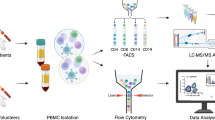
Abstract
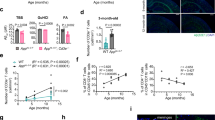
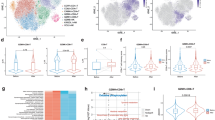
Amyloid-related imaging abnormalities (ARIA) are the principal safety concern limiting anti-amyloid therapies for Alzheimer’s disease, yet their biology remains unclear. Here we show, through multi-omic profiling of peripheral blood from three ARIA+ patients and matched controls, that ARIA is associated with coordinated reprogramming of CD8 + T cells. CD8+ effector memory (TEM) and terminally differentiated (TEMRA) subsets were expanded, clonally enriched, and transcriptionally primed for cytotoxicity and vascular trafficking. Transcription factor inference and metabolomics converged on glycolytic reprogramming favoring short-lived effector function. Ligand-receptor modeling revealed enhanced monocyte-to-T cell signaling through antigen presentation, adhesion, and chemokine axes, while integration with a cerebrovascular atlas confirmed that ARIA-associated TEMRAs are transcriptionally “addressed” for vascular engagement. Together, these findings identify a peripheral immune signature linking metabolic reprogramming, clonal CD8+ expansion, and altered intercellular communication to ARIA, with implications for biomarker development and risk mitigation pending validation in larger cohorts.
Similar content being viewed by others
Data availability
The single-cell RNA sequencing, CITE-seq, and V(D)J data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE316096. Processed count matrices are publicly available; raw sequencing data are not available due to patient privacy considerations. Interactive data browsers for this study can be accessed at https://www.morgantilab.com/datasets. The metabolomics data generated in this study have been deposited in Metabolomics Workbench under accession code ST004524. The brain single-nucleus RNA sequencing data from lecanemab-treated patients used for cross-modal integration in this study are available in the GEO database under accession code GSE263079. Source data are provided with this paper.
Code availability
Customized analysis scripts used in this study are available from the corresponding author upon reasonable request.
References
van Dyck, C. H. et al. Lecanemab in Early Alzheimer’s Disease. N Engl J Med 388, 9–21 (2023).
Honig, L. S. et al. Updated safety results from phase 3 lecanemab study in early Alzheimer’s disease. Alzheimers Res Ther 16, 105 (2024).
Cummings, J. et al. Lecanemab: Appropriate Use Recommendations. J Prev Alzheimers Dis 10, 362–377 (2023).
Kane, M. in Medical Genetics Summaries (eds V. M. Pratt et al.) (2012).
Cogswell, P. M. et al. Alzheimer Disease Anti-Amyloid Immunotherapies: Imaging Recommendations and Practice Considerations for Monitoring of Amyloid-Related Imaging Abnormalities. American Journal of Neuroradiology 46, 24–32 (2025).
Reish, N. J. et al. Multiple Cerebral Hemorrhages in a Patient Receiving Lecanemab and Treated with t-PA for Stroke. N Engl J Med 388, 478–479 (2023).
Solopova, E. et al. Fatal iatrogenic cerebral beta-amyloid-related arteritis in a woman treated with lecanemab for Alzheimer’s disease. Nat Commun 14, 8220 (2023).
Shields, L. B. E. et al. Initial Experience with Lecanemab and Lessons Learned in 71 Patients in a Regional Medical Center. J Prev Alzheimers Dis 11, 1549–1562 (2024).
van Olst, L. et al. Microglial mechanisms drive amyloid-β clearance in immunized patients with Alzheimer’s disease. Nature Medicine 31, 1604–1616 (2025).
Sallusto, F., Lenig, D., Forster, R., Lipp, M. & Lanzavecchia, A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999).
Wherry, E. J. & Kurachi, M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15, 486–499 (2015).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018).
Chang, C. H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Cao, J. et al. Effects of altered glycolysis levels on CD8(+) T cell activation and function. Cell Death Dis 14, 407 (2023).
McGettrick, A. F. & O’Neill, L. A. J. The Role of HIF in Immunity and Inflammation. Cell Metab 32, 524–536 (2020).
Trapani, J. A. & Smyth, M. J. Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol 2, 735–747 (2002).
Intlekofer, A. M. et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 6, 1236–1244 (2005).
Ingley, E. Functions of the Lyn tyrosine kinase in health and disease. Cell Commun Signal 10, 21 (2012).
Menten, P., Wuyts, A. & Van Damme, J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev 13, 455–481 (2002).
Gate, D. et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020).
Kasmani, M. Y. et al. Clonal lineage tracing reveals mechanisms skewing CD8+ T cell fate decisions in chronic infection. J Exp Med 220 https://doi.org/10.1084/jem.20220679 (2023).
Machado-Santos, J. et al. The compartmentalized inflammatory response in the multiple sclerosis brain is composed of tissue-resident CD8+ T lymphocytes and B cells. Brain 141, 2066–2082 (2018).
Pereira, B. I. et al. Senescent cells evade immune clearance via HLA-E-mediated NK and CD8(+) T cell inhibition. Nat Commun 10, 2387 (2019).
Jordao, M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363 https://doi.org/10.1126/science.aat7554 (2019).
Goddery, E. N. et al. Microglia and Perivascular Macrophages Act as Antigen Presenting Cells to Promote CD8 T Cell Infiltration of the Brain. Front Immunol 12, 726421 (2021).
Romagnoli, P. A., Premenko-Lanier, M. F., Loria, G. D. & Altman, J. D. CD8 T cell memory recall is enhanced by novel direct interactions with CD4 T cells enabled by MHC class II transferred from APCs. PLoS One 8, e56999 (2013).
Ley, K., Laudanna, C., Cybulsky, M. I. & Nourshargh, S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol 7, 678–689 (2007).
Groom, J. R. & Luster, A. D. CXCR3 in T cell function. Exp Cell Res 317, 620–631 (2011).
Jorfi, M. et al. Infiltrating CD8(+) T cells exacerbate Alzheimer’s disease pathology in a 3D human neuroimmune axis model. Nat Neurosci 26, 1489–1504 (2023).
van Olst, L. et al. Adaptive immune changes associate with clinical progression of Alzheimer’s disease. Mol Neurodegener 19, 38 (2024).
Chen, X. et al. Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy. Nature 615, 668–677 (2023).
Akbar, A. N. & Henson, S. M. Are senescence and exhaustion intertwined or unrelated processes that compromise immunity?. Nat Rev Immunol 11, 289–295 (2011).
Henson, S. M., Riddell, N. E. & Akbar, A. N. Properties of end-stage human T cells defined by CD45RA re-expression. Curr Opin Immunol 24, 476–481 (2012).
Pulko, V. et al. Human memory T cells with a naive phenotype accumulate with aging and respond to persistent viruses. Nat Immunol 17, 966–975 (2016).
Ranasinghe, S. et al. Antiviral CD8(+) T Cells Restricted by Human Leukocyte Antigen Class II Exist during Natural HIV Infection and Exhibit Clonal Expansion. Immunity 45, 917–930 (2016).
Wang, Z. et al. Clonally diverse CD38(+)HLA-DR(+)CD8(+) T cells persist during fatal H7N9 disease. Nat Commun 9, 824 (2018).
Zhou, X. et al. MHC class II regulation of CD8(+) T cell tolerance and implications in autoimmunity and cancer immunotherapy. Cell Rep 42, 113452 (2023).
Ramakrishnan, A. et al. Epigenetic dysregulation in Alzheimer’s disease peripheral immunity. Neuron 112, 1235–1248 e1235 (2024).
van Nierop, G. P. et al. Phenotypic and functional characterization of T cells in white matter lesions of multiple sclerosis patients. Acta Neuropathol 134, 383–401 (2017).
Smolders, J. et al. Tissue-resident memory T cells populate the human brain. Nat Commun 9, 4593 (2018).
Taylor, X. et al. Amyloid-beta (Abeta) immunotherapy induced microhemorrhages are linked to vascular inflammation and cerebrovascular damage in a mouse model of Alzheimer’s disease. Mol Neurodegener 19, 77 (2024).
Gross, C. C. et al. CD8(+) T cell-mediated endotheliopathy is a targetable mechanism of neuro-inflammation in Susac syndrome. Nat Commun 10, 5779 (2019).
Doedens, A. L. et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol 14, 1173–1182 (2013).
Pearce, E. L. Metabolism as a driver of immunity. Nat Rev Immunol 21, 618–619 (2021).
Pearce, E. L. & Pearce, E. J. Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643 (2013).
Winford, E. et al. Terminally differentiated effector memory T cells associate with cognitive and AD-related biomarkers in an aging-based community cohort. Immun Ageing 21, 36 (2024).
O’Sullivan, D. & Pearce, E. L. Targeting T cell metabolism for therapy. Trends Immunol 36, 71–80 (2015).
Scharping, N. E. et al. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity 45, 374–388 (2016).
Springer, T. A. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell 76, 301–314 (1994).
Groom, J. R. & Luster, A. D. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 89, 207–215 (2011).
Acknowledgements
We are deeply grateful to the patients and their caregivers who generously participated in this study. Their willingness to contribute time and effort, often under challenging circumstances, made this work possible. We thank them for their trust and commitment to advancing our understanding of Alzheimer’s disease and treatment-related complications. This work was supported by the National Institutes of Health, National Institute on Aging (R01AG081421 (LAJ), R01AG080589 (LAJ)), National Institute of Neurological Disorders and Stroke (RF1NS118558 (JMM)), National Center for Advancing Translational Sciences (TL1TR001997 (AVP)), the CNS Metabolism COBRE P20GM148326 (JMM, LAJ), and the Alzheimer’s Association ABA-25-1376140 (LAJ, JMM).
Author information
Authors and Affiliations
Contributions
L.A.J.: Conceptualization; Methodology; Investigation; Supervision; Writing-original draft; Writing-review & editing; Funding acquisition. K.S.: Formal analysis; Software; Visualization; Data curation; Writing-review & editing. A.V.P.: Investigation; Data curation; Writing-review & editing. J.L.F.: Investigation; Data curation; Writing-review & editing. A.R.E.: Resources; Investigation; Project administration; Writing-review & editing. C.L.S.: Resources; Investigation; Project administration; Writing-review & editing. D.A.H.: Methodology; Investigation; Data curation; Writing-review & editing. N.J.N.: Investigation; Data curation; Writing-review & editing. L.C.M.: Investigation; Data curation; Writing-review & editing. L.J.V.E.: Resources; Writing-review & editing. D.W.F.: Methodology; Writing-review & editing. G.E.C.: Conceptualization; Resources; Project administration; Writing-review & editing. J.M.M.: Conceptualization; Methodology; Investigation; Data curation; Formal analysis; Visualization; Writing-original draft; Writing-review & editing; Supervision; Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The Authors have no competing interests to disclose.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Johnson, L.A., Saito, K., Pallerla, A.V. et al. Clonal expansion of cytotoxic CD8⁺ T cells in lecanemab-associated ARIA. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68921-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68921-3