Abstract
The ability of plants to sense toxic and nutrient ions is critical for their growth and survival, yet how this ability is regulated remains largely unknown. We previously identified the receptor-like kinase PSKR1/ALR1 (ALR1) in Arabidopsis as a receptor that senses phytotoxic aluminium (Al) ions, which cause severe crop yield loss and forest decline on acidic soils widely distributed over the world. Here we further show that the phosphorylation status of specific Ser residues in ALR1(Ser696/698) controls plant Al-sensing ability. ALR1(Ser696/698) phosphorylation levels are rapidly reduced by Al ions, and the dephosphorylation promotes the interaction and inter-phosphorylation of ALR1 and the BAK1 coreceptor, thereby activating STOP1-dependent Al signalling and resistance. We next identify a clade of PP2C-type phosphatases (PP2CH1 and PP2CH2) that mediate the dephosphorylation of ALR1(Ser696/698). We show that Al ions rapidly increase the protein accumulation of PP2CH1/2 and promote their interaction with ALR1. The lack of both PP2CHs notably increases the phosphorylation levels of ALR1(Ser696/698), therefore reducing the strength of Al signalling. Additionally, we found a receptor-like cytoplasmic kinase, PBL27, responsible for phosphorylating ALR1(Ser696/698) and playing a negative role in the regulation of ALR1-mediated Al signalling. These findings uncover a phosphatase/kinase-mediated phosphorylation switching mechanism of ALR1 that controls plant Al-sensing ability, providing insights into ion-sensing mechanisms in living organisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data generated and analysed in this study are available in the article and its Supplementary Information file. Source data are provided with this paper.
References
Uexküll, H. R. V. & Mutert, E. Global extent, development and economic impact of acid soils. Plant Soil 171, 1–15 (1995).
Sade, H. et al. Toxicity and tolerance of aluminum in plants: tailoring plants to suit to acid soils. Biometals 29, 187–210 (2016).
Godbold, D. L., Fritz, E. & Hüttermann, A. Aluminum toxicity and forest decline. Proc. Natl Acad. Sci. USA 85, 3888–3892 (1988).
Kochian, L. V., Pineros, M. A., Liu, J. & Magalhaes, J. V. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 571–598 (2015).
Ma, J. F., Ryan, P. R. & Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 6, 273–278 (2001).
Ma, J. F., Zheng, S. J., Matsumoto, H. & Hiradate, S. Detoxifying aluminium with buckwheat. Nature 390, 569–570 (1997).
Magalhaes, J. V. et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39, 1156–1161 (2007).
Hoekenga, O. A. et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl Acad. Sci. USA 103, 9738–9743 (2006).
Liu, J., Magalhaes, J. V., Shaff, J. & Kochian, L. V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–399 (2009).
Iuchi, S. et al. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl Acad. Sci. USA 104, 9900–9905 (2007).
Yamaji, N. et al. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21, 3339–3349 (2009).
Ohyama, Y. et al. Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol. 162, 1937–1946 (2013).
Fan, W. et al. Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. N. Phytol. 208, 456–468 (2015).
Kundu, A. et al. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 21, 35–44 (2019).
Zhang, H., Tao, X., Fan, X., Zhang, S. & Qin, G. PpybZIP43 contributes to sucrose synthesis in pear fruits by activating PpySPS3 expression and interacts with PpySTOP1. Physiol. Plant. 174, e13732 (2022).
Zhang, Y. et al. F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl Acad. Sci. USA 116, 319–327 (2019).
Fang, Q., Zhou, F., Zhang, Y., Singh, S. & Huang, C. F. Degradation of STOP1 mediated by the F-box proteins RAH1 and RAE1 balances aluminum resistance and plant growth in Arabidopsis thaliana. Plant J. 106, 493–506 (2021).
Guo, J. et al. Mutation of HPR1 encoding a component of the THO/TREX complex reduces STOP1 accumulation and aluminium resistance in Arabidopsis thaliana. N. Phytol. 228, 179–193 (2020).
Zhu, Y. F., Guo, J., Zhang, Y. & Huang, C. F. The THO/TREX complex component RAE2/TEX1 is involved in the regulation of aluminum resistance and low phosphate response in Arabidopsis. Front. Plant Sci. 12, 698443 (2021).
Fang, Q. et al. Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell 32, 3921–3938 (2020).
Fang, Q., Zhang, J., Yang, D. L. & Huang, C. F. The SUMO E3 ligase SIZ1 partially regulates STOP1 SUMOylation and stability in Arabidopsis thaliana. Plant Signal. Behav. 16, 1899487 (2021).
Xu, J. et al. SIZ1 negatively regulates aluminum resistance by mediating the STOP1–ALMT1 pathway in Arabidopsis. J. Integr. Plant Biol. 63, 1147–1160 (2021).
Zhou, F. et al. The MEKK1–MKK1/2–MPK4 cascade phosphorylates and stabilizes STOP1 to confer aluminum resistance in Arabidopsis. Mol. Plant 16, 337–353 (2022).
Cao, H. et al. The RAE1–STOP1–GL2–RHD6 module regulates the ALMT1-dependent aluminum resistance in Arabidopsis. Nat. Commun. 15, 6294 (2024).
Matsubayashi, Y., Ogawa, M., Kihara, H., Niwa, M. & Sakagami, Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 142, 45–53 (2006).
Wang, J. et al. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525, 265–268 (2015).
Ding, Z. J. et al. The LRR receptor-like kinase ALR1 is a plant aluminum ion sensor. Cell Res. 34, 281–294 (2024).
Rodriguez, M. C., Petersen, M. & Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649 (2010).
Bi, G. et al. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell 30, 1543–1561 (2018).
Li, B. et al. The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat. Commun. 10, 4996 (2019).
Chen, J. et al. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proc. Natl Acad. Sci. USA 113, E5519–E5527 (2016).
Xiao, D. et al. SENESCENCE-SUPPRESSED PROTEIN PHOSPHATASE directly interacts with the cytoplasmic domain of SENESCENCE-ASSOCIATED RECEPTOR-LIKE KINASE and negatively regulates leaf senescence in Arabidopsis. Plant Physiol. 169, 1275–1291 (2015).
Hartmann, J., Linke, D., Bonniger, C., Tholey, A. & Sauter, M. Conserved phosphorylation sites in the activation loop of the Arabidopsis phytosulfokine receptor PSKR1 differentially affect kinase and receptor activity. Biochem. J. 472, 379–391 (2015).
Schweighofer, A., Hirt, H. & Meskiene, I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9, 236–243 (2004).
Ho, C. H., Lin, S. H., Hu, H. C. & Tsay, Y. F. CHL1 functions as nitrate sensor in plants. Cell 138, 1184–1194 (2009).
Liu, K. H. et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 377, 1419–1425 (2022).
Kobayashi, T. et al. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4, 2792 (2013).
Lilay, G. H. et al. Arabidopsis bZIP19 and bZIP23 act as zinc sensors to control plant zinc status. Nat. Plants 7, 137–143 (2021).
Jiang, Z. et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346 (2019).
Kaufmann, C., Motzkus, M. & Sauter, M. Phosphorylation of the phytosulfokine peptide receptor PSKR1 controls receptor activity. J. Exp. Bot. 68, 1411–1423 (2017).
Ardito, F., Giuliani, M., Perrone, D., Troiano, G. & Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (review). Int. J. Mol. Med. 40, 271–280 (2017).
Bhaskara, G. B., Wong, M. M. & Verslues, P. E. The flip side of phospho-signalling: regulation of protein dephosphorylation and the protein phosphatase 2Cs. Plant Cell Environ. 42, 2913–2930 (2019).
Wang, H., Chevalier, D., Larue, C., Ki Cho, S. & Walker, J. C. The protein phosphatases and protein kinases of Arabidopsis thaliana. Arabidopsis Book 5, e0106 (2007).
Kerk, D., Templeton, G. & Moorhead, G. B. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 146, 351–367 (2008).
Singh, A. et al. A protein phosphatase 2C, AP2C1, interacts with and negatively regulates the function of CIPK9 under potassium-deficient conditions in Arabidopsis. J. Exp. Bot. 69, 4003–4015 (2018).
Leran, S. et al. Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 8, ra43 (2015).
Bai, Z. et al. A kinase–phosphatase–transcription factor module regulates adventitious root emergence in Arabidopsis root–hypocotyl junctions. Mol. Plant 13, 1162–1177 (2020).
Xie, W. et al. PP2C.D phosphatase SAL1 positively regulates aluminum resistance via restriction of aluminum uptake in rice. Plant Physiol. 192, 1498–1516 (2023).
Chen, Q. et al. COP1 promotes ABA-induced stomatal closure by modulating the abundance of ABI/HAB and AHG3 phosphatases. N. Phytol. 229, 2035–2049 (2021).
Kong, L. et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 6, 8630 (2015).
Liang, X. & Zhou, J. M. Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 69, 267–299 (2018).
Lu, D. et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl Acad. Sci. USA 107, 496–501 (2010).
Zhang, J. et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301 (2010).
Li, L. et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338 (2014).
Chung, E. H., El-Kasmi, F., He, Y., Loehr, A. & Dangl, J. L. A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe 16, 484–494 (2014).
Shinya, T. et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79, 56–66 (2014).
Sui, J. X., Tian, H. Y., Ding, Z. J. & Kong, X. P. Crop designs: the ideal root architecture for future crop breeding. N. Crops 1, 100030 (2024).
Sun, R. Q. et al. Genome-wide characterization, identification, and isolation of auxin response factor (ARF) gene family in maize. N. Crops 2, 100053 (2025).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Allen, J. J. et al. A semisynthetic epitope for kinase substrates. Nat. Methods 4, 511–516 (2007).
Acknowledgements
This study was supported by the National Key Research and Development Program of China (grant no. 2022YFA1303402 to Z.J.D. and S.J.Z.), the National Natural Science Foundation of China (grant nos. 31970272 to Z.J.D. and 32400209 to C.X.), the Postdoctoral Fellowship Program of CPSF (grant no. GZC20241505 to C.X.), the Ministry of Education and Bureau of Foreign Experts of China (grant no. B14027 to S.J.Z.), the ZJU Tang Scholar Foundation (to Z.J.D.), and the Fundamental Research Funds for the Central Universities (grant no. 226-2024-00102 to Z.J.D.).
Author information
Authors and Affiliations
Contributions
Z.J.D., S.J.Z. and C.X. designed the research. C.X., K.K.G., M.Q.C., Y.X.W. and Z.Y.C. performed the experiments. C.X., J.Y.Y. and G.X.L. were involved in plant material generation. C.X., Z.J.D., J.M.X. and Y.R.W. analysed the data. W.N.D. and C.W.J. discussed part of the experimental design. C.X., Z.J.D., S.J.Z. and M.B. contributed to writing the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Steffen Abel and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
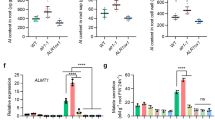
Extended Data Fig. 1 Phosphorylation of ALR1 Ser696/698 reduces Al resistance in STOP1-dependent pathway.
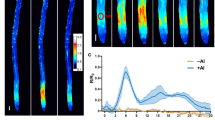
(a) Expression analysis of ALR1 in 1-week-old seedlings of indicated genotypes by RT-qPCR (n = 3 biological repeats). (b) GFP-STOP1 fluorescence signals in roots of 5-day-old STOP1p:GFP-STOP1 lines in genetic background of WT and ALR1S2D/alr1 under control and 75 µM AlCl3 treatment for 3 h (bar = 100 µm). (c) Quantification of GFP-STOP1 fluorescence intensity in root apices following Al treatment in (b) (n = 20 seedlings). (d, e) Detection of ALMT1 expression (d) and root malate secretion (e) in 1-week-old seedlings of indicated genotypes treated with or without 75 µM (d) and 25 µM (e) AlCl3 (n = 3 biological repeats). (f) Morin staining in roots of WT, ALR1S2A/alr1 and ALR1S2D/alr1 seedlings with or without 20 µM Al treatment for 1 h (bar = 100 µm). (g) H2DCF-DA staining showing ROS production in root apices of indicated genotypes under control and 75 µM Al treatment for 15 min (bar = 100 µm). (h) Phosphorylation of the recombinant N-terminal of RbohD (RbohDN) by the ALR1CD and its variant mutants (ALR1CD-S2A, ALR1CD-S2D) in an pS39-dependent in vitro phosphorylation assay. CBS indicates coomassie blue staining. (i) BiFC assay showing the interaction of BAK1 with ALR1, ALR1S2A and ALR1S2D. (bar = 20 µm). (j) Relative fluorescence intensity in (i) was calculated with ImageJ (n = 10-20 biological repeats). (k) Pull-down assay of the interaction between BAK1KD and ALR1 variant mutants (ALR1CD, ALR1CD-S2A and ALR1CD-S2D). GST-tagged ALR1 variant mutants were used as the bait, GST was used as the control, and TF-tagged BAK1KD was used as the prey. Error bars represent means ± SD. All experiments were repeated at least three independent times with similar results. All data were analyzed by unpaired two-tailed t test (c, j) and ordinary one-way ANOVA (d, e).
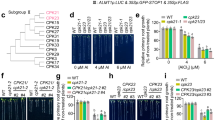
Extended Data Fig. 2 The screening of PP2Cs subfamilies interacting with ALR1.
(a) Screening of the interaction of the representative PP2Cs from different subfamilies with ALR1 in BiFC assays. Bar = 100 µm. (b) Phylogenetic tree analysis of the PP2C H subfamily. (c) Split-LUC assay showing the interaction of ALR1 with PP2CH1/2 (bar = 1 cm). All experiments were repeated at least three independent times with similar results.
Extended Data Fig. 3 The subcellular and tissue expression patterns of PP2CH1/2.
(a) PP2CH1-YFP and PP2CH2-YFP fluorescence signals in wild type (WT) mesophyll protoplasts showing the subcellular localization of PP2CHs. ALMT1-mCherry RFP was used as a plasma membrane marker. Bar = 20 µm. (b) PP2CH1pro:GUS and PP2CH2pro:GUS reporter expression in different organs. Columns 1–4 indicate cotyledons (bar = 1 mm), roots (bar = 200 μm), embryos (bar = 1 mm), and flowers (bar = 2 mm). All experiments were repeated at least three independent times with similar results.
Extended Data Fig. 4 Identification and phenotypic analysis of pp2ch mutants.
(a) Mutations in the pp2ch1 single and pp2ch1h2 double mutants. For pp2ch1h2#2, a single G base-pair deletion between nucleotides 69 and 70 from the ATG introduces a frameshift and an early stop codon in the coding region of PP2CH1 (AT1G09160). A 22-bp deletion from nucleotides 264 to 287 from the ATG introduces a frameshift and an early stop codon in PP2CH2 (AT1G47380). For pp2ch1h2#7, a single A base-pair insertion between nucleotides 69 to 70 from the ATG introduces a frameshift and an early stop codon in the coding region of PP2CH1 (AT1G09160). A single G base-pair insertion from nucleotides 283 to 284 from the ATG introduces a frameshift and an early stop codon in PP2CH2 (AT1G47380). For pp2ch1#1/#9, a single G base-pair deletion between nucleotides 61 to 62 and a single G base-pair insertion from nucleotides 68 to 69 from the ATG introduce the frameshift and early stop codon in the coding region of PP2CH1 (AT1G09160), respectively. Homozygous pp2ch2 T-DNA insertion mutant was identified by three-primers method. (b, d) Root growth of indicated genotypes under control and 1.2 mM Al treatment for 7 days (bar = 1 cm). (c, e) Quantification of relative root growth in (b, d) (n = 20 seedlings). (f, g) Expression analysis of PP2CH1/2 in PP2CH1/2 overexpression lines by RT-qPCR (n = 3 biological repeats). Error bars represent means ± SD. All experiments were repeated at least three independent times with similar results. All data were analyzed by two-tailed unpaired t test (e, f, g) and ordinary one-way ANOVA (c).
Extended Data Fig. 5 Analysis of Al content and ROS production in WT and pp2ch1h2 mutants.
(a) Morin staining in roots of WT and pp2ch1h2 mutant seedlings with or without 20 µM Al treatment for 1 h (bar = 100 µm). (b) Al content in root tips of 1-week-old WT and pp2ch1h2 mutant under Al (25 µM) treatment for 24 h (n = 4 biological repeats). (c) Hematoxylin staining in roots of WT and pp2ch1h2 mutant seedlings with or without 20 µM Al treatment for 1 h (bar = 100 µm). (d) H2DCF-DA staining showing ROS production in root apices of indicated genotypes under control and Al (75 µM) treatment for 15 min (bar = 100 µm). Error bars represent means ± SD. All experiments were repeated at least three independent times with similar results. All data were analyzed by two-tailed unpaired t test (b).
Extended Data Fig. 6 PP2CH1/2 and ALR1 regulate Al resistance in the same signaling pathway.
(a) Root growth of indicated genotypes under control and 1.2 mM Al treatment for 7 days (bar = 1 cm). (b) Quantification of relative root growth in (a) (n = 20 seedlings). (c, d) Morin and hematoxylin staining in roots of WT, alr1, pp2ch1h2 and alr1pp2ch1h2 seedlings with or without 20 µM Al treatment for 1 h (bar = 100 µm). (e, f) Expression analysis of Al-responsive genes ALMT1 (e) and MATE (f) in roots of 1-week-old seedlings treated with or without 75 µM AlCl3 for 6 h (n = 3 biological repeats). (g) Malate secretion from roots following 24 h control and Al (25 µM) treatments (n = 4 biological repeats). Error bars represent means ± SD. All experiments were repeated at least three independent times with similar results. All data were analyzed by ordinary one-way ANOVA (b, e, f and g).
Extended Data Fig. 7 Loss of PP2CHs function affects plant PSK response.
(a) Root growth of indicated genotypes following 7-day control and 100 nM PSK peptide treatments (bar = 1 cm). (b) Quantification of relative root growth in (a) calculated by dividing the root length under PSK condition by that under control treatment (bar = 1 cm, n = 10 seedlings). All experiments were repeated at least three independent times with similar results. All data were analyzed by ordinary one-way ANOVA (b).
Extended Data Fig. 8 Al-induced PP2CH1 expression is STOP1-independent.
(a, b) Expression analysis of PP2CH1/2 in 1-week-old WT and stop1 mutant roots under control and 75 μM Al (1 h, 2 h) treatments (n = 3 biological repeats). (c) FLAG-PP2CH1 proteins in 35S:FLAG-PP2CH1 root apices with or without 100 µM Al, 100 µM La and 100 µM Fe treatments for 1 h. Protein levels were detected with α-Flag antibody. Error bars represent means ± SD. All data were analyzed by two-tailed unpaired t test (a, b).
Extended Data Fig. 9 Mass spectra identifying the ALR1CD phosphorylation sites by PBL27.
Serine 696 and 698 (S696/698) were identified in the phosphopeptides of the ALR1CD.
Extended Data Fig. 10 ALR1S696/698 phosphorylation synergize with Al binding.
(a,b) Pull-down assay of the interaction between BAK1KD and ALR1 variants (ALR1CD, ALR1CD-S2A, ALR1CD-S2AC4A, ALR1CD-S2D and ALR1CD-S2DC4A) under 100 nM Al treatments (in binding buffer). GST-tagged ALR1 variants were used as the baits, GST was used as the control, and TF-tagged BAK1KD was used as the prey. (c, d) Pull-down assay of the interaction between RbohDN and ALR1 variants (ALR1CD, ALR1CD-S2A, ALR1CD-S2AC4A, ALR1CD-S2D and ALR1CD-S2DC4A). GST-tagged ALR1 variants were used as the baits, GST was used as the control, and TF-tagged RbohDN was used as the prey. (e, f) Inter-phosphorylation of ALR1CD variants (ALR1CD, ALR1CD-C4A, ALR1CD-S2A, ALR1CD-S2AC4A, ALR1CD-S2D and ALR1CD-S2DC4A, TF-tagged) and BAK1KD (GST-tagged) with or without 100 nM Al treatments in vitro. (g, h) Phosphorylation of the recombinant N-terminal of RbohD (RbohDN) by the ALR1 variants (ALR1CD, ALR1CD-C4A, ALR1CD-S2A, ALR1CD-S2AC4A, ALR1CD-S2D and ALR1CD-S2DC4A) in an pS39-dependent in vitro phosphorylation assay with or without 100 nM Al treatments. CBS indicates coomassie blue staining. All experiments were repeated at least three independent times with similar results.
Supplementary information
Supplementary Tables
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data and unprocessed western blots.
Source Data Fig. 2
Statistical source data and unprocessed western blots.
Source Data Fig. 3
Statistical source data and unprocessed western blots.
Source Data Fig. 4
Statistical source data and unprocessed western blots.
Source Data Fig. 5
Statistical source data and unprocessed western blots.
Source Data Fig. 6
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 1
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data and unprocessed western blots.
Source Data Extended Data Fig. 10
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, C., Gao, K.K., Cui, M.Q. et al. The PP2CH- and PBL27-mediated phosphorylation switch of aluminium ion receptor PSKR1/ALR1 controls plant aluminum sensing ability. Nat. Plants 11, 1074–1088 (2025). https://doi.org/10.1038/s41477-025-01983-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-01983-1