Abstract
Calcium (Ca2+) is an essential macronutrient for plant growth and defence, yet the molecular mechanisms regulating its uptake from soil remain largely undefined. Through bioinformatics and electrophysiological screening, we identified a group of plant-specific proteins, named the IONIC CURRENT FAMILY A (ICAs), which confer Ca2+-permeable non-selective cation channel (CNCC) activities in heterologous systems. In Arabidopsis thaliana, AtICA1, AtICA2, AtICA3 and AtICA4 are predominantly expressed in root cells, and their proteins localize to the plasma membrane. Under either limited or excessive external Ca2+ conditions, ica1/2/3/4 quadruple mutants display hypersensitivity or reduced sensitivity, respectively, as evidenced by altered root length. In addition, these mutants show increased sensitivity to various abiotic and biotic stresses under normal Ca2+ conditions. The ica mutants lack the previously characterized CNCC-mediated currents in roots that facilitate cellular Ca2+ uptake, resulting in lower Ca2+ levels compared with wild-type (WT) plants. Our findings suggest that AtICA1/2/3/4 may function as components of CNCCs, mediating Ca2+ uptake crucial for broad environmental stress tolerance under normal Ca2+ conditions. This study provides molecular insight into the mechanisms governing Ca2+ uptake in plant roots and expands our understanding of how plants maintain Ca2+ homeostasis under varying environmental conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
White, P. J. & Broadley, M. R. Calcium in plants. Ann. Bot. 92, 487–511 (2003).
Thor, K. Calcium—nutrient and messenger. Front. Plant Sci. 10, 440 (2019).
Wang, C. & Luan, S. Calcium homeostasis and signaling in plant immunity. Curr. Opin. Plant Biol. 77, 102485 (2024).
Feng, D. et al. Exogenous calcium: its mechanisms and research advances involved in plant stress tolerance. Front. Plant Sci. 14, 1143963 (2023).
Kim, S. A., Kwak, J. M., Jae, S. K., Wang, M. H. & Nam, H. G. Overexpression of the gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol. 42, 74–84 (2001).
Wang, C. et al. Mechanisms of calcium homeostasis orchestrate plant growth and immunity. Nature 627, 382–388 (2024).
Edel, K. H., Marchadier, E., Brownlee, C., Kudla, J. & Hetherington, A. M. The evolution of calcium-based signalling in plants. Curr. Biol. 27, R667–R679 (2017).
Luan, S. & Wang, C. Calcium signaling mechanisms across kingdoms. Annu. Rev. Cell Dev. Biol. 37, 311–340 (2021).
Catterall, W. A. Voltage-gated calcium channels. Cold Spring Harb. Perspect. Biol. 3, a003947 (2011).
Julius, D. TRP channels and pain. Annu. Rev. Cell Dev. Biol. 29, 355–384 (2013).
Schmid, R. & Evans, R. J. ATP-gated P2X receptor channels: molecular insights into functional roles. Annu. Rev. Physiol. 81, 43–62 (2019).
Zelman, A. K., Dawe, A., Gehring, C. & Berkowitz, G. A. Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front. Plant Sci. 3, 95 (2012).
DeFalco, T. A., Moeder, W. & Yoshioka, K. Opening the gates: insights into cyclic nucleotide-gated channel-mediated signaling. Trends Plant Sci. 21, 903–906 (2016).
Chiu, J. C. et al. Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol. Biol. Evol. 19, 1066–1082 (2002).
Wudick, M. M., Michard, E., Nunes, C. O. & Feijó, J. A. Comparing plant and animal glutamate receptors: common traits but different fates? J. Exp. Bot. 69, 4151–4163 (2018).
Yuan, F. et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371 (2014).
Hou, C. C. et al. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 24, 632–635 (2014).
Förderer, A., Yu, D. L., Li, E. T. & Chai, J. J. Resistosomes at the interface of pathogens and plants. Curr. Opin. Plant Biol. 67, 102212 (2022).
Gao, Q. F. et al. A receptor-channel trio conducts Ca2+ signalling for pollen tube reception. Nature 607, 534–539 (2022).
Jacott, C. N., Ridout, C. J. & Murray, J. D. Unmasking Mildew Resistance Locus O. Trends Plant Sci. 26, 1006–1013 (2021).
Demidchik, V. & Tester, M. Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol. 128, 379–387 (2002).
Demidchik, V. et al. Arabidopsis thaliana root non-selective cation channels mediate calcium uptake and are involved in growth. Plant J. 32, 799–808 (2002).
Demidchik, V., Davenport, R. J. & Tester, M. Nonselective cation channels in plants. Annu. Rev. Plant Biol. 53, 67–107 (2002).
Schwacke, R. & Flügge, U. I. Identification and characterization of plant membrane proteins using ARAMEMNON. Methods Mol. Biol. 1696, 249–259 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Tammaro, P., Shimomura, K. & Proks, P. Xenopus oocytes as a heterologous expression system for studying ion channels with the patch-clamp technique. Methods Mol. Biol. 491, 127–139 (2008).
Tsubaki, M., Takeuchi, F. & Nakanishi, N. Cytochrome b561 protein family: expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim. Biophys. Acta 1753, 174–190 (2005).
Asard, H., Barbaro, R., Trost, P. & Bérczi, A. Cytochromes b561: ascorbate-mediated trans-membrane electron transport. Antioxid. Redox Signal. 19, 1026–1035 (2013).
Aravind, L. DOMON: an ancient extracellular domain in dopamine β-monooxygenase and other proteins. Trends Biochem. Sci. 26, 524–526 (2001).
Preger, V. et al. Auxin-responsive genes AIR12 code for a new family of plasma membrane b-type cytochromes specific to flowering plants. Plant Physiol. 150, 606–620 (2009).
Clúa, J. et al. A CYBDOM protein impacts iron homeostasis and primary root growth under phosphate deficiency in Arabidopsis. Nat. Commun. 15, 423 (2024).
Maniero, R. A. et al. Ferric reduction by a CYBDOM protein counteracts increased iron availability in root meristems induced by phosphorus deficiency. Nat. Commun. 15, 422 (2024).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Lu, P. L. et al. Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase. Proc. Natl Acad. Sci. USA 111, 1813–1818 (2014).
Vargas, J. D. et al. Stromal cell derived receptor 2 and cytochrome b561 are functional ferric reductases. Biochim. Biophys. Acta 1651, 116–123 (2003).
Schroeder, B. C., Cheng, T., Jan, Y. N. & Jan, L. Y. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 134, 1019–1029 (2008).
Tian, W. et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135 (2019).
Tzounopoulos, T., Maylie, J. & Adelman, J. P. Induction of endogenous channels by high levels of heterologous membrane proteins in Xenopus oocytes. Biophys. J. 69, 904–908 (1995).
Kuruma, A., Hirayama, Y. & Hartzell, H. C. A hyperpolarization- and acid-activated nonselective cation current in Xenopus oocytes. Am. J. Physiol. Cell Physiol. 279, C1401–C1413 (2000).
Pan, Y. J. et al. Dynamic interactions of plant CNGC subunits and calmodulins drive oscillatory Ca2+ channel activities. Dev. Cell 48, 710–725 (2019).
Fischer, M. et al. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 419, 259–262 (1997).
Charpentier, M. et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105 (2016).
Thor, K. et al. The calcium-permeable channel OSCA1.3 regulates plant stomatal immunity. Nature 585, 569–573 (2020).
Deng, P. et al. OsCYBDOMG1, a cytochrome b561 domain-containing protein, regulates salt tolerance and grain yield in rice. Theor. Appl. Genet. 136, 76 (2023).
Zipfel, C. & Oldroyd, G. E. D. Plant signalling in symbiosis and immunity. Nature 543, 328–336 (2017).
Yang, J., Punshon, T., Guerinot, M. L. & Hirschi, K. D. Plant calcium content: ready to remodel. Nutrients 4, 1120–1136 (2012).
Xu, J. et al. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360 (2006).
Li, L. G., Kim, B. G., Cheong, Y. H., Pandey, G. K. & Luan, S. A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl Acad. Sci. USA 103, 12625–12630 (2006).
Yang, Z. J. et al. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 10, 1199 (2019).
Li, J. F. et al. Phosphatidic acid-regulated SOS2 controls sodium and potassium homeostasis under salt stress. EMBO J. 42, e112401 (2023).
Xu, T. F., Niu, J. F. & Jiang, Z. H. Sensing mechanisms: calcium signaling mediated abiotic stress in plants. Front. Plant Sci. 13, 925863 (2022).
Monshausen, G. B., Messerli, M. A. & Gilroy, S. Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 147, 1690–1698 (2008).
Liu, K. H. et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545, 311–316 (2017).
Hu, Y. C. & Schmidhalter, U. Drought and salinity: a comparison of their effects on mineral nutrition of plants. J. Plant Nutr. Soil Sci. 168, 541–549 (2005).
Munns, R. & Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008).
Jekabsone, A., Andersone-Ozola, U., Karlsons, A., Romanovs, M. & Ievinsh, G. Effect of salinity on growth, ion accumulation and mineral nutrition of different accessions of a crop wild relative legume species, Trifolium fragiferum. Plants 11, 797 (2022).
Yamaguchi, H., Matsushita, M., Nairn, A. C. & Kuriyan, J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol. Cell 7, 1047–1057 (2001).
Chubanov, V., Köttgen, M., Touyz, R. M. & Gudermann, T. TRPM channels in health and disease. Nat. Rev. Nephrol. 20, 175–187 (2024).
Wang, J. J. & Chen, H. D. A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. aBIOTECH 1, 6–14 (2020).
Chen, J. et al. Amyloplast sedimentation repolarizes LAZYs to achieve gravity sensing in plants. Cell 186, 4788–4802 (2023).
Tian, W. et al. A molecular pathway for CO2 response in guard cells. Nat. Commun. 6, 6057 (2015).
Song, J. L. et al. Two new members of CsFEXs couple proton gradients to export fluoride and participate in reducing fluoride accumulation in low-fluoride tea cultivars. J. Agr. Food Chem. 68, 8568–8579 (2020).
McKie, A. T. et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 291, 1755–1759 (2001).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Caterina, M. J. et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824 (1997).
Ali, R., Zielinski, R. E. & Berkowitz, G. A. Expression of plant cyclic nucleotide-gated cation channels in yeast. J. Exp. Bot. 57, 125–138 (2006).
Jefferson, R. A. The GUS reporter gene system. Nature 342, 837–838 (1989).
Ren, Z. J. et al. A chloride efflux transporter, BIG RICE GRAIN 1, is involved in mediating grain size and salt tolerance in rice. J. Integr. Plant Biol. 63, 2150–2163 (2021).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Pasternak, T., Paponov, I. A. & Kondratenko, S. Optimizing protocols for Arabidopsis shoot and root protoplast cultivation. Plants 10, 375 (2021).
Veronese, P. et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273 (2006).
He, Y. X. et al. The Arabidopsis pleiotropic drug resistance transporters PEN3 and PDR12 mediate camalexin secretion for resistance to Botrytis cinerea. Plant Cell 31, 2206–2222 (2019).
Wang, W. B. et al. SHOU4/4L link cell wall cellulose synthesis to pattern-triggered immunity. New Phytol. 238, 1620–1635 (2023).
Arnon, D. I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15 (1949).
Tang, R. J. et al. A calcium signalling network activates vacuolar K+ remobilization to enable plant adaptation to low-K environments. Nat. Plants 6, 384–393 (2020).
Meng, S. F. et al. Four plasma membrane-localized MGR transporters mediate xylem Mg loading for root-to-shoot Mg translocation in. Mol. Plant 15, 805–819 (2022).
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R. & Gilroy, S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl Acad. Sci. USA 111, 6497–6502 (2014).
Wang, X. H. et al. A transceptor-channel complex couples nitrate sensing to calcium signaling in Arabidopsis. Mol. Plant 14, 774–786 (2021).
Acknowledgements
We thank S. Gilroy (University of Wisconsin-Madison) for providing YC3.6-expressing Arabidopsis seeds; J. Sheen (Harvard Medical School) for providing GCaMP6s-expressing Arabidopsis seeds; H. Chen (Tsinghua University) for providing CRISPR/Cas9 vectors; H. Wang (Capital Normal University) for assistance with HEK293T cell culture; Y. Liu (Zhongguancun Xuyue, Non-invasive Micro-test Technology Industrial Alliance) and L. Zheng [Bapu (Shanghai) Information Sci. & Tech. Co., Ltd.] for technical assistance in the Ca2+ flux measurements. This work was supported by grants from the National Key R&D Program of China (2021YFA1300702) to W.T., the National Natural Science Foundation of China (32270326) to W.T., the Key Program of the National Natural Science Foundation of China (31930010) to L.L., the National Natural Science Foundation of China (32270265) to L.L., and startup funds from the School of Advanced Agricultural Sciences at Peking University.
Author information
Authors and Affiliations
Contributions
Z.R. and W.T. conceived the project; Q.G., C.H., S.L., L.L. and W.T. supervised the project. Z.R. conducted most of the experiments. Z.L. performed TEVC experiments on Xenopus oocytes. Y.X. performed patch-clamp experiments in protoplasts. Y.D. conducted the pathogen inoculation experiments. L.G. performed Ca2+ imaging assays in HEK293T cells. Z.R. and W.T. wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Alex Costa, Erwan Michard and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sequence features, predicted structure, and phylogenetic analysis of AtICA proteins.
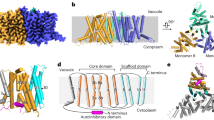
a, Plasma membrane localization of AtICA1(At4g12980), AtACYB-1, and HsSDR2 in Xenopus laevis oocytes. Scale bars, 100 μm. Experiments were repeated three times with similar results. b, AtICA1 protein contains the DOMON and cytochrome b561 domains. AtICA1 topology and transmembrane domains as predicted by the TMHMM - 2.0 software (https://services.healthtech.dtu.dk/services/TMHMM-2.0/). c, AlphaFold-predicted structures of the dimer, trimer and tetramer of AtICA1 protein (https://alphafoldserver.com/), with the dimeric configuration yielded the highest predicted Template Modeling (pTM) score of 0.34. d, Phylogenetic tree and nomenclature of ICA proteins within Arabidopsis thaliana, Oryza sativa, Physcomitrium patens, Medicago sativa, Zea mays, Triticum aestivum, Solanum tuberosum, and Glycine max. The sequences of AtICA1 homologues were retrieved from NCBI GenBank. The phylogeny of these homologues was analysed using MEGA 11 using ClustalW. At, Arabidopsis thaliana; Os, Oryza sativa; Pp, Physcomitrium patens; Mt, Medicago sativa L. Zm, Zea mays L. Ta, Triticum aestivum, St, Solanum tuberosum. Gm, Glycine max.
Extended Data Fig. 2 Electrophysiological characterization of currents associated with AtICA expression in Xenopus laevis oocytes and assessment of ferric-chelate reductase function.
a, Representative current traces from oocytes expressing AtICAs recorded in a bath solution containing 10 mM CaCl2, with additional recordings made in the presence of 1 mM DIDS or 100 µM LaCl3. b, Representative current traces from oocytes expressing AtICAs recorded in bath solutions containing BaCl2, MgCl2, KCl, NaCl, or TEA-Cl. c, AlphaFold-predicted structures of AtICA1, AtICA3, and AtICA6/HYP1 highlighting conserved histidine residues. Structural models of AtICA1, AtICA3, and AtICA6/HYP1 predicted by AlphaFold2. Conserved histidine residues implicated in potential reductase activity (AtICA1: H221, H290; AtICA3: H211, H280; AtICA6/HYP1: H221, H278) are marked by red arrows in the figure. d, Quantification of ferric-chelate reductase activity in Xenopus oocytes expressing FRO2, AtICA6/HYP1, AtICA6/HYP1H221L/H278L, AtICA1, AtICA1H221L/H290L, AtICA3, AtICA3H211L/H280L. Water-injected oocytes served as a negative control. e, Statistical analysis of current amplitudes recorded at -140 mV from oocytes expressing FRO2, AtICA6/HYP1, AtICA6/HYP1H221L/H278L, AtICA1, AtICA1H221L/H290L, AtICA3, AtICA3H211L/H280L or from water-injected controls. Bath solution contained 10 mM CaCl2. n = 10 for water-injected, n = 11 for FRO2, n = 13 for AtICA6/HYP1, n = 8 for AtICA6/HYP1H221L/H278L, n = 11 for AtICA1, n = 16 for AtICA1H221L/H290L, n = 12 for AtICA3 and AtICA3H211L/H280L (d); n = 7 for water-injected and FRO2, n = 6 for AtICA6/HYP1, AtICA6/HYP1H221L/H278L, AtICA1, AtICA1H221L/H290L, AtICA3, AtICA3H211L/H280L (e). Box plots show all points, min to max. P values were determined using two-tailed Student’s t-tests.
Extended Data Fig. 3 Ionic currents conferred by ICA expression are not significantly affected by anions in bath solutions and are dependent on cation concentrations.
a-d, Current-voltage (I-V) relationships for water-injected oocytes and oocytes expressing AtICA1 (a), AtICA2 (b), AtICA3 (c), and AtICA4 (d) were measured in solutions with different concentrations of Ba2+ (0, 2, 10, and 50 mM). n = 8 (a,b,d). n = 8 for 0 mM Ba2+, n = 7 for 2 mM Ba2+, n = 6 for 10 mM Ba2+ and 50 mM Ba2+ (c). e-h, I-V relationships for water-injected oocytes and those expressing AtICA1 (e), AtICA2 (f), AtICA3 (g), and AtICA4 (h) were evaluated in solutions with varying concentrations of K+ (0, 2, 10, and 50 mM). n = 8 for 0 and 10 mM K+, n = 9 for 2 mM K+, and 50 mM K+ (e); n = 8 for 0 and 2 mM K+, n = 9 for 10 mM K+, n = 10 for 50 mM K+ (f); n = 8 for 0 mM K+, n = 9 for 2 mM and 50 mM K+, n = 6 for 10 mM K+ (g); n = 8 for 0 and 50 mM K+, n = 6 for 2 mM K+, n = 9 for 10 mM K+ (h); i-l, I-V relationships for water-injected oocytes and oocytes expressing AtICA1 (i), AtICA2 (j), AtICA3 (k), and AtICA4 (l) were measured while perfused with solutions containing CaCl2, Ca(NO3)2, or Ca-(Glu)2, respectively. n = 8 (i-l). m-p, I-V relationships for water-injected oocytes and oocytes expressing AtICA1 (m), AtICA2 (n), AtICA3 (o), and AtICA4 (p) were measured in solutions containing KCl, KNO3, or K-Glu, respectively. n = 8 (m-p). q, I-V relationships for oocytes expressing AtICA1 were measured in solutions containing 10 mM CaCl2 with pH 5.6 or pH 7.4. n = 6. All data are mean ± s.d. Similar results were obtained from more than three independent batches of oocytes.
Extended Data Fig. 4 Expression patterns of AtICA1, AtICA2, AtICA3, and AtICA4 in Arabidopsis.
a, GUS reporter expression driven by AtICA promoters in Arabidopsis seedlings. Representative images showing GUS staining patterns in whole seedlings of transgenic lines expressing ProAtICA1:GUS, ProAtICA2:GUS, ProAtICA3:GUS, ProAtICA4:GUS, ProAtICA5:GUS, and ProAtICA6:GUS. Scale bars, 1 mm. b, GUS expression patterns in various tissues of mature plants. GUS staining was performed on 20-day-old plants and 40-day-old flowers grown in soil, with observations shown for the entire plant (first column), leaf (second column), and flowers (third and fourth columns). Scale bars are 1 cm (first column), 0.5 cm (second column), 1 cm (third column), and 200 µm (fourth column). c,d, Subcellular localization of AtICA-YFP fusion proteins in roots. YFP fluorescence signals were captured in the root tips (c) and mature root zone (d) of 7-day-old transgenic seedlings expressing ProAtICA1:AtICA1-YFP, ProAtICA2:AtICA2-YFP, ProAtICA3:AtICA3-YFP, and ProAtICA4:AtICA4-YFP. Scale bar, 100 µm (c), 50 µm (d). Experiments were repeated three times with similar results. e, Experimental setup for flg22 treatment for qRT-PCR analysis of AtICA1 expression. Schematic illustrating the treatment of 7-day-old WT seedlings grown in liquid 1/2 MS medium, followed by transfer to liquid 1/2 MS medium supplemented with 200 nM flg22 for the indicated time points. f, AtICA1 transcript abundance is induced by flg22. qRT-PCR analysis of AtICA1 transcript levels in WT seedlings at various time points following treatment with 200 nM flg22, as described in (e). g, Experimental setup for NaCl treatment for qRT-PCR analysis of AtICA1 expression. Schematic illustrating the treatment of WT seedlings grown on solid 1/2 MS medium for 7 days, followed by transfer to solid 1/2 MS medium containing 85 mM NaCl for the indicated time points. h, AtICA1 transcript abundance is induced by NaCl. qRT-PCR analysis of AtICA1 transcript levels in WT seedlings at various time points following transfer to medium containing 85 mM NaCl, as described in (g). Data are mean ± s.d. n = 3. P values were determined using two-tailed Student’s t-tests and indicate significant differences between 0 hour and other time pionts.
Extended Data Fig. 5 Characterization of Arabidopsis ica mutant alleles.
a, Schematic representation of sgRNA target sites and resulting mutations in AtICA genes. Diagrams illustrating the genomic regions of AtICA1, AtICA2, AtICA3, AtICA4, AtICA5, and AtICA6/HYP1 targeted by CRISPR-Cas9. The locations of single guide RNAs (sgRNAs) are indicated. Below each gene model, the sequences of the corresponding mutated alleles identified in the generated mutant lines are shown. Deleted bases are indicated by red dashes, inserted bases are shown in red, and substituted bases are highlighted in blue. b, List of Arabidopsis ica mutant lines and their respective alleles used in this study. A summary list detailing the names of the specific Arabidopsis ica mutant lines and the corresponding alleles they carry, as characterized in (a).
Extended Data Fig. 6 Phenotypic analysis of high-order ica mutants under various conditions.
a, ica1/2/3/4-1 and ica1/2/3/4-2 mutants exhibited no significant morphological differences compared to WT under standard growth conditions. Photographs were taken after 21 and 50 days of growth in the greenhouse. b,c, Ca2+ concentration-dependence of currents associated with AtICA1 and AtICA3 expression in Xenopus laevis oocytes. Statistical analysis was performed on current amplitudes at -140 mV measured in oocytes expressing AtICA1 (b) and AtICA3 (c), using solutions with different concentrations of Ca2+ (0.05, 0.1, 1, 5, 10, and 18 mM). d, Triple mutants ica1/3/4, ica2/3/4, ica1/2/3, and ica1/2/4 showed no obvious morphological differences compared to WT under standard growth conditions, with photographs taken at 21 and 50 days. e, Triple mutants ica1/3/4, ica2/3/4, ica1/2/3, and ica1/2/4 displayed similar behaviors to WT in response to NaCl treatment. Photographs were taken after seedlings were grown in 1/2 MS or 1/2 MS supplemented with 100 mM NaCl for 12 days. f,g, Quantification of primary root lengths for seedlings grown in 1/2 MS (f) or in 1/2 MS plus 100 mM NaCl (g) from the experiments described in (e). h, Triple mutants ica1/3/4, ica2/3/4, ica1/2/3, and ica1/2/4 were comparable to WT in response to mannitol treatment, with photographs taken after 12 days of growth in 1/2 MS or 1/2 MS with 250 mM mannitol. i,j, Primary root lengths of seedlings grown in 1/2 MS (i) or in 1/2 MS plus 250 mM mannitol (j) were quantified from the experiments in (h). k, Triple mutants similarly responded to flg22 treatment as WT, with photographs taken after 12 days of growth in 1/2 MS or 1/2 MS plus 200 nM flg22. l,m, Quantification of primary root lengths for seedlings grown in 1/2 MS (l) or in 1/2 MS plus 200 nM flg22 (m) from the experiments described in (k). n, ica1/3/4, ica2/3/4, ica1/2/3, and ica1/2/4 triple mutants displayed comparable sensitivity to Botrytis cinerea infection, with disease symptoms photographed 48 h post-inoculation with Botrytis cinerea spores. o, Lesion diameters were measured at 48 h post-inoculation. p, ica1/2/3/4/5/6 sextuple mutants exhibited similar sensitivity to NaCl treatment compared to the ica1/2/3/4 quadruple mutants, with photographs taken after 12 days of growth in 1/2 MS or in 1/2 MS plus 100 mM NaCl. q, Quantification of primary root length for seedlings from the experiments described in (p). Data are presented as mean ± s.d. (b,c,q). Box plots show all points, min to max (f,g,i,j,l,m,o). P values indicate significant differences between WT and the mutants and are derived from two-sided Student’s t-tests.
Extended Data Fig. 7 Abiotic stress sensitivity of ica quadruple mutants in iron- and vitamin-deficient medium, and complementation of the salt-hypersensitive phenotype.
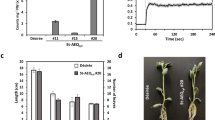
a, b, ica quadruple mutants display hypersensitivity to high salt treatment. Photographs of WT, ica1/2/3/4-1, and ica1/2/3/4-2 plants grown in 1/2 MS, 1/2 MS + 85 mM NaCl, or 1/2 MS + 100 mM NaCl for 12 days are shown in (a). Scale bars, 1 cm. Quantification of primary root lengths corresponding to (a) is presented in (b). c,d, ica quadruple mutants exhibit hypersensitivity to mannitol treatment. Photographs of WT, ica1/2/3/4-1, and ica1/2/3/4-2 plants grown in 1/2 MS, 1/2 MS + 250 mM mannitol, or 1/2 MS + 300 mM mannitol for 12 days are presented in (c). Scale bars, 1 cm. The quantification of primary root lengths from (c) is shown in (d). e,f, ica quadruple mutants demonstrate hypersensitivity to heat stress. 5-day-old WT, ica1/2/3/4-1, and ica1/2/3/4-2 seedlings were subjected to 37 °C for 3 days or 45 °C for 50 minutes. Photographs were taken after either a 10-day or a 7-day recovery period in the greenhouse (e). Scale bars, 1 cm. Quantitative analysis of primary root lengths from (e) is depicted in (f). Seedlings were grown in 1/2 MS basal salts without adding iron and vitamins (a-f). g,h, The expression of AtACYB-1 does not restore normal salt tolerance in the ica1/2/3/4 mutants. Six independent transgenic lines expressing proAtICA3:ACYB-1 in the ica1/2/3/4 background exhibit hypersensitivity to high salt stress, comparable to that of the ica1/2/3/4 mutants. The quantification of primary root lengths is presented in (g) and (h). i, The expression of AtICA1 successfully complements the hypersensitive phenotype of the ica1/2/3/4 mutants under high salt stress. The quantification of primary root length is shown. j, The expression of AtICA3 complements the hypersensitivity of the ica1/2/3/4 mutants to high salt stress. The quantification of primary root lengths is presented. Data are mean ± s.d. (b,d,f). Box plots show all points, min to max (g,h,i,j). P values were derived from two-sided Student’s t-tests and indicate significant differences between WT and the mutants.
Extended Data Fig. 8 ica1/2/3/4 quadruple mutants exhibit defective whole-cell Na⁺ currents in root epidermal cells and reduced Ca2+ influx in root surface.
a-d, ica1/2/3/4 quadruple mutants exhibit defective whole-cell Na⁺ currents in root epidermal cells. Typical whole-cell recordings of Na⁺ currents in WT, ica1/2/3/4-1, and ica1/2/3/4-2 root epidermal cells under normal conditions (a). The current-voltage (I-V) relationships derived from these recordings are shown in (b). Similar analyses were conducted for WT, ica1/2/3/4-1, and ica1/2/3/4-2 root epidermal cells under NaCl stress (c and d). e-h, The whole-cell K⁺ currents in WT, ica1/2/3/4-1, and ica1/2/3/4-2 root epidermal cells are comparable. Typical whole-cell recordings of K⁺ currents in WT, ica1/2/3/4-1, and ica1/2/3/4-2 root epidermal cells under normal conditions (e). The current-voltage (I-V) relationships derived from these recordings are shown in (f). Similar analyses were conducted for WT, ica1/2/3/4-1, and ica1/2/3/4-2 root epidermal cells under NaCl stress (g and h). Data are mean ± s.e.m. (b,d,f,h), n = 8 cells. Three biological repeats were performed with similar results.
Extended Data Fig. 9 Ion content analysis of ica1/2/3/4 quadruple mutants under control and salt stress conditions.
a-g, Ca (a), K (b), Na (c), Mg (d), Fe (e), Cu (f) and Zn (g) content was measured in 12-day-old seedlings grown in 1/2 MS or 1/2 MS supplemented with 85 mM NaCl. Data are mean ± s.d., n = 3 (a), n = 5 (b), n = 3 in 1/2 MS; n = 4 in 1/2 MS + NaCl (c), n = 5 (d,e), n = 5 in 1/2 MS; n = 5 for WT and ica1/2/3/4-1, n = 4 for ica1/2/3/4-2 in 1/2 MS + NaCl (f), n = 5 (g). P values were determined using two-tailed Student’s t-tests. Experiments were repeated three times with similar results.
Extended Data Fig. 10 Basal [Ca2+]cyt level and stimulus-induced [Ca2+]cyt elevations are largely comparable between WT and ica1/2/3/4 mutants.
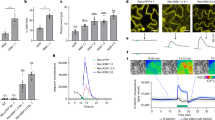
a-d, [Ca2+]cyt responses to NaCl and mannitol in root epidermal and cortical cells of plants expressing the FRET-based Ca2+ sensor YC3.6. Measurements were performed in root epidermal and cortical cells of WTYC3.6 and ica1/2/3/4YC3.6 seedlings. Roots were imaged every 2 s. Time course of YC3.6 emission ratio changes in response to local application of 200 mM NaCl (a). Basal (pre-stimulus at 12 s) and peak YC3.6 emission ratios derived from experiments as in (a) were shown in (b). Time course of YC3.6 emission ratio changes in response to local application of 600 mM mannitol (c). Basal (pre-stimulus) and peak YC3.6 emission ratios derived from experiments as in (c) were shown in (d). e-g, High salt-induced local Ca2+ signals were similar in WTGCaMP6s and ia1/2/3/4GCaMP6s roots. Measurements were performed in WTGCaMP6s and ica1/2/3/4GCaMP6s seedlings. Roots were imaged every 1 s following local application of 200 mM NaCl. Representative GCaMP6s fluorescence images of roots from WTGCaMP6s and ica1/2/3/4GCaMP6s before treatment (0 s), and at 40 s and 180 s after application of 200 mM NaCl (e). Red circles indicate the local region of interest (ROI) analyzed in (f). Scale bars, 100 µm. Time course of GCaMP6s fluorescence changes at the local ROI (f). Peak GCaMP6s fluorescence changes from (f) were shown in (g). h-j, High salt-induced propagation of Ca2+ waves were similar in WTGCaMP6s and ia1/2/3/4GCaMP6s roots. Representative GCaMP6s fluorescence images (as in e) with the red circle marking the distal ROI for wave analysis (h). Time course of GCaMP6s fluorescence changes in the distal ROI (i). Peak GCaMP6s fluorescence changes from (i) were shown in (j). Data are mean ± s.e.m. (a,c,f,i), mean ±s.d. (b,d,g,j). P values were determined using two-tailed Student’s t-tests. Experiments were repeated three times with similar results.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ren, Z., Liu, Z., Xi, Y. et al. Arabidopsis IONIC CURRENT FAMILY A proteins facilitate environmental calcium acquisition essential for stress tolerance. Nat. Plants 12, 125–139 (2026). https://doi.org/10.1038/s41477-025-02179-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-02179-3