Abstract
Late blight, caused by Phytophthora infestans (P. infestans), is one of the most destructive diseases in potato production. Yet, the interaction landscape between potato and the late blight pathogen at single-cell and spatial resolution remains elusive. Here we utilize Stereo-seq to present the spatiotemporal transcriptome atlas in potato leaves inoculated with P. infestans at single-cell resolution. We retrieve the major cell types of potato leaves, highlighting the coordination and specialization of immune responses among distinct cell types. We also conduct a comprehensive analysis to elucidate the multifaceted infection strategies employed by P. infestans to facilitate successful host colonization. Furthermore, we distinguish pathogen-targeted cells (PTCs) and surrounding PTC cells (SPCs) on the basis of pathogen distribution, thereby revealing the cellular and spatial heterogeneity of plant immune responses. Specifically, transcriptional profiling indicates that PTCs are mainly involved in cell wall reinforcement and regulation of redox homeostasis, whereas SPCs probably play a role in coordinating systemic immune signalling. Collectively, our findings elucidate a regulatory paradigm wherein PTCs and SPCs integrate with the cell-type-specific responses and dynamic expression patterns of P. infestans, orchestrating a complex and finely tuned host microenvironment during the interaction between potato and P. infestans. These findings provide a resource for advancing our understanding of the dynamic and heterogeneous nature of plant–pathogen interactions, offering novel insights to enhance crop disease resistance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All datasets supporting the findings of this study are available in the main text or Extended Data. All raw sequence data are available in CNSA at https://db.cngb.org/data_resources/project/CNP0007678 (ref. 99). The biological materials used in this study are available from S.D. on reasonable request. Source data are provided with this paper.
Code availability
The codes related to analysis are available in GitHub at https://github.com/jkfo002/Potato-Phytophthora-Spatial-Alts (ref. 100).
References
Haverkort, A. J. et al. Societal costs of late blight in potato and prospects of durable resistance through cisgenic modification. Potato Res. 51, 47–57 (2008).
Kamoun, S. et al. The top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434 (2015).
Fry, W. E. et al. Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology 105, 966–981 (2015).
Haverkort, A. J., Struik, P. C., Visser, R. G. F. & Jacobsen, E. Applied biotechnology to combat late blight in potato caused by Phytophthora infestans. Potato Res. 52, 249–264 (2009).
Hardham, A. R. Cell biology of plant–oomycete interactions. Cell Microbiol. 9, 31–39 (2007).
Boevink, P. C., Birch, P. R. J., Turnbull, D. & Whisson, S. C. Devastating intimacy: the cell biology of plant–Phytophthora interactions. New Phytol. 228, 445–458 (2020).
Whisson, S. C., Boevink, P. C., Wang, S. & Birch, P. R. The cell biology of late blight disease. Curr. Opin. Microbiol. 34, 127–135 (2016).
Zhu, J., Moreno-Pérez, A. & Coaker, G. Understanding plant pathogen interactions using spatial and single-cell technologies. Commun. Biol. 6, 814 (2023).
Faulkner, C. & Robatzek, S. Plants and pathogens: putting infection strategies and defence mechanisms on the map. Curr. Opin. Plant Biol. 15, 699–707 (2012).
Ngou, B. P. M., Ding, P. & Jones, J. D. G. Thirty years of resistance: zig-zag through the plant immune system. Plant Cell 34, 1447–1478 (2022).
Ngou, B. P. M., Jones, J. D. G. & Ding, P. Plant immune networks. Trends Plant Sci. 27, 255–273 (2022).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Dodds, P. N. & Rathjen, J. P. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548 (2010).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Jones, J. D. G., Staskawicz, B. J. & Dangl, J. L. The plant immune system: from discovery to deployment. Cell 187, 2095–2116 (2024).
Li, X., Kapos, P. & Zhang, Y. NLRs in plants. Curr. Opin. Immunol. 32, 114–121 (2015).
Jones, J. D., Vance, R. E. & Dangl, J. L. Intracellular innate immune surveillance devices in plants and animals. Science https://doi.org/10.1126/science.aaf6395 (2016).
Bigeard, J., Colcombet, J. & Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539 (2015).
Couto, D. & Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016).
Monte, I. Jasmonates and salicylic acid: evolution of defense hormones in land plants. Curr. Opin. Plant Biol. 76, 102470 (2023).
Spoel, S. H. & Dong, X. Salicylic acid in plant immunity and beyond. Plant Cell 36, 1451–1464 (2024).
Peng, Y., Yang, J., Li, X. & Zhang, Y. Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol. 72, 761–791 (2021).
Zhang, J., Coaker, G., Zhou, J. M. & Dong, X. Plant immune mechanisms: from reductionistic to holistic points of view. Mol. Plant 13, 1358–1378 (2020).
Yu, X., Liu, Z. & Sun, X. Single-cell and spatial multi-omics in the plant sciences: technical advances, applications, and perspectives. Plant Commun. 4, 100508 (2023).
Zhu, J. et al. Single-cell profiling of Arabidopsis leaves to Pseudomonas syringae infection. Cell Rep. 42, 112676 (2023).
Delannoy, E. et al. Cell specialization and coordination in Arabidopsis leaves upon pathogenic attack revealed by scRNA-seq. Plant Commun. 4, 100676 (2023).
Tang, B., Feng, L., Hulin, M. T., Ding, P. & Ma, W. Cell-type-specific responses to fungal infection in plants revealed by single-cell transcriptomics. Cell Host Microbe 31, 1732–1747.e5 (2023).
Nobori, T. et al. A rare PRIMER cell state in plant immunity. Nature https://doi.org/10.1038/s41586-024-08383-z (2025).
O’Connell, R. J. et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44, 1060–1065 (2012).
Fawke, S., Doumane, M. & Schornack, S. Oomycete interactions with plants: infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 79, 263–280 (2015).
Giacomello, S. A new era for plant science: spatial single-cell transcriptomics. Curr. Opin. Plant Biol. 60, 102041 (2021).
Seyfferth, C. et al. Advances and opportunities in single-cell transcriptomics for plant research. Annu. Rev. Plant Biol. 72, 847–866 (2021).
Depuydt, T., De Rybel, B. & Vandepoele, K. Charting plant gene functions in the multi-omics and single-cell era. Trends Plant Sci. 28, 283–296 (2023).
Cao, Y. et al. Single-cell RNA sequencing profiles reveal cell type-specific transcriptional regulation networks conditioning fungal invasion in maize roots. Plant Biotechnol. J. 21, 1839–1859 (2023).
Liang, X. et al. Single-cell transcriptomic analyses reveal cellular and molecular patterns of rubber tree response to early powdery mildew infection. Plant Cell Environ. 46, 2222–2237 (2023).
Birnbaum, K. D. Power in numbers: single-cell RNA-seq strategies to dissect complex tissues. Annu. Rev. Genet. 52, 203–221 (2018).
Denyer, T. & Timmermans, M. C. P. Crafting a blueprint for single-cell RNA sequencing. Trends Plant Sci. 27, 92–103 (2022).
Zhang, C. et al. Genome design of hybrid potato. Cell 184, 3873–3883.e12 (2021).
Xia, K. et al. The single-cell stereo-seq reveals region-specific cell subtypes and transcriptome profiling in Arabidopsis leaves. Dev. Cell 57, 1299–1310.e4 (2022).
Liu, Y. et al. Three-step biosynthesis of salicylic acid from benzoyl-CoA in plants. Nature https://doi.org/10.1038/s41586-025-09185-7 (2025).
Anjali et al. Role of plant secondary metabolites in defence and transcriptional regulation in response to biotic stress. Plant Stress 8, 100154 (2023).
Vranová, E., Coman, D. & Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700 (2013).
Krattinger, S. G. et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363 (2009).
Kurt, F. An insight into Oligopeptide Transporter 3 (OPT3) family proteins. Protein Pept. Lett. 28, 43–54 (2021).
Ma, Z. et al. A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072 (2015).
Sabbadin, F. et al. Secreted pectin monooxygenases drive plant infection by pathogenic oomycetes. Science 373, 774–779 (2021).
Turella, S. et al. Enzymatic oxidation of galacturonides from pectin breakdown contributes to stealth infection by Oomycota phytopathogens. Nat. Commun. 16, 3467 (2025).
Kubicek, C. P., Starr, T. L. & Glass, N. L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 52, 427–451 (2014).
Abrahamian, M., Ah-Fong, A. M., Davis, C., Andreeva, K. & Judelson, H. S. Gene expression and silencing studies in Phytophthora infestans reveal infection-specific nutrient transporters and a role for the nitrate reductase pathway in plant pathogenesis. PLoS Pathog. 12, e1006097 (2016).
Fernandez, J., Marroquin-Guzman, M. & Wilson, R. A. Mechanisms of nutrient acquisition and utilization during fungal infections of leaves. Annu. Rev. Phytopathol. 52, 155–174 (2014).
Birch, P. R. et al. Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11, 373–379 (2008).
Wang, S., McLellan, H., Boevink, P. C. & Birch, P. R. J. RxLR effectors: master modulators, modifiers and manipulators. Mol. Plant Microbe Interact. 36, 754–763 (2023).
Vleeshouwers, V. G. et al. Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol. 49, 507–531 (2011).
Huang, J. et al. Phytophthora effectors modulate genome-wide alternative splicing of host mRNAs to reprogram plant immunity. Mol. Plant 13, 1470–1484 (2020).
Jiang, R. H. & Tyler, B. M. Mechanisms and evolution of virulence in oomycetes. Annu. Rev. Phytopathol. 50, 295–318 (2012).
Kaschani, F. et al. An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154, 1794–1804 (2010).
He, Q. et al. Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proc. Natl Acad. Sci. USA 115, E7834–E7843 (2018).
Wang, H. et al. Uptake of oomycete RXLR effectors into host cells by clathrin-mediated endocytosis. Plant Cell 35, 2504–2526 (2023).
Oliveira-Garcia, E. et al. Clathrin-mediated endocytosis facilitates the internalization of Magnaporthe oryzae effectors into rice cells. Plant Cell 35, 2527–2551 (2023).
Wang, H. et al. Filamentous pathogen effectors enter plant cells via endocytosis. Trends Plant Sci. 28, 1214–1217 (2023).
Petre, B. et al. Host-interactor screens of Phytophthora infestans RXLR proteins reveal vesicle trafficking as a major effector-targeted process. Plant Cell 33, 1447–1471 (2021).
Liang, Q., Solis Soto, L., Haymaker, C. & Chen, K. LSGI: interpretable spatial gradient analysis for spatial transcriptomics data. Genome Biol. 26, 238 (2025).
Ringli, C., Keller, B. & Ryser, U. Glycine-rich proteins as structural components of plant cell walls. Cell. Mol. Life Sci. 58, 1430–1441 (2001).
Molina, A. et al. Plant cell wall-mediated disease resistance: current understanding and future perspectives. Mol. Plant 17, 699–724 (2024).
Elhanani, O., Ben-Uri, R. & Keren, L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell 41, 404–420 (2023).
Javed, T. & Gao, S. J. WRKY transcription factors in plant defense. Trends Genet. 39, 787–801 (2023).
Wu, Y. et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290 (2009).
Chen, L. et al. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. J. Exp. Bot. 72, 1473–1489 (2021).
Yazaki, K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 580, 1183–1191 (2006).
Chai, L. X. et al. A putative nuclear copper chaperone promotes plant immunity in Arabidopsis. J. Exp. Bot. 71, 6684–6696 (2020).
Lee, H. Y. et al. Plasma membrane-localized plant immune receptor targets H+-ATPase for membrane depolarization to regulate cell death. New Phytol. 233, 934–947 (2022).
Zuluaga, A. P. et al. Transcriptional dynamics of Phytophthora infestans during sequential stages of hemibiotrophic infection of tomato. Mol. Plant Pathol. 17, 29–41 (2016).
Bronkhorst, J. et al. An actin mechanostat ensures hyphal tip sharpness in Phytophthora infestans to achieve host penetration. Sci. Adv. 8, eabo0875 (2022).
Nobori, T. & Ecker, J. R. Yet uninfected? Resolving cell states of plants under pathogen attack. Cell Rep. Methods 3, 100538 (2023).
Choi, S. J., Lee, Z., Kim, S., Jeong, E. & Shim, J. S. Modulation of lignin biosynthesis for drought tolerance in plants. Front. Plant Sci. 14, 1116426 (2023).
Muro-Villanueva, F., Mao, X. & Chapple, C. Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr. Opin. Biotechnol. 56, 202–208 (2019).
Xie, M. et al. Regulation of lignin biosynthesis and its role in growth–defense tradeoffs. Front. Plant Sci. 9, 1427 (2018).
Pinto, L., Soler-López, L., Serrano, A. & Sánchez-Rodríguez, C. Between host and invaders: the subcellular cell wall dynamics at the plant–pathogen interface. Annu. Rev. Plant Biol. 76, 255–284 (2025).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Xu, Z. et al. STOmicsDB: a comprehensive database for spatial transcriptomics data sharing, analysis and visualization. Nucleic Acids Res. 52, D1053–D1061 (2024).
Zhang, B. et al. Generating single-cell gene expression profiles for high-resolution spatial transcriptomics based on cell boundary images. GigaByte 2024, gigabyte110 (2024).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792.e21 (2022).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods 19, 171–178 (2022).
Qiu, X. et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Amezquita, R. A. et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145 (2020).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Qiu, C. et al. Systematic reconstruction of cellular trajectories across mouse embryogenesis. Nat. Genet. 54, 328–341 (2022).
Van de Sande, B. et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020).
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J. & Gao, G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113 (2020).
Frith, M. C., Li, M. C. & Weng, Z. Cluster-Buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 31, 3666–3668 (2003).
Moerman, T. et al. GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics 35, 2159–2161 (2019).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Conesa, A. & Gotz, S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832 (2008).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Li, Y. et al. Single-cell spatiotemporal transcriptome atlas of potato-phytophthora infestans interaction. CNSA https://db.cngb.org/data_resources/project/CNP0007678 (2025).
Dai, J. et al. Potato-Phytophthora spatiotemporal transcriptome. GitHub https://github.com/jkfo002/Potato-Phytophthora-Spatial-Alts (2025).
Acknowledgements
We thank S. Raffaele, T. Zhang, X. Yang, X. Guo, K. Ye, L. Wang, F. Zhang and H. Shu for project discussion and critical comments; Y. Xia, H. Qin, J. Chen and Q. Zhong for supporting cell segmentation and database construction. This work was supported by Guangdong Major Project of Basic and Applied Basic Research (2021B0301030004), the National Natural Science Foundation of China (32488302 and 32001601), the Joint Research Program of State Key Laboratory of Agricultural and Forestry Biosecurity (SKLJRP2506), the Guangdong Provincial Genomics Data Center (2021B1212100001) and the Guangdong Laboratory of Lingnan Morden Agriculture (AGIS-ZDXM202203).
Author information
Authors and Affiliations
Contributions
S.H. and S.D. conceived the project. S.H., S.D., Y.L. and J.D. designed the study. Y.L., J.D. and Z.D. performed data analyses. Y.L., J.D. and W.S. performed experiments. X.L., Y.C., X.X., T.Y., X.M. and L.K. gave technical support and conceptual advice. Y.L. and J.D. wrote the draft manuscript. Y.L., J.D., S.D. and S.H. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Doil Choi, Edouard Evangelisti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Experimental design and Stereo-Seq at single-cell resolution.
a, Inoculation of potato leaves with P. infestans strain 88069td. Morphological observations of P. infestans at different developmental stages. c: cyst; gt: germ tube; a: appressorium; h: haustoria; m: mycelium. Scale bars presented in each time point. All images are representative of results repeated three times with similar results. b, Representative images showing fluorescent brightener (FB) staining of the plant cell walls (left panel) and Qubit ssDNA staining of oligonucleotide (right panel) on the chip to visualize cell-cell boundary. All images are representative of results repeated four times with similar results. c, Representative images showing single-cell segmentations based on the cell wall boundary and nucleus. Purple dots represent the corresponding leaf cells as showing in this paper. d, Statistics of cell numbers (left panel), gene counts (middle panel) and UMI (right panel) of each cell type. UEC, upper epidermal cell; PMC, palisade mesophyll cell; SMC, spongy mesophyll cell; LEC, lower epidermal cell; GC, guard cell; VC, vascular related cell. The box plots displaying the 25th to 75th percentiles, with a center line representing the median and whiskers extending to the minimum and maximum values within 1.5 times the interquartile range. The sample sizes for these box plots are provided in Supplementary Table 6. e, Spatial visualization of all leaf samples at four time points classified by cell type. Each dot represented a segmented cell and the color indicated cell type.
Extended Data Fig. 2 Spatiotemporal transcriptomic atlas of mock-treated potato leaves.
a, Spatial visualization of representative marker genes from each cell type of mock-treated sample. Scale bar, 100 μm. b, Dot plots showing the expression of selected marker genes for each cell type. The dot size suggests the percentage of cells expressing the corresponding marker genes, color intensity indicates the relative expression level. c, The UMAP visualization of the unsupervised clustering results for cells from mock-treated sample, each dot represented a segmented cell and the color indicated the cell type. d, Recall and precision rates of cell type segmentation. UEC, upper epidermal cell; PMC, palisade mesophyll cell; SMC, spongy mesophyll cell; LEC, lower epidermal cell; VC, vascular related cell; GC, guard cell.
Extended Data Fig. 3 Spatiotemporal transcriptomic atlas of all samples.
a, The UMAP visualization (upper panel) and spatial visualization (lower panel) of the unsupervised clustering results for cells from each separated time point, each dot represented a segmented cell and the color indicated cluster identity. b, Statistics of UMI (left panel) and gene counts (right panel) of each cluster. The box plots displaying the 25th to 75th percentiles, with a center line representing the median and whiskers extending to the minimum and maximum values within 1.5 times the interquartile range. The sample sizes for these box plots are provided in Supplementary Table 6. c, Spatial visualization of clusters showcasing the align with experimental design. Clusters were labeled as infected or non-infected based on their location within the actual P. infestans zoospore droplet-treated area. Each dot represented a segmented cell, with red, blue and green colors indicating clusters from inoculated, non-inoculated and mock regions, respectively. d, Statistics of cell number in each cluster. e, UMAP visualization presenting the clusters categorized as Inoculated, Non-inoculated and Mock-treated parts (left panel) and cell types (right panel). Each dot represented a segmented cell, red color: Inoculated cells; blue color: Non-inoculated cells; green: Mock-treated cells.
Extended Data Fig. 4 Coordination gene expression patterns responding to pathogen infection in potato leaf.
a, Bubble plot showing the enriched GO terms of the commonly upregulated (left panel) and down-regulated (right panel) genes across different cell types. The size of bubble represents the counts of genes within indicated GO terms, and the color indicated the P value adjusted with Benjamini-Hochberg method. Enrichment of GO terms were assessed using two-tailed Fisher’s exact test implemented in the clusterProfiler package. b, c, Line graph (b) and spatial visualization (c) showing the representative commonly up and down-regulated genes across different cell types. Scale bar, 100 μm. SMC, spongy mesophyll cell; UEC, upper epidermal cell; LEC, lower epidermal cell; VC, vascular related cell; PMC, palisade mesophyll cell.
Extended Data Fig. 5 Coordination gene expression patterns across all major cell types.
a, b, Line graph (a) and spatial visualization (b) showing the representative genes expression pattern across different cell types. Scale bar, 100 μm. SMC, spongy mesophyll cell; UEC, upper epidermal cell; LEC, lower epidermal cell; VC, vascular related cell; PMC, palisade mesophyll cell.
Extended Data Fig. 6 Spatial visualization of the expression patterns of P. infestans gene modules.
Spatial visualization of the expression patterns of the GHs and AAs (a), transporters (b), RxLRs and other pathogenic factors (c). Scale bar, 100 μm. GH: Glycoside Hydrolase; AA: Auxiliary Activity; RxLR: Arg-X-Leu-Arg effector.
Extended Data Fig. 7 Representative gene expression profiles associated with PTCs.
a, The images showing the NMF factors across potato leaf cells at each time point. Arrows indicate the direction of the gradients, and the color intensity in the cells reflects the loading values of the NMF factors, corresponding to the levels of gene expression in individual cells. Representative enlarged images are shown in Fig. 4c. b, Spatial visualization showing the representative genes expression pattern across infected potato leaves. The right side of the leaf indicated the inoculated side. Scale bar, 100 μm. c, The scatterplot showcasing the pseudo-temporal expression pattern of representative genes, with smooth expression curve represented by the red line.
Extended Data Fig. 8 Spatial dynamic profiles of representative clusters in potato leaves at 6hpi.
a, Neighborhood enrichment analysis between PTCs and cell clusters in spatial coordinates. Colors indicated the enrich scores. b, Heat map showing Pearson correlation coefficients between PTCs and cell clusters. The hierarchical clustering results shows in the top. c, Slingshot showing the pseudotime trajectories of the representative clusters. Each dot represented a segmented cell and colored based on clusters. d, Pseudotime trajectory analysis of representative clusters at 6 hpi. Cells are colored by pseudotime or cell type. UEC, upper epidermal cell; SMC, spongy mesophyll cell; PMC, palisade mesophyll cell; LEC, lower epidermal cell; GC, guard cell. e, Box plot showing immune scores of all major cell clusters. Colors of the box indicate the different cell clusters. The box plots displaying the 25th to 75th percentiles, with a center line representing the median and whiskers extending to the minimum and maximum values within 1.5 times the interquartile range. The sample sizes for these box plots are provided in Supplementary Table 6.
Extended Data Fig. 9 Dynamic changes of gene expression pattern across potato leaves.
a, Scatter plot showing the expression patterns of StPR1, StBCB, StWRKY41, StWRKY70, StLHCA, and StLHCB at 6hpi. b, Upset plot showing a comparison of genes in two adjacent gene clusters at 6hpi related to (Fig. 5e). c, Venn diagram showing overlapping DEGs between induced expressed genes and TFs in the middle stage of the pseudotime trajectory at 6hpi. d, Scatter plot displaying the correlated expression of StWRKY75 with representative regulated genes. Cells are colored based on clusters.
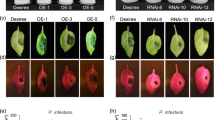
Extended Data Fig. 10 StWRKY75 is induced in host microenvironment of potato leaves and contributes to resistance by modulating multi-layered pathway.
a, The representative images showing spatial visualization of StWRKY75. The white dotted lines represent the boundary between the inoculated and the non-inoculated regions as defined by the spatial distribution of zoospore droplets. Scale bar, 100 μm. b, Box plot showing the expression level of StWRKY75 in Mock, PTCs and SPCs. Pairwise comparisons within each time point were performed using two-tailed Mann-Whitney U test (ns: not significant, *P < 0.05, **P < 0.01, ***P < 0.001). c, d, Transcript levels of StWRKY75-RNAi (c) and overexpression plants (d) were measured by RT-qPCR and normalized with endogenous StELF1. Bars represent the mean ± s.d. (n = 3 biological replicates). The asterisks indicate significant differences as assessed by one-way ANOVA (with Tukey’s test) (****P < 0.0001). e, f, Relative biomass of P. infestans was determined by qPCR of P. infestans genomic DNA normalized to potato genomic DNA at 4 dpi. Bars represent the mean ± s.d. (n = 3 biological replicates). The asterisks indicate significant differences as assessed by one-way ANOVA (with Tukey’s test) (***P < 0.001, ****P < 0.0001). This experiment was repeated three times with similar results. g, h, P. infestans infection assay on StWRKY75-RNAi (g) and overexpression plants (h). Plants were photographed at 4 dpi. Scale bar, 1.5 cm. i, Volcano plot depicting the DEGs resulting from the overexpression of StWRKY75 compared with WT. j, Bulk RNA-seq of Desiree WT and StWRKY75 overexpression plants treated with water (Mock) or P. infestans strain 88069 at 12 hpi. DEGs (adjusted P < 0.05, |log2-adjusted fold change| > 1) between Desiree WT and StWRKY75 overexpression after infection are shown. k, Transcript levels of indicate genes were measured by RT-qPCR and normalized with endogenous StELF1. Bars represent the mean ± s.d. (n = 3 biological replicates). The asterisks indicate significant differences as assessed by one-way ANOVA (with Tukey’s test) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). In b–f and k, exact P values are shown in source data with 95% confidence intervals. In i and j, Adjusted P value (Benjamini-Hochberg method) was calculated with two-tailed Wald test implement in DESeq2 package.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7.
Supplementary Table 1
Lists of cell type-specific expressed marker genes or cluster-specific expressed marker genes.
Supplementary Table 2
Gene lists and GO term analysis for coordinated and heterogeneous cell-type expression patterns.
Supplementary Table 3
Lists of genes for dynamic expression of P. infestans or host vesicle trafficking and CME.
Supplementary Table 4
Lists of genes for dynamic expression in PTCs and SPCs.
Supplementary Table 5
Lists of genes for intercellular spatial expression patterns across leaf tissue.
Supplementary Table 6
The cell counting used in this study.
Supplementary Table 7
Primers used in this study.
Source data
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Dai, J., Dong, Z. et al. Host microenvironment in potato–Phytophthora infestans interaction revealed by single-cell spatiotemporal transcriptome. Nat. Plants 12, 241–257 (2026). https://doi.org/10.1038/s41477-025-02181-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-02181-9