Abstract
As drought severely threatens the stability of crop yields, it is crucial to develop cultivars with enhanced drought resilience. Here we demonstrate that natural variation in ZmDapF1, encoding a putative diaminopimelate epimerase, contributes to maize drought-stress resistance without compromising grain yield. ZmDapF1 inhibits the activity of ZmMDH6, a chloroplast NADP-dependent malate dehydrogenase. ZmDapF1 gene knockout mutants exhibited significantly enhanced seedling viability and grain yield under drought stress, while maintaining high yields under normal field conditions. Natural variations in the ZmDapF1 promoter increase its binding affinity to a MYB transcription factor, ZmMYB121, which represses ZmDapF1 expression under drought. Therefore, ZmMYB121 plays a positive role in drought resistance. Knocking out ZmDapF1 resulted in increased ZmMDH6 activity, enhanced photosynthetic rate and reduced reactive oxygen species accumulation under drought, which may confer the enhanced drought resilience. Thus, genetic engineering targeting ZmDapF1 holds great potential for developing maize varieties with improved drought resilience.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq data sets are available via the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA1222190 at http://www.ncbi.nlm.nih.gov/sra/. Source data are provided with this paper.
References
Gupta, A., Rico-Medina, A. & Caño-Delgado, A. I. The physiology of plant responses to drought. Science 368, 266–269 (2020).
Lobell, D. B., Deines, J. M. & Tommaso, S. D. Changes in the drought sensitivity of US maize yields. Nat. Food 1, 729–735 (2020).
Simpkins, G. Maize sensitivity to drought. Nat. Rev. Earth Environ. 1, 625 (2020).
Liu, B. et al. Manipulating ZmEXPA4 expression ameliorates the drought-induced prolonged anthesis and silking interval in maize. Plant Cell 33, 2058–2071 (2021).
Wang, X. et al. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48, 1233–1241 (2016).
Tian, T. et al. Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm. Nat. Genet. 55, 496–506 (2023).
Dwivedi, S. L., Reynolds, M. P. & Ortiz, R. Mitigating tradeoffs in plant breeding. iScience https://doi.org/10.1016/j.isci.2021.102965 (2021).
Gao, H. et al. Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize. Mol. Plant 15, 1558–1574 (2022).
Morran, S. et al. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249 (2011).
Yang, Z. & Qin, F. The battle of crops against drought: genetic dissection and improvement. J. Integr. Plant Biol. 65, 496–525 (2023).
Mittler, R., Zandalinas, S. I., Fichman, Y. & Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 23, 663–679 (2022).
Edreva, A. Generation and scavenging of reactive oxygen species in chloroplasts: a submolecular approach. Agric. Ecosyst. Environ. 106, 119–133 (2005).
Flexas, J. & Medrano, H. Drought-inhibition of photosynthesis in C3 plants: stomatal and non-stomatal limitations revisited. Ann. Bot. 89, 183–189 (2002).
Gill, S. S. & Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930 (2010).
Pompelli, M. F. et al. Photosynthesis, photoprotection and antioxidant activity of purging nut under drought deficit and recovery. Biomass Bioenergy 34, 1207–1215 (2010).
Metzler, M. C., Rothermel, B. A. & Nelson, T. Maize NADP-malate dehydrogenase: cDNA cloning, sequence, and mRNA characterization. Plant Mol. Biol. 12, 713–722 (1989).
Kagawa, T. & Bruno, P. L. NADP-malate dehydrogenase from leaves of Zea mays: purification and physical, chemical, and kinetic properties. Arch. Biochem. Biophys. 260, 674–695 (1988).
Selinski, J. & Scheibe, R. Malate valves: old shuttles with new perspectives. Plant Biol. 21, 21–30 (2018).
Hatch, M. D. & Slack, C. R. NADP-specific malate dehydrogenase and glycerate kinase in leaves and evidence for their location in chloroplasts. Biochem. Biophys. Res. Commun. 34, 589–593 (1969).
Yang, N. et al. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat. Genet. 51, 1052–1059 (2019).
Pillai, B. et al. Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis. J. Mol. Biol. 385, 580–594 (2009).
Wiseman, J. S. & Nichols, J. S. Purification and properties of diaminopimelic acid epimerase from Escherichia coli. J. Biol. Chem. 259, 8907–8914 (1984).
Urao, T., Yamaguchi-Shinozaki, K., Urao, S. & Shinozaki, K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5, 1529–1539 (1993).
Biedenkapp, H., Borgmeyer, U., Sippel, A. E. & Klempnauer, K.-H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature 335, 835–837 (1988).
Liu, S. et al. Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol. 21, 163 (2020).
Chen, L., Hu, B., Qin, Y., Hu, G. & Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 136, 178–187 (2019).
Dubos, C. et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581 (2010).
Farrington, J. A., Ebert, M., Land, E. J. & Fletcher, K. Bipyridylium quaternary salts and related compounds. V. Pulse radiolysis studies of the reaction of paraquat radical with oxygen. Implications for the mode of action of bipyridyl herbicides. Biochim. Biophys. Acta 314, 372–381 (1973).
Shapiguzov, A. et al. Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors. Elife https://doi.org/10.7554/eLife.43284 (2019).
Miginiac-Maslow, M. et al. Light-activation of NADP-malate dehydrogenase: a highly controlled process for an optimized function. Physiol. Plant. 110, 322–329 (2000).
Ruelland, E. et al. An internal cysteine is involved in the thioredoxin-dependent activation of sorghum leaf NADP-malate dehydrogenase. J. Biol. Chem. 272, 19851–19857 (1997).
Sagong, H.-Y. & Kim, K.-J. Structural basis for redox sensitivity in Corynebacterium glutamicum diaminopimelate epimerase: an enzyme involved in l-lysine biosynthesis. Sci. Rep. 7, 42318 (2017).
Yang, N. et al. Two teosintes made modern maize. Science https://doi.org/10.1126/science.adg8940 (2023).
Chen, L. et al. Genome sequencing reveals evidence of adaptive variation in the genus Zea. Nat. Genet. 54, 1736–1745 (2022).
Ferreira, R. R., Varisi, V. A., Meinhardt, L. W., Lea, P. J. & Azevedo, R. A. Are high-lysine cereal crops still a challenge?. Braz. J. Med Biol. Res. 38, 985–994 (2005).
Wang, Q. et al. DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli. Nucleic Acids Res. 46, 6880–6892 (2018).
Kandoi, D., Mohanty, S. & Tripathy, B. C. Overexpression of plastidic maize NADP-malate dehydrogenase (ZmNADP-MDH) in Arabidopsis thaliana confers tolerance to salt stress. Protoplasma 255, 547–563 (2018).
Taniguchi, M. & Miyake, H. Redox-shuttling between chloroplast and cytosol: integration of intra-chloroplast and extra-chloroplast metabolism. Curr. Opin. Plant Biol. 15, 252–260 (2012).
Gietl, C. Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim. Biophys. Acta 1100, 217–234 (1992).
Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant. 120, 21–26 (2004).
Lu, D., Grant, M. & Lim, B. L. NAD(H) and NADP(H) in plants and mammals. Mol. Plant 18, 938–959 (2025).
Droux, M. et al. Ferredoxin-thioredoxin reductase: a catalytically active dithiol group links photoreduced ferredoxin to thioredoxin functional in photosynthetic enzyme regulation. Arch. Biochem. Biophys. 256, 372–380 (1987).
Ferte, N., Jacquot, J.-P. & Meunier, J.-C. Structural, immunological and kinetic comparisons of NADP-dependent malate dehydrogenases from spinach (C3) and corn (C4) chloroplasts. Eur. J. Biochem. 154, 587–595 (1986).
Jiang, M. et al. Rice NADP-dependent malate dehydrogenase gene OsMDH8.2 is involved in heat tolerance. Fundam. Res. https://doi.org/10.1016/j.fmre.2023.12.010 (2023).
Bradbury, P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88, 76–82 (2011).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Yin, B., Li, T., Zhang, S., Li, Z. & He, P. Sensitive analysis of 33 free amino acids in serum, milk, and muscle by ultra-high performance liquid chromatography-quadrupole-orbitrap high resolution mass spectrometry. Food Anal. Methods 9, 2814–2823 (2016).
Ogata, K. et al. Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl Acad. Sci. USA 89, 6428–6432 (1992).
Jin, H. & Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585 (1999).
Yoo, S.-D., Cho, Y.-H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016).
Tian, T. et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129 (2017).
Tang, D. et al. SRplot: a free online platform for data visualization and graphing. PLoS One 18, e0294236 (2023).
Kauss, D., Bischof, S., Steiner, S., Apel, K. & Meskauskiene, R. FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg++-branch of this pathway. FEBS Lett. 586, 211–216 (2011).
Shen, B.-R. et al. An optimized transit peptide for effective targeting of diverse foreign proteins into chloroplasts in rice. Sci. Rep. https://doi.org/10.1038/srep46231 (2017).
Keryer, E., Collin, V., Lavergne, D., Lemaire, S. & Issakidis-Bourguet, E. Characterization of Arabidopsis mutants for the variable subunit of ferredoxin:thioredoxin reductase. Photosynth. Res. 79, 265–274 (2004).
Chen, W. et al. Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 375, eabg7985 (2022).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Acknowledgements
We thank the Center for Crop Functional Genomics and Molecular Breeding at China Agricultural University for their excellent support in transgenic maize construction and propagation. This research was supported by Biological Breeding-National Science and Technology Major Project (2023ZD0407101), the National Key Research and Development Program of China (2022YFF1001602), Chinese Universities Scientific Fund (2025TC135, 2025TC148), the National Natural Science Foundation of China (32430010, 32272024, 32171940) and the Beijing Nova Program (20250484919).
Author information
Authors and Affiliations
Contributions
F.Q. designed and supervised the study and revised the manuscript. Y. Lian performed the experiments, analysed the data and drafted the manuscript. S.Y. conducted gene association and nucleotide diversity analysis. T.T. processed the raw sequencing reads and quantified the gene expression levels in RNA-seq analysis. Z.Y., C.W., Y.S. and X.Y. provided important materials and suggestions for the work. X.F., C.L., S.L., T.Z., Y.W., Z.W. and Y.B. assisted in the experiment of gene cloning and plant drought resistance analysis. Y. Li, Y.Z. and X.W. helped in the yield test in the field.
Corresponding author
Ethics declarations
Competing interests
Two patent applications related to this work have been submitted by F.Q., Y. Lian, S.L. and Z.Y. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
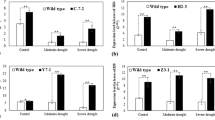
Extended Data Fig. 1 Manhattan plot for the GWAS and drought resistance assays.
a, Results of the GWAS for maize drought tolerance. The SNPs located within ZmDapF1 are highlighted as red dots. b, Relative expression levels of ZmDapF1 in ZmDapF1 (SNP530 = G)-OE lines. Data were obtained from three independent replicates, and for each replicate three plants of each genotype were mixed and analyzed. c, Drought resistance assays of ZmDapF1 (SNP530 = G)-OE lines. Left panel: the representative photos taken before and after the drought treatment. Right panel: survival rates of WT and ZmDapF1 (SNP530 = G)-OE lines after drought stress. Fifteen plants of each genotype were tested in each experiment, and five replicates were conducted. Scale bar, 5 cm. d, e, Water loss rate of detached leaves at the indicated time points. Four leaves of each genotype of 18-day old seedlings were detached and dehydrated on a clean bench. The weights of leaves were measured over the course of 3 h. Data are means ± s.d. from three biological replicates. Statistical significance was determined by a two-sided t-test in b-e.
Extended Data Fig. 2 ZmDapF1-Hap2 locus co-segregated with drought resistance.
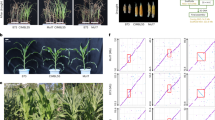
Comparison of drought resistance between the plants harboring the homozygous ZmDapF1-Hap1 and ZmDapF1-Hap2 allele from two F2 population derived from the crosses of CML118×Chang7-2 (a) and CML118×H35 (b), respectively. Left panel: representative images taken before and after drought treatment; right panel: statistical analysis of survival rates. Survival rates were compared between genotypes in at least five replicates. Fifteen plants of each genotype were tested in each experiment. Values represent means ± s.d., and statistical significance was determined by a two-sided t-test. Scale bar, 5 cm.
Extended Data Fig. 3 ZmDapF1 exhibits diaminopimelate epimerase activity in vitro but does not affect lysine content in maize plants.
a, Schematic illustration of the diaminopimelate epimerase activity assay for ZmDapF1. b, Schematic diagrams of the ZmDapF1 protein domain. The red line indicates the position of the non-synonymous variants caused by SNP530. c, In vitro diaminopimelate epimerase activity assay of two haplotypes of ZmDapF1. Data represent the means ± s.d., based on four replicated experiments. Significance was determined by one-way ANOVA (Tukey’s test). d, Analysis of free amino acid contents in the leaves of 18-day old plants of WT and ZmDapF1 (SNP530 = G)-OE lines under well-watered (WW) and water stressed (WS) conditions. Data are means ± s.d. from two biological replicates and the leaves from four plants of each kind of plants were sampled in each biological replicate.
Extended Data Fig. 4 Expression patterns of ZmMYB genes under drought and multiple sequence alignment of representative MYB proteins from different clades.
a, Heat map of gene expression levels of ZmMYBs in B73 in response to drought treatment, based on RNA-sequencing analysis quantified by FPKM (fragments per kilobase million). Drought treatment was applied until relative leaf water content reached ~70% (WS1) and ~58% (WS2). b, Reverse transcirption-quantitative PCR (RT-qPCR) analysis of expression levels of drought-induced ZmMYBs in B73 under well-watered and drought-stress conditions. Data were obtained from three independent replicates, and for each replicate leaves from four plants of each genotype were mixed and analyzed. Data represent the means ± s.d., and statistical significance was determined by a two-sided t-test. c, Phylogenetic relationships of the ZmMYB and AtMYB proteins. The phylogenetic tree was constructed using the Neighbor-Joining method with MEGA 7 software. d, ZmMYB121 and ZmMYB131 protein contain the C2 motifs, which are characteristic of most R2R3-MYB repressors. The conserved R2 and R3 MYB domains are indicated and the C2 motifs are framed.
Extended Data Fig. 5 ZmMYB121 preferentially binds to ZmDapF1-Hap2 promoter.
a, No suppression of cell growth was observed on the yeast cells transformed with ZmMYB121-mut compared with those transformed with the AD empty vector. b, Other five ZmMYB proteins did not show differently inhibition activity on the ZmDapF1-Hap1 and ZmDapF1-Hap2 promoters in yeast one-hybrid assays. c, Competition test in EMSA showing that the ZmDapF1-Hap2 sequence strongly competes with the ZmDapF1-Hap1 sequence for ZmMYB121 binding at both MBS1 (MYB binding site 1) (up) and MBS2 (down). The binding intensities were quantified using ImageJ software, and relative values are shown on the images. Similar results were obtained in three independent experiments.
Extended Data Fig. 6 ZmMYB121 is located in nucleus.
a, Confocal microscopy images of the subcellular localization of ZmMYB121-GFP in maize mesophyll protoplasts. AtAHL22-RFP was used as a nucleus marker. Scale bar, 5 μm. b, Subcellular location of ZmMYB121-GFP in tobacco leaves. Scale bar, 50 μm. Images were taken by laser confocal scanning microscope ZEISS710. Similar results were obtained in three independent experiments (a, b).
Extended Data Fig. 7 ZmMYB121 confers drought resistance through regulation of ZmDapF1.
a, Drought resistance test of ZmMYB121-OE and zmdapf1-KO plants. Left panel: representative images taken before and after drought treatment; right panel: statistical analysis of survival rates. Twelve plants of each genotype were tested in each experiment, and six replicates were conducted. Significance was determined by one-way ANOVA (Tukey’s test). Scale bar, 5 cm. b, Schematic diagram illustrating the CRISPR-targeted mutations in zmmyb121-KO plants. c, Representative images taken before and after drought treatment in drought resistance test of zmmyb121-KO plants. Scale bar, 5 cm. d, Statistical analysis of survival rates in drought resistance test of zmmyb121-KO plants. Fifteen plants of each genotype were tested in each experiment, and four replicates were conducted. e, Relative expression levels of ZmDapF1 in zmmyb121-KO plants under WW and WS conditions. Data were obtained from three independent replicates; for each replicate, leaves from three plants of each genotype were mixed and analyzed. Values represent means ± s.d., and statistical significance was determined by a two-sided t-test (d-e).
Extended Data Fig. 8 Subcellular location of ZmDapF1-GFP and ZmMDH6-GFP in maize mesophyll protoplasts.
a, Subcellular localization of ZmDapF1-GFP in maize protoplasts. Scale bar, 5 μm. b, List of proteins identified in the affinity-purified products of ZmDapF1-GFP but not GFP control by mass spectrometry analysis. The score reflected the degree of protein matching. A higher score indicated a greater possibility of the protein being correctly identified. c, Subcellular localization of ZmMDH6-GFP in maize protoplasts. Scale bar, 5 μm. In a, c, GFP signals and chloroplast autofluorescence were visualized by laser confocal scanning microscope ZEISS710. Similar results were obtained in three independent experiments (a, c).
Extended Data Fig. 9 Amino acid sequence alignments of NADP-MDH and DapF proteins from different species.
a, Amino acid sequence alignments of NADP-MDH proteins from Arabidopsis thaliana (At), Oryza sativa (Os), Zea mays (Zm) and Sorghum bicolor (Sb). Identical and highly conserved residues are presented in dark blue and pink, respectively. b, Amino acid sequence alignments of DapF proteins from Escherichia coli (E. coli), Corynebacterium glutamicum (Cg), Arabidopsis thaliana (At), Zea mays (Zm) and Oryza sativa (Os). Cysteine residues involved in the disulfide bond formation are marked with red arrows.
Extended Data Fig. 10 Plant morphology and seed lysine content analysis of different transgenic maize.
a, Representative photos of each genotype in 2023. Scale bar, 50 cm. b, c, Plant heights of WT, zmdapf1-KO plants (b) and ZmMYB121-OE lines (c) under WW and WS conditions in 2023. Data represent the means ± s.d. from four plots, for each plot 18-24 plants were quantified. Statistical significance was determined by a two-sided t-test. d, Analysis of lysine content in the seeds of different transgenic maize harvested from Zhangye (left) and Hainan (right) in 2022. Data are means ± s.d. from three biological replicates, and six seeds of each kind of plants were sampled in each biological replicate. Statistical significance was determined by a two-sided t-test.
Supplementary information
Supplementary Tables
Supplementary Tables 1 and 2.
Source data
Source Data Figs. 1–6 and Extended Data Figs. 1–4, 7, 8 and 10
Statistical source data.
Source Data Figs. 2 and 4 and Extended Data Fig. 5
Unprocessed western blots and EMSA images.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lian, Y., Yang, S., Tian, T. et al. Natural variation in ZmDapF1 enhances maize drought resilience. Nat. Plants 11, 2381–2394 (2025). https://doi.org/10.1038/s41477-025-02141-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-02141-3