efflux. Rather, TMEM65 overexpression greatly enhanced Na+- and Li+-dependent mitochondrial Ca2+ efflux. Indeed, its silencing blunted the Na+-dependent component of organellar Ca2+ extrusion (Fig. 3a-d, Supplementary Fig 5a-b). To further support this finding, we took advantage of the specific pharmacological inhibitor of mitochondria Na+/Ca2+ exchange, the benzothiazepine CGP-3715734. This compound causes an increase in agonist-induced [Ca2+]mt peaks in control cells due to the inhibition of organelle Ca2+ efflux35. Strikingly, CGP-37157 was able to abrogate the effect elicited by TMEM65 overexpression, restoring normal mitochondrial Ca2+ dynamics (Fig. 3 e-g). Overall, these results indicate that i) mitochondrial Ca2+ efflux strongly counteracts both in resting conditions and in all phases of agonist-evoked responses the action of MCU and ii) TMEM65 is a key contributor to the process of Na+-dependent mitochondrial Ca2+ efflux.
a–d [Ca2+]mt measurements in digitonin-permeabilized HeLa cells. Representative [Ca2+]mt traces of control, TMEM65 overexpressing and silenced cells in the absence (a) or presence (b) of 10 mM Na+. Histograms (mean ± s.d.) of maximal Na+-independent (c) and Na+-dependent (d) mitochondrial Ca2+ influx and efflux rates. Data were analyzed by one-way ANOVA followed by Holm–Sidak’s multiple comparisons test. * indicates p < 0.05. e–g [Ca2+]mt measurements in control and TMEM65 overexpressing intact HeLa cells treated with either vehicle (DMSO) or 20 μM CGP-37157. e Representative [Ca2+]mt traces. f Histograms (mean ± s.d.) of histamine-induced [Ca2+]mt peak values. g Histograms (mean ± s.d.) of normalized mitochondrial Ca2+ efflux. Data were analyzed by two-way ANOVA followed by Holm–Sidak’s multiple comparisons test. * indicates p < 0.001, ns=not significant. Individual data points, descriptive statistics and exact p values are included in Source Data Table.
TMEM65 acts independently of NCLX
The potential relationship between TMEM65 and the mitochondrial Na⁺/Ca²⁺ exchanger NCLX warrants investigation. Although the TMEM65 sequence shows no similarity to that of known exchangers—suggesting a possible modulatory role—functional analyses indicate that its overexpression and silencing strongly enhance and suppress, respectively, Na⁺-dependent mitochondrial Ca²⁺ extrusion. This positions TMEM65 as the molecular rate-limiting factor for the process. Conflicting reports in the literature36,37 prompted us to test whether TMEM65 activity depends on the sole characterized mitochondrial Na⁺/Ca²⁺ exchanger. In cells acutely silenced for NCLX (Supplementary Fig 3f), TMEM65 overexpression markedly reduced [Ca²⁺]mt peaks (Fig. 4a, b), indicating functional independence. To rule out compensation from residual NCLX, we generated both polyclonal and monoclonal NCLX knockout (KO) cell lines. In a consistent manner, TMEM65 overexpression lowered agonist-induced [Ca²⁺] peaks (Fig. 4c) and increased Ca²⁺ extrusion rates in intact (Fig. 4e) and permeabilized cells (Fig. 4f, g), confirming that NCLX is dispensable for TMEM65 function. In the same models, the Na⁺/Ca²⁺ exchange inhibitor CGP-37157 not only abrogated the stimulatory effect of TMEM65 overexpression (Fig. 4b) but also remained active in NCLX-silenced and NCLX-KO cells (Fig. 4b; Supplementary Fig 6). Together, these results demonstrate that TMEM65 operates independently of NCLX, and indicate TMEM65 as one of the molecular targets of CGP-37157.
a–e [Ca2+]mt measurements in intact HeLa cells expressing an aequorin-based probe. a Representative [Ca2+]mt traces showing the effect of TMEM65 overexpression in control and NCLX-silenced cells. b Histograms (mean ± s.d.) of histamine-induced [Ca2+]mt peak values. Where indicated, cells were treated with 20 μM CGP-37157. Data were analyzed by three-way ANOVA followed by Holm–Sidak’s multiple comparisons test. * indicates p < 0.01. c Representative [Ca2+]mt traces showing the effect of TMEM65 overexpression in control and NCLX-KO cells. d Histograms (mean ± s.d.) of histamine-induced [Ca2+]mt peak values. Data were analyzed by two-way ANOVA followed by Holm–Sidak’s multiple comparisons test. * indicates p < 0.001. e Histograms (mean ± s.d.) of normalized mitochondrial Ca2+ extrusion rate. Data were analyzed by two-way ANOVA followed by Holm–Sidak’s multiple comparisons test. * indicates p < 0.001. f, g [Ca2+]mt measurements in digitonin-permeabilized HeLa cells. f Representative [Ca2+]mt traces showing the effect of TMEM65 overexpression in control and NCLX-KO cells. g Histograms (mean ± s.d.) of maximal Na+-dependent mitochondrial Ca2+ efflux rates. Data were analyzed by two-way ANOVA followed by Holm–Sidak’s multiple comparisons test. * indicates p < 0.001. Individual data points, descriptive statistics and exact p values are included in Source Data Table.
TMEM65 deficiency causes necrotic lesions in vivo in C. elegans
To better understand the physiological relevance of TMEM65 in vivo, we decided to employ the nematode Caenorhabditis elegans. Based on protein alignment (BLAST) using human TMEM65 as a query, we found that C33H5.19/tag-321 and C03B8.3 genes encode two homologs (here referred to also as CeTMEM65-1 and CeTMEM65-2, respectively) (Fig. 5a, Supplementary Fig 7a-c), one of which (TAG-321) with a higher percent identity and a predicted AlphaFold 3D structure relatively similar to the human protein variant (Fig. 5b). First, we tested the expression and functionality of CeTMEM65 in mammalian cells. Western blot shows that CeTMEM65-1 is efficiently expressed also in HeLa cells, while CeTMEM65-2 fails to be detected, probably due to instability of the protein at 37°C (Supplementary Fig 7d). In terms of activity, the effect of CeTMEM65-1 on mitochondrial Ca2+ handling is comparable to its human counterpart, indicating that their biochemical function is maintained through evolution (Supplementary Fig 7e). Next, we performed CRISPR/Cas9 gene editing and generated deletions of the promoters and exons within tag-321 and C03B8.3 genes. Although both tag-321(bon103) and C03B8.3(bon104) were (null) alleles (Supplementary Fig 7f), homozygous mutants did not display any obvious difference compared to wt (N2) nematodes at normal growth conditions (20°C, E. coli B-type strain as a bacterial food). When we generated C03B8.3;tag-321 double (null) mutants, we observed a significant reduction of hatched larvae compared to wt, tag-321(bon103) and C03B8.3(bon104) mutants at 20 °C (Fig. 5c). Lifespan assays at 20 °C showed no differences between wt and mutant nematodes, whereas there was a trend toward a minor decreased median lifespan for CeTMEM65 single and double mutants compared to wt at 25 °C (Fig. 5d-e and Supplementary tables ST1-3). Intriguingly, while at 20°C we could find larvae in plates from wt and CeTMEM65 single mutants, at 25 °C the large majority of C03B8.3;tag-321(null) mutant animals laid eggs that did not hatch. Motivated by this intriguing observation, we decided to assess the number of larvae that could develop and survive at 25 °C (Fig. 5f). Compared to wild type animals, only ~5% of all C03B8.3(bon104);tag-321(bon103) mutants hatched (Fig. 5g), with most of the degenerating embryos at gastrula/comma stage showing large vacuoles (Fig. 5h). Of note, C03B8.3(null) mutants had a hatching rate similar to wt, whereas tag-321 (null) animals showed a modest, although significant increased number of larvae that could hatch at 25 °C (Fig. 5g). Given the resemblance of the large vacuoles to necrotic structures, we reasoned that mcu-1(lof) and consequent inhibition of mitochondrial Ca2+ influx would restore normal hatching of CeTMEM65(null) mutants at 25 °C (Fig. 5i). We first attempted to generate C03B8.3;mcu-1;tag-321 triple mutants using the already existing alleles. However, since mcu-1 and tag-321 genes are both on chromosome IV at a very close distance (less than 1 cM), we could not introduce the mcu-1(ju1154) allele into the existing C03B8.3(null);tag-321(null) background. To overcome this problem, we performed another gene editing in mcu-1(ju1154) animals and generated an equivalent lesion within tag-321 gene. DNA sequencing confirmed that the newly generated tag-321(bon105) allele was matching the previous tag-321(bon103) mutation (Supplementary Fig 7g-h). We backcrossed the new strain, generated C03B8.3(bon104);mcu-1(ju1154);tag-321(bon105) triple mutants and assessed animal hatching. Remarkably, we found that mcu-1(lof) completely suppressed the degeneration of C03B8.3;tag-321 double mutant embryos at 25 °C (Fig 5j, k), indicating a genetic interaction between mcu-1 and CeTMEM65 and further supporting the prominent role of TMEM65 as a key regulator of mitochondrial Ca2+ efflux. To confirm our findings and rule out any eventual egg-laying defects of individual hermaphrodites, we performed a population-based hatching assay in which eggs were extracted from hundreds of gravid young adult hermaphrodites and plated on bacteria-seeded NGM plates (Fig. 5l). Consistent with our previous findings, while the large majority of C03B8.3;tag-321 double mutant embryos did not hatch at 25 °C, C03B8.3(bon104);mcu-1(ju1154);tag-321(bon105) triple mutants hatched and developed at a comparable rate of wt animals (Fig. 5m). Using this experimental paradigm, CeTMEM65 single mutants did not show any difference compared to wt (Fig. 5n).). Of note, the hatching rate of mcu-1(ju1154) was comparable to wt (Supplementary Fig 7i). Together, our in vivo evidence demonstrates that TMEM65 deficiency may facilitate mitochondrial Ca2+ overload when cells are exposed to environmental challenges (e.g., restrictive temperatures) that may induce degenerative processes (necrosis-like death). Finally, we tested whether NCLX loss of function could recapitulate the same phenotype observed in C03B8.3;tag-321 double mutants (Fig. 5o). It was previously suggested that NCX-9 is the putative NCLX-type Na+/Ca2+ exchanger in C. elegans38. Using two ncx-9(lof) alleles, we carried out our population-based hatching assay and found that NCX-9 loss did not alter C. elegans hatching at 25 °C (Fig. 5p). Although we cannot rule out that other NCLX-type exchangers may compensate for NCX-9 deficiency and/or contribute to mitochondrial Ca2+ extrusion, our data report unique developmental defects associated with CeTMEM65 dysfunction that causes death in an MCU-dependent manner.
a BLAST analysis of human TMEM65 against the C. elegans WormBase database. The figure shows the hits ranked by sequence similarity. Below, the sequence alignments of human TMEM65 (Hs TMEM65, in blue) vs C. elegans TAG-321 (Ce TAG-321, green) and C03B8.3 (Ce C03B8.3, red). b AlphaFold-predicted structures of human TMEM65 and C. elegans orthologs. The models highlight domains with conserved fold and organization. c Hatching assay of wt (grey), tag-321(bon103) (green), C03B8.3(bon104) (red) and C03B8.3(bon104);tag-321(bon103) (blue) grown at 20°C. Lifespan assays at (d) 20 °C and (e) 25 °C. The means (±SEM) of the median survival are reported below the graphs. Genotypes are indicated by colored dashes. f Schematic representation of the hatching assay. Created in BioRender. Bano, D. (2025) https://BioRender.com/asudllhg Hatching assay of wt (grey), tag-321(bon103) (green), C03B8.3(bon104) (red) and C03B8.3(bon104);tag-321(bon103) (blue) larvae from eggs incubated at 25 °C. h Representative DIC images of eggs extracted from gravid adult animals and maintained at 25 °C. Yellow arrowhead indicates a vacuole. i Schematic representation of mitochondrial Ca2+ transport. Genetic inhibition results in MCU complex and TMEM65 loss (as grey cartoons). Created in BioRender. Bano, D. (2025) https://BioRender.com/asudllhj Representative DIC images of eggs that were extracted from gravid adult animals and maintained at 25 °C. Yellow arrowhead indicates a vacuole. k Hatching assay of wt, C03B8.3(bon104);tag-321(bon103) and C03B8.3(bon104);mcu-1(ju1154);tag-321(bon105) at 25 °C. l Schematic representation of a population-based hatching assay paradigm. Created in BioRender. Bano, D. (2025) https://BioRender.com/asudllhm Hatching assay of wt, C03B8.3(bon104);tag-321(bon103) and C03B8.3(bon104);mcu-1(ju1154);tag-321(bon105) at 25 °C using the paradigm described in (5 l). n Hatching assay of wt, tag-321(bon103) and C03B8.3(bon104) at 25 °C using the paradigm described in (5 l). o Schematic representation of NCLX genetic inhibition. Created in BioRender. Bano, D. (2025) https://BioRender.com/asudllhp Hatching assay of wt, ncx-9(gk234237) and ncx-9(gk773708) at 25 °C using the paradigm described in (5 l). All data are presented as mean ± s.d., unless otherwise specified. Data from panels c, g, k, m and n were analyzed by one-way ANOVA followed by Holm–Sidak’s multiple comparisons test. Data from panel p were analyzed using an unpaired two-sample t-test. **p < 0.01, ****p < 0.0001, ns=not significant. Individual data points, descriptive statistics and exact p values are included in Source Data Table.
In conclusion, we confirmed the critical function of TMEM65 in regulating mitochondrial Ca²⁺ fluxes. TMEM65 is both sufficient to enhance Na⁺-dependent mitochondrial Ca²⁺ extrusion, as shown by overexpression experiments, and necessary to prevent organellar Ca²⁺ overload, as evidenced by siRNA-mediated knockdown in vitro and in vivo in in C. elegans mutants. Importantly, TMEM65 activity does not require the presence of NCLX, which has long been considered the bona fide mitochondrial Na⁺/Ca²⁺ exchanger. This unexpected result aligns with recent data showing that TMEM65 alone, when reconstituted in liposomes, exhibits intrinsic Ca²⁺ transport activity. These findings establish TMEM65 as an autonomous molecular entity with Na⁺/Ca²⁺ exchange capacity and prompt a critical reassessment of the genuine contribution of NCLX to mitochondrial Ca²⁺ homeostasis. More importantly, our findings also shed light on the pathogenic mechanism underlying a reported human neuromuscular syndrome associated with TMEM65 deficiency, where mitochondrial dysfunction emerges as a central feature. Based on our in vivo experimental paradigm, MCU inhibition may represent a viable strategy to mitigate the deleterious effects of TMEM65 loss. Considering that excessive mitochondrial Ca²⁺ accumulation is implicated in numerous diseases, our work lays the foundation for therapeutic approaches aimed at restoring mitochondrial Ca²⁺ balance.
Methods
Chemicals, cell culture and transfection
All chemicals were purchased from Sigma-Aldrich, unless otherwise specified. All the experiments were performed in HeLa cells (ATCC Number: CCL-2) cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco #41966052, Thermo Fisher Scientific), supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific), containing penicillin (100 U/ml) and streptomycin (100 μg/ml) (Euroclone). NCLX-KO cells were purchased from SyntheGo (now EditCo). Knockout cells were generated using Crispr-Cas9 technology by targeting exon 2 of the SLC8B1 gene using the following guide RNA sequence: ACACCUGAAGCUGGAAACUG. When needed, cells were seeded on either glass coverslips (onto 13- or 24-mm in diameter) or 96-well plates and allowed grow to 50-80% confluence before transfection. Transfection of siRNA was performed using RNAiMAX (Thermo Fisher Scientific) according to manufacturer’s instructions. Transfection of plasmid DNA was performed using polyethylenimine (PEI MAX, Polysciences).
Plasmids and siRNAs
Plasmids encoding C-term 3xFlag tagged human TMEM65 (NM_194291.2), C. elegans TAG-321 (NM_068883.5), and C. elegans C03B8.3 (NM_066278.5) were purchased from VectorBuilder. Untagged human TMEM65 was amplified using the following primer: fw 5’- ATTAGCTAGCACCACCATGTCCCGGCTG-3’, rv 5’- ATTAGAATTCTTAACTTTTCGTTTCCAGTTTTTCATCTTCTTC-3’. The PCR fragment was cloned into pcDNA3.1 (Thermo Fisher Scientific) using NheI and BamHI sites.
Mutagenesis was performed using In-Fusion cloning (Takara) using the following primer pairs:
D132N | fw: 5’-CTTTTTGAATAATGCAATTATGATTGTTGCTGG |
rv: 5’-GCATTATTCAAAAAGCCAAACCCTATGAAAGG | |
G140L | fw: 5’-TGTTGCTTTAACCCATATTGAAATGTCTATTGG |
rv: 5’-TGGGTTAAAGCAACAATCATAATTGCATTATCC | |
E144Q | fw: 5’-CCATATTCAAATGTCTATTGGAATTATTTTGGGAA |
rv: 5’-GACATTTGAATATGGGTTCCAGCAACAATCA | |
D167N | fw: 5’-TGTGTCAAACCTAGCTGGACTTGGACTTGC |
rv: 5’-GCTAGGTTTGACACAAGATTTCCCAAAGCAGC | |
G170L | fw: 5’-TCTAGCTCTTCTTGGACTTGCAGGCTACG |
rv: 5’-CCAAGAAGAGCTAGATCTGACACAAGATTTCCC | |
E178Q | fw: 5’-CTACGTTCAAGCATTGGCTTCCAGGTTAGG |
rv: 5’-AATGCTTGAACGTAGCCTGCAAGTCC | |
Q195E | fw: 5’-ACCAAAGGAAGTTGACATGTGGCAAACACGTC |
rv: 5’-TCAACTTCCTTTGGTGTGAGATCAGGAATTGAC | |
ΔPolyAc | fw: 5’-GAGGAGGTAAACTGGAAACGAAAAGTGACTAC |
rv: 5’-CCAGTTTACCTCCTCCAAAGAAAATTAAAGG | |
GZ dead | fw: 5’-TGCTGTGACTATTGCATGCATTCTAGCTATGTTTCCTTTAATTTTCTTTGGAG |
rv: 5’-GCAATAGTCACAGCAACAGCTTTTGCCAAATGTGTACTAAGACGTGTTTG |
All constructs were verified by Sanger sequencing.
The following siRNAs were purchased from Sigma-Aldrich: esiRNA-RLuc (cat no EHURLUC), esiRNA-TMEM65 (cat no EHU042431), esiRNA-SLC8B1 (cat no EHU154091), siRNA Universal Negative Control #2 (cat no SIC002), siRNA-TMEM65#1 (cat no SASI_Hs01_00147974), siRNA-TMEM65#2 (cat no SASI_Hs02_00371420), siRNA-TMEM65#3 (cat no SASI_Hs01_00147977), siRNA-TMEM65#4 (SASI_Hs01_00147978).
Antibodies, SDS-PAGE and western blot
Cells were lysed in RIPA buffer (150 mM NaCl, 25 mM Tris-Cl pH 8, 1 mM EGTA-Tris, 1% Triton X-100, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with Complete EDTA-free protease inhibitor mixture (Roche Applied Science) and PhosStop (Roche Applied Science) for 30 minutes on ice. Crude extracts were centrifuged 15000 g for 10 minutes to remove debris, and proteins in the supernatant were quantified using the BCA Protein Assay Kit (Pierce). Approximately 30 μg of proteins were dissolved in LDS sample buffer (Life Technologies) supplemented with 10 mM dithiothreitol, heated for 5 minutes at 95°C and loaded on 4-12% Bis-Tris NuPage gels (Thermo Fisher Scientific). After electrophoretic separation, proteins were transferred onto nitrocellulose membranes by semidry transfer (BioRad). Membranes were blocked for 1 hour at RT with 5% non-fat dry milk in TBS-T (50 mM Trizma, 150 mM NaCl and 0.1% Tween) and probed with the indicated primary antibodies over night at 4 °C. Isotype matched, horseradish peroxidase-conjugated secondary antibodies (1:10000, BioRad) were used followed by detection by chemiluminescence (SuperSignal Pico, Pierce). The following primary antibodies were used: anti-GRP75 (1:1000, SCBT sc-133137), anti-TMEM65 (1:1000, Thermo Fisher Scientific PA5-112762), anti-Flag (1:1000, Cell Signaling #2368), anti SDHA (1:1000, Cell Signaling #5839), anti-PDHA1 (1:1000, Cell Signaling #2784), and anti-MITOK (1:10000, Sigma HPA010980), anti-NCLX (Proteintech 21430-1-AP, Thermo PA5-114330 and Abcam ab83551). Western blots are representative of at least three independent preparations. For Western blot analyses of C. elegans samples, adult animals were collected in water and snap-frozen in liquid nitrogen. C. elegans pellets were then sonicated in RIPA buffer supplemented with protease and phosphatase inhibitors (Roche). Lysates were centrifuged as described above and protein concentrations were quantified via Bradford assay (Sigma-Aldrich). Approximately 20-30 μg of protein was resolved on 8% polyacrylamide gels and transferred onto nitrocellulose membranes using semi-dry Trans-Blot TurboTM (Bio-Rad). Following antibody incubation (anti-GFP, 1:1000, Roche #118144600001), membranes were developed in ECL and imaged using the Chemidoc imaging system (Bio-Rad). Uncropped images of Western blots used for the assembly of final figures are provided as Supplementary Figs.
RNA extraction, reverse transcription, and qPCR
Total RNA was extracted from HeLa cells using the RNeasy Plus Micro Kit (Qiagen) according to the manufacturer’s instructions. RNA concentration was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific). For cDNA synthesis, 1 μg of RNA from each sample was reverse transcribed using the SuperScript II cDNA synthesis kit (Thermo Fisher Scientific), following the manufacturer’s protocol. Oligo(dT)12–18 primer (Thermo Fisher Scientific) was used for first-strand cDNA synthesis, along with dNTPs (deoxynucleotide triphosphates, Thermo Fisher Scientific) and reverse transcriptase.
The resulting cDNA was analyzed by quantitative PCR (qPCR) using TaqMan probe-based chemistry to assess relative gene expression levels. qPCR reactions were carried out in 384-well plates using TaqMan Gene Expression Master Mix (Applied Biosystems) and gene-specific TaqMan probes. Amplification was performed on a QuantStudio 5 real-time PCR system (Thermo Fisher Scientific) under standard cycling conditions. Relative gene expression was calculated using the ΔΔCt method, with TOMM20 or ACTIN serving as the endogenous control, as indicated in figure legend. The following probes were used: TOMM20-VIC (cat no Hs00740685_sH), ACTB-VIC (cat no Hs01060665_g1), MCU-FAM (cat no Hs00293548_m1), MCU-FAM (cat no Hs00293548_m1), SLC8B1-FAM (cat no Hs00905331_m1).
Bioinformatic analyses
To evaluate conservation profiles of MCU and TMEM65, reference proteomes of eukaryotic species were retrieved from Uniprot as fasta files (Release 2022_02, 25-May-2022). Only species containing at least 3000 entries in main (canonical) set were considered for further analysis. Homology searches were performed using phmmer (HMMER version 3.3.2)39. Human MCU and TMEM65 protein sequences were used as input against reference proteomes using BLOSUM62 as scoring matrix. Hits with an E-value below 0.05 were considered as homologs. Multiple sequence alignments (ClustalW) and phylogenetic trees (RAxML) were performed using MegAlign Pro (DNASTAR version 17.3.1). Visualization of protein topology was generated using Protter40. Prediction of transmembrane domains was performed using TMHMM41. The presence of mitochondrial targeting peptides was evaluated using TPP342, MitoFates43 and TargetP44. Highly conserved amino acids were identified by performing a multiple sequence alignment across selected species and selecting residues with a sequence logo score ≥ 3.5 bits (log₂-scaled; maximum score = 4.3 bits for 20 amino acids). To evaluate tissue expression of TMEM65, RNA-seq expression values (TPM) were retrieved from GTEx portal45. To normalize to mitochondrial abundance, we calculated the average expression of a large subset of broadly expressed mitochondrial proteins, and normalized TMEM65 expression to these values.
Mitochondrial isolation and proteinase K protection assay
Mitochondria were isolated from HeLa cells through differential centrifugation as previously described46. Mitoplasts were obtained through osmotic swelling by incubating mitochondrial fraction in 20 mM Tris-Cl pH 7.4 for 20 minutes on ice. An equal amount of mitoplasts was treated with proteinase K (100 µg/ml) at room temperature for the indicated time and the proteolytic reaction was quenched by PMSF addition. Samples were then loaded on SDS-PAGE and processed for Western blot as described above. Results are representative of at least three different proteolytic reactions.
Immunofluorescence and confocal imaging
HeLa cells expressing TMEM65-3xFlag were grown on 24 mm coverslips until 50% confluence. Cells were then washed with PBS, fixed in 4% formaldehyde for 10 minutes and quenched with 50 mM NH4Cl in PBS. Cells were permeabilized for 10 minutes with 0.1% Triton X-100 in PBS and blocked in PBS containing 2% BSA for 1 hour. Cells were then incubated with primary antibodies for 3 hours at room temperature and washed 3 times with 0.1% Triton X-100 in PBS. The following primary antibodies were used: anti-SDHA (1:100, Cell Signaling #5839), anti-Flag (1:100, Cell Signaling #2368), The appropriate isotype matched AlexaFluor-conjugated secondary antibodies (1:500, Thermo Fisher Scientific) were incubated for 1 hour at RT and coverslips were mounted with ProLong Diamond Antifade reagent (Thermo Fisher Scientific). Images were acquired on a Leica TCS-SP5-II-RS-WLL equipped with a 100x, 1.4 N.A. Plan-apochromat objective. AlexaFluor488 was excited by the 488 nm laser line and images were collected in the 495-535 nm range. AlexaFluor555 was sequentially excited with the 543 nm laser line and signal was collected in the 555-600 nm range. Pixel size was set below 100 nm to meet the Nyquist criterion. For each image, a z-stack of the whole cell was acquired, with a step size of 130 nm. Images are presented as maximum projections of the whole stack and were generated using the Fiji image processing package based on ImageJ47. Images are representative of at least three independent transfections.
[Ca2+] measurements
For experiments in intact cells using bioluminescent probes, HeLa cells were grown on 96-well plates and cotransfected with an Aequorin based probe48,49 together with the indicated constructs. siRNAs were delivered through RNAiMAX-mediated reverse transfection 72 hours before experiments. Plasmids were delivered through PEI-mediated forward transfection 36/48 hours before experiments. Untargeted siRNA (esiRNA-RLuc or universal negative siRNA) and/or pcDNA3.1 were used as controls. For aequorin reconstitution, cells were incubated for 1 hour at 37 °C with 5 μM coelenterazine in KRB (Krebs-Ringer modified buffer: 125 mM NaCl, 5 mM KCl, 1 mM Na3PO4, 1 mM MgSO4, 5.5 mM glucose, 20 mM HEPES, pH 7.4) supplemented with 1 mM CaCl2. After reconstitution, cells were washed and placed in 70 μl of KRB and luminescence was recorded on a PerkinElmer Envision plate reader equipped with a two-injector unit. Each well was monitored for 50 seconds during which histamine was first injected (final concentration is 100 μM) to activate calcium transients, and then a hypotonic, Ca2+-rich, digitonin-containing solution was added to discharge the remaining aequorin pool. Output data were analyzed and calibrated with a custom made macro-enabled Excel workbook. Where indicated, cells were pre-treated with either vehicle (DMSO) or 20 μM CGP-37157 (Tocris Bioscience).
For experiments in permeabilized cells using bioluminescent probes, HeLa cells were grown on 13 mm round glass coverslips at 50-80% confluence and cotransfected with a low-affinity mitochondrial-targeted aequorin-based probe together with the indicated constructs. For aequorin reconstitution, cells were incubated for 1-2 hours at 37 °C with 5 μM coelenterazine in KRB supplemented with 1 mM CaCl2 and then transferred to the perfusion chamber of a custom-built luminometer49,50. Experiments in permeabilized cells were performed in an intracellular-like buffer (IB: 130 mM KCl, 2 mM K2HPO4, 5 mM glutamic acid, 2 mM malic acid, 1 mM MgCl2, 20 mM HEPES, pH 7.0), supplemented with additional 10 mM KCl, 10 mM NaCl or 10 mM LiCl as specified. Plasma membrane was permeabilized by perfusing cells for 1 min with 100 μM digitonin dissolved in IB containing 500 μM EGTA. Mitochondrial Ca2+ uptake in permeabilized cells was initiated by perfusing IB containing 5 μM Ca2+, and influx was blocked by adding 10 μM ruthenium red (RuR). The experiments were terminated by lysing the cells with 100 µM digitonin in a hypotonic Ca2+-rich solution (10 mM CaCl2 in H2O), thus discharging the remaining aequorin pool. The light signal was collected and calibrated into [Ca2+] values by an algorithm based on the Ca2+ response curve of aequorin at physiological conditions of pH, [Mg2+] and ionic strength51,52.
For experiments in intact cells using fluorescent probes, HeLa cells were grown on 24 mm round glass coverslips at 30-50% confluence and cotransfected with mitoGCaMP6 together with the indicated constructs. Cells were transferred to an imaging chamber and incubated in KRB supplemented with 1 mM CaCl2. Images were acquired on an Olympus IX-73 microscope equipped with a 40x/1.3 N.A. oil-immersion semiapochromat objective (UPLFLN40XO). Excitation light was selected with a Deltaram V high speed monochromator (Photon Technology International) equipped with a 75 W Xenon Arc lamp. MitoGCaMP6 emission was collected through a 525/50 filter (Chroma) mounted on an OptoSpin25 wheel (Cairn research). Images were captured with a Kinetix22 sCMOS camera (Photometrics). The system is controlled by Metamorph 7.10 (Molecular devices) and was assembled by Crisel Instruments. MitoGCaMP6 was used in excitation ratiometric mode, where cells are alternatively illuminated every second at 485 and 410 nm32. Typical exposure time was 100 milliseconds. After background correction, 485/410 fluorescence ratio was calculated for each cell. Where indicated, 100 μM histamine was added, followed by addition of 3 μM ionomycin and 5 mM CaCl2 to check for homogenous probe saturation. Analysis was performed with the Fiji distribution of ImageJ47. Data are presented as fluorescence ratio (R, 485/410 nm), and each data point represents one coverslip (containing approximately 1 to 5 individual cells).
ΔΨm measurements
The measurement of mitochondrial membrane potential is based on the distribution of the mitochondrion-selective lipophilic cation dye tetramethylrhodamine, methyl ester (TMRM, Thermo Fisher Scientific). Cells were loaded with 20 nM TMRM for 30 minutes at 37 °C and then transferred to the imaging system. Images were acquired on a Zeiss Axiovert 200 microscope equipped with a 40x/1.3 N.A. PlanFluor objective. Excitation was performed with a Deltaram V high speed monochromator (Photon Technology International) equipped with a 75 W Xenon Arc lamp. Images were captured with a Evolve 512 Delta EMCCD (Photometrics). The system is controlled by Metamorph 7.5 and was assembled by Crisel Instruments. TMRM excitation was performed at 560 nm and emission was collected through a 590–650 nm bandpass filter. On each coverslips images of 5 random fields were acquired with a fixed 200 milliseconds exposure time. At the end of each experiment, 10μM CCCP was added to collapse ΔΨm. data are presented as average background-corrected fluorescence in resting conditions. Data were obtained from at least three independent preparations. All analyses were performed with the Fiji distribution of ImageJ.
Caenorhabditis elegans strains and maintenance
Nematode cultures were maintained at 20 °C unless differently stated. C. elegans was grown on nematode growth media (NGM) plates seeded with a bacterial lawn of OP50 E. coli following standard culture methods. The following strains were used in this study: wild type N2 (Bristol), BAN544 tag-321(bon103)IV, BAN545 C03B8.3(bon104)III, BAN547 C03B8.3(bon104)III;tag-321(bon103)IV, BAN580 C03B8.3(bon104)III;mcu-1(ju1154)IV;tag-321(bon105)IV, CZ19982 mcu-1(ju1154)IV. We are grateful to Dr. Damien M. O’Halloran (Institute for Neuroscience, George Washington University, Washington, D. C, USA) for kindly providing ncx-9(gk234237) and ncx-9(gk773708). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
CRISPR/Cas9 gene editing
Young adult C. elegans hermaphrodites were injected with customized injection mix of Cas9 protein (pre-complexed with sgRNAs), target-specific sgRNAs, target donor homology repair template (ssODN) (InVivo Biosystem, Eugene, Oregon, USA) and a plasmid encoding [myo-2p::GFP]. F1 offspring were screened for GFP expression in the pharynx, while F2 were genotyped using specific oligonucleotides upstream and downstream the expected deletions. DNA sequencing was used to confirm the mutation. Selected positive mutants were backcrossed at least five times with wt (N2) males.
DIC microscopy
Eggs from adult nematodes were extracted via bleaching, washed twice with water and then transferred onto glass slides. Images were acquired with an EPI-SCOPE1 Apotome microscope equipped with a 63x oil immersion objective (Zeiss).
Hatching assays
Two independent experimental paradigms were used to assess hatching of larvae. In the first one, animals were grown at 20 °C, collected at adulthood and bleached with a hypochlorite solution. The extracted eggs were transferred onto bacteria-seeded NGM plates and incubated at either 20°C (for maintenance) or 25 °C. Individual young adults were then transferred on freshly bacteria-seeded plates and the hatched progeny was quantified over time (up to 48 hours). For each experiment, between 8 and 15 young adult nematodes per genotype were used. The second hatching assay was a population-based analysis. Animals were grown at 20 °C, collected at adulthood and bleached with a hypochlorite solution. Eggs were plated in freshly prepared bacteria-seed plates and transferred at 25 °C until hatched animals reached adulthood. Then, synchronized gravid hermaphrodites were collected and bleached with a hypochlorite solution. The extracted eggs were seeded on plates, counted and incubated at 25 °C. Hatched larvae were quantified using a stereomicroscope.
Lifespan assay
Gravid adult animals grown at 20 °C were collected and bleached using a NaClO/NaOH solution to extract eggs. Synchronized populations were then placed in an incubator at either 20 °C or at 25 °C, according to the experimental paradigm. Animals were transferred every 2 days and scored at least every other day for touch-provoked movement until death. Animals that died abnormally (e.g., internal hatching, vulva protrusions) were scored as censored. Survival curves were generated using GraphPad Prism software (GraphPad Software Inc., San Diego, USA).
Statistics and reproducibility
No statistical method was used to predetermine sample size. All experiments were repeated independently at least 3 times with similar results. For the analysis of [Ca2+] dynamics, representative traces are shown together with bar graphs. No data were excluded from the analyses. Experiments were not randomized, and investigators were not blinded to allocation during experiments or outcome assessment. Bar graphs display mean ± standard deviation (s.d.), with individual data points represented by dots unless otherwise indicated. Variance was assessed using one-way, two-way, or three-way ANOVA, as specified in the figure legends. Comparisons between two groups were performed using unpaired two-sample t-tests. For multiple comparisons, Holm–Šidák post hoc tests were applied. Where applicable, raw data and exact p-values are provided in the source data tables. All statistical analyses were conducted using OriginPro (OriginLab), Prism (GraphPad), or Excel (Microsoft).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Additional unprocessed and processed data supporting the findings are available in the Supplementary Information or Source Data files. Source data are provided with this paper.
Code availability
No custom code or mathematical algorithms were used in this study. All analyses were conducted using established methods and standard software, as indicated in the “Methods” section.
References
Berridge, M. J., Lipp, P. & Bootman, M. D. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol 1, 11–21 (2000).
Montero, M. et al. Chromaffin-cell stimulation triggers fast millimolar mitochondrial Ca2+ transients that modulate secretion. Nat Cell Biol 2, 57–61 (2000).
Rizzuto, R., De Stefani, D., Raffaello, A. & Mammucari, C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol 13, 566–578 (2012).
Csordás, G., Weaver, D. & Hajnóczky, G. Endoplasmic Reticulum–Mitochondrial Contactology: Structure and Signaling Functions. Trends Cell Biol 28, 523–540 (2018).
Denton, R. M. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim Biophys Acta 1787, 1309–1316 (2009).
Baumgartner, H. K. et al. Calcium elevation in mitochondria is the main Ca2+ requirement for mitochondrial permeability transition pore (mPTP) opening. J Biol Chem 284, 20796–20803 (2009).
Garbincius, J. F. & Elrod, J. W. Mitochondrial calcium exchange in physiology and disease. Physiol Rev 102, 893–992 (2022).
Bernardi, P. et al. Identity, structure, and function of the mitochondrial permeability transition pore: controversies, consensus, recent advances, and future directions. Cell Death & Differentiation 2023 30, 1869–1885 (2023).
Hoth, M., Button, D. C. & Lewis, R. S. Mitochondrial control of calcium-channel gating: a mechanism for sustained signaling and transcriptional activation in T lymphocytes. Proc Natl Acad Sci USA 97, 10607–10612 (2000).
Zeviani, M. & Viscomi, C. Mitochondrial Neurodegeneration. Cells 11, 637–682 (2022).
Zeviani, M. & Carelli, V. Mitochondrial disorders. Curr Opin Neurol 20, 564–571 (2007).
Saris, N. E. & Carafoli, E. A historical review of cellular calcium handling, with emphasis on mitochondria. Biochemistry (Mosc) 70, 187–194 (2005).
Bernardi, P. Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol Rev 79, 1127–1155 (1999).
Kamer, K. J. & Mootha, V. K. The molecular era of the mitochondrial calcium uniporter. Nat Rev Mol Cell Biol 16, 545–553 (2015).
Palty, R. et al. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci USA 107, 436–441 (2010).
Jiang, D., Zhao, L. & Clapham, D. E. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science (1979) 326, 144–147 (2009).
Tsai, M.-F. et al. Functional reconstitution of the mitochondrial Ca2+/H+ antiporter Letm1. J Gen Physiol 143, 67–73 (2013).
Austin, S. et al. TMBIM5 is the Ca2+ /H+ antiporter of mammalian mitochondria. EMBO Rep 23, e54978–e54997 (2022).
Patron, M. et al. Regulation of mitochondrial proteostasis by the proton gradient. EMBO J 41, e110476–e110500 (2022).
Nazli, A. et al. A mutation in the TMEM65 gene results in mitochondrial myopathy with severe neurological manifestations. European Journal of Human Genetics 2017 25, 744–751 (2017).
De Stefani, D. et al. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340 (2011).
Sharma, P. et al. Evolutionarily conserved intercalated disc protein Tmem65 regulates cardiac conduction and connexin 43 function. Nat Commun 6, 8391–8404 (2015).
Teng, A. C. T. et al. Tmem65 is critical for the structure and function of the intercalated discs in mouse hearts. Nat Commun 13, 6166–6184 (2022).
Rath, S. et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res 49, D1541–D1547 (2021).
Morgenstern, M. et al. Quantitative high-confidence human mitochondrial proteome and its dynamics in cellular context. Cell Metab 12, 2464–2483 (2021).
Hansen, F. M. et al. Mitochondrial phosphoproteomes are functionally specialized across tissues. bioRxiv 2022 03, 485457 (2022).
Nishimura, N., Gotoh, T., Oike, Y. & Yano, M. TMEM65 is a mitochondrial inner-membrane protein. PeerJ 2, e349 (2014).
Rhee, H. W. et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science (1979) 339, 1328–1331 (2013).
Tosatto, A., Rizzuto, R. & Mammucari, C. Ca2+ Measurements in Mammalian Cells with Aequorin-based Probes. Bio Protoc 7, e2155–e2162 (2017).
Kim, S. et al. Transmembrane glycine zippers: Physiological and pathological roles in membrane proteins. Proc Natl Acad Sci USA 102, 14278–14283 (2005).
Yang, D. et al. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci USA 99, 9942–9947 (2002).
Hill, J. M. M. et al. Measuring baseline ca(2+) levels in subcellular compartments using genetically engineered fluorescent indicators. Methods Enzymol 543, 47–72 (2014).
Carafoli, E., Tiozzo, R., Lugli, G., Crovetti, F. & Kratzing, C. The release of calcium from heart mitochondria by sodium. J Mol Cell Cardiol 6, 361–371 (1974).
Cox, D. A., Conforti, L., Sperelakis, N. & Matlib, M. A. Selectivity of inhibition of Na(+)-Ca2+ exchange of heart mitochondria by benzothiazepine CGP-37157. J Cardiovasc Pharmacol 21, 595–599 (1993).
Hernández-SanMiguel, E. et al. The mitochondrial Na+/Ca2+ exchanger plays a key role in the control of cytosolic Ca2+ oscillations. Cell Calcium 40, 53–61 (2006).
Garbincius, J. F. et al. TMEM65 regulates and is required for NCLX-dependent mitochondrial calcium efflux. Nature Metabolism 2025 7, 714–729 (2025).
Zhang, J. L. et al. TMEM65 functions as the mitochondrial Na+/Ca2+ exchanger. Nat Cell Biol 27, 1301–1310 (2025).
Sharma, V., Roy, S., Sekler, I. & O’Halloran, D. M. The NCLX-type Na+/Ca2+ Exchanger NCX-9 Is Required for Patterning of Neural Circuits in Caenorhabditis elegans. J Biol Chem 292, 5364–5377 (2017).
Eddy, S. R. Accelerated Profile HMM Searches. PLoS Comput Biol 7, e1002195 (2011).
Omasits, U., Ahrens, C. H., Müller, S. & Wollscheid, B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 (2014).
Krogh, A., Larsson, B., Von Heijne, G. & Sonnhammer, E. L. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580 (2001).
Savojardo, C., Martelli, P. L., Fariselli, P. & Casadio, R. TPpred3 detects and discriminates mitochondrial and chloroplastic targeting peptides in eukaryotic proteins. Bioinformatics 31, 3269–3275 (2015).
Fukasawa, Y. et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics 14, 1113–1126 (2015).
Armenteros, J. J. A. et al. Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance 2, e201900429–e201900443 (2019).
Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–585 (2013).
Frezza, C., Cipolat, S. & Scorrano, L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2, 287–295 (2007).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012).
Granatiero, V., Patron, M., Tosatto, A., Merli, G. & Rizzuto, R. Using targeted variants of aequorin to measure Ca2+ levels in intracellular organelles. Cold Spring Harb Protoc 2014, 86–93 (2014).
Granatiero, V., Patron, M., Tosatto, A., Merli, G. & Rizzuto, R. The use of aequorin and its variants for Ca2+ measurements. Cold Spring Harb Protoc 2014, 9–16 (2014).
Bonora, M. et al. Subcellular calcium measurements in mammalian cells using jellyfish photoprotein aequorin-based probes. Nat Protoc 8, 2105–2118 (2013).
Alonso, M. T., Rodríguez-Prados, M., Navas-Navarro, P., Rojo-Ruiz, J. & García-Sancho, J. Using aequorin probes to measure Ca 2+ in intracellular organelles. Cell Calcium 64, 3–11 (2017).
Greotti, E. & De Stefani, D. Biosensors for detection of calcium. in Methods in Cell Biology vol. 155 337–368 (Academic Press Inc., 2020).
Acknowledgements
The authors are grateful to Luca Barbato (UniPD) for the help with mutagenesis, Ms. Christiane Bartling-Kirsch (DZNE) and Mr Amal John Mathew (DZNE) for their technical assistance. Moreover, we would like to thank Dr. Christopher E. Hopkins and Dr. Ben Jussila (InVivo Biosystems) for their technical support with CRISPR/Cas9 in C. elegans. This work was supported by grants from the University of Padova (STARS@UNIPD), the Italian Ministry of University and Research (PRIN 2017YF9FBS_002, 2020RRJP5L_002 and PRIN 2020R2BP2E_002), European Union (Next Generation EU CN00000041), the Italian Association for Cancer Research (AIRC 5 per mille 2019 - ID 22759), CARIPARO (ID 59583), CARIPLO, Telethon (GJC21054) and AFM-Telethon (ID ID29104) foundations. DB is a member of: the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy – EXC2151 – 390873048, Excellence Cluster ImmunoSensation2; the ETERNITY project consortium, funded by the European Union through Horizon Europe Marie Skłodowska-Curie Actions Doctoral Networks (MSCA-DN) under the grant number 101072759; the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – SPP2453 project number 541647076. BX was supported by a Chinese Scholarship Council (CSC) fellowship (grant number 202108110050). MSH is supported by the ETERNITY project consortium (101072759).
Author information
Authors and Affiliations
Contributions
Methodology: M.V., A.K., E.S., D.B., D.D.S. Validation: M.V., L.W., A.K., V.C., B.X., M.S., M.S.H., D.B., D.D.S. Investigation: M.V., L.W., A.K., V.C., E.S., B.X., M.S., M.S.H., D.D.S. Formal analysis: M.V., L.W., A.K., V.C., E.S., D.B., D.D.S. Visualization: L.W., E.S., D.B., D.D.S. Resources: D.E. Funding acquisition: D.E., R.R., D.B., D.D.S. Conceptualization: R.R., D.B., D.D.S. Writing-Original Draft: R.R., D.B., D.D.S. Writing—review and editing: D.B., D.D.S. Supervision: R.R., D.B., D.D.S. Project administration: R.R., D.B., D.D.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Elizabeth Jonas, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vetralla, M., Wischhof, L., Kahsay, A. et al. TMEM65-dependent Ca2+ extrusion safeguards mitochondrial homeostasis. Nat Commun 17, 923 (2026). https://doi.org/10.1038/s41467-025-67647-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67647-y