Abstract
Segatella is a prevalent genus within individuals’ gut microbiomes worldwide, especially in non-Western populations. Although metagenomic assembly and genome isolation have shed light on its genetic diversity, the lack of available isolates from this genus has resulted in a limited understanding of how members’ genetic diversity translates into phenotypic diversity. Within the confines of a single gut microbiome, we have isolated 63 strains from diverse lineages of Segatella. We performed comparative analyses that exposed differences in cellular morphologies, preferences in polysaccharide utilization, yield of short-chain fatty acids, and antibiotic resistance across isolates. We further show that exposure to Segatella isolates either evokes strong or muted transcriptional responses in human intestinal epithelial cells. Our study exposes large phenotypic differences within related Segatella isolates, extending this to host-microbe interactions.
Similar content being viewed by others
Introduction
The microbiomes of individuals residing in non-industrialized countries are dominated by a single clade within the Bacteroidota phylum, members of the Segatella complex1,2. The most abundant and prevalent gut Segatella species, S. copri (formerly known as Prevotella copri3,4), has been associated with various health outcomes, such as glucose intolerance, rheumatoid arthritis and low-grade systemic inflammation with HIV infection5,6,7,8,9. However, these associations often conflict, prompting debates regarding the specific role S. copri plays within the host intestinal environment. Difficulties in culturing this organism have impeded further exploration into the impact of Segatella on the gut ecosystem and host health. It is an obligate anaerobe with understudied nutritional preferences, and, despite the tremendous amount of genetic diversity reported within this clade1,3,10,11, until recently, there has been only one strain available from public strain collections. Furthermore, this strain has been recalcitrant to genetic modification and to colonization in mice12,13. Overall, these constraints have led to large gaps in experimental evidence supporting these purported roles.
Within the human gut microbiome, each species is often represented by numerous strains exhibiting genomic and functional diversity14,15,16,17. Lineages of S. copri have been identified throughout the world and assembled into metagenomic assembled genomes (MAGs). Using these and available genomes, Blanco-Míguez et al. have expanded the S. copri complex into 13 distinct species1,3,18. Against this backdrop, we asked to what extent the genomic diversity translates into functional and phenotypical variation, relevant both for the bacterium itself and for human health.
In contrast to certain related oral Segatella species, Segatella within the gut poses challenges in cultivation and manipulation. It was not until very recently that people have successfully engineered a small subset of S. copri complex12. Efforts have also been made to use mouse models to study S. copri complex, but due to limited understanding of its growth preferences and the absence of S. copri complex as a natural member of the mouse gut microbiome, colonization has posed challenges. Gellman et al. found that supplementing the mice with plant-derived microbiota-accessible carbohydrates enables colonization and maintenance of Segatella strains in mice13. In practice, this complicates experimental setups and may not be universally applicable for all research purposes.
Given the challenges of genetic modification and murine colonization, comparative genomics and phenotypic characterization offer an alternative approach to study diverse Segatella clades belong to S. copri complex. In this study, we obtained 63 isolates from a fecal sample of Fijian origin. There was considerable genomic diversity among the set of clades inhabiting this single individual. Considering that these clades may co-exist because of niche separation, the set of closely related clades provides an interesting vantage point to study the phenotypic diversity across clades. Additionally, we find significant differences in their metabolic preferences and production of short-chain fatty acids (SCFAs), in addition to their overall effects on host intestinal cells.
Results
Diverse Segatella clades isolated from a single individual’s gut microbiome
To explore the diversity of Segatella, we employed a refined culturing method on samples enriched in Segatella, as determined by metagenomic sequencing. Study participants of the Fiji Microbiome Community Project (FijiCOMP) had high overall burdens of Segatella (roughly thirty percent on average)19. We selected a 40-year-old female whose microbiome had the highest abundance of Segatella (73.9% of the known taxa, as determined by MetaPhlAn220) (Supplementary Fig. 1A). By applying our isolation procedures followed by whole genome sequencing, we obtained 63 isolates with high-quality genomes, per field standards21, barring 12 isolates that lack full-length 16S rRNA sequences.
The isolates derived from this single individual were remarkably diverse. Despite possessing nearly identical 16S rRNA sequences, the isolate genomes were clustered into six large clades, based on their genomic content and average nucleotide identity (ANI) (Fig. 1A, Supplementary Fig. 1B). Phylogenetic analysis yielded the same results (Supplementary Fig. 1C). The core genome, defined as those genes represented by over 95% of the isolate genomes, makes up only 423 genes, or 16.8% of each genome on average. Inter-clade ANIs fall below 95%, indicating higher-order relationships beyond the species level (Supplementary Fig. 1D). Whereas the isolates obtained from Clade I are likely isogenic (average ANI over 0.999) and may include mutations that occurred during culture, isolates from Clades III and IV were sparser and showed high intra-clade diversity. Interestingly, Clade I isolates were the easiest to isolate, despite their relatively low abundance within the metagenomic sample, 40% of which consists of Clade IV isolates (Fig. 1B). The discrepancy between relative abundance and our ability to culture members of each clade hints at different traits regarding oxygen tolerance and nutritional preferences, resulting in the enrichment of certain clades over the others during isolation. Based on the comparative phylogenetic analysis with isolates from Blanco-Míguez et al. (2023), the clades were identified as: Clade I (S. sanihominis), Clade II (S. sinica), Clade III (S. sinensis), Clade IV (S. copri), Clade V (S. brunsvicensis), Clade VI (S. brasiliensis) (Supplementary Fig. 1A, Supplementary Table 1).
A A cluster map of the genes within the 63 genomes isolated from the Fiji_W2.48.ST FijiCOMP gut microbiome, the genome of S. copri DSM18205 downloaded from RefSeq, and the 94 isolate genomes from previous studies. Core genes are defined as those genes appearing in >95% of all isolate genomes. The source of each genome (this study or others’) is indicated in red and gray. The 63 genomes from this study and 94 from others’ studies are indicated in red and gray, respectively. B The proportion of isolates from our isolate collection (63 in total) belong to each clade is plotted, in addition to the estimated relative abundance of each clade in the metagenome, as estimated using DiTASiC, which is showed as the percentage of total Segatella abundance. C Prevalence of Segatella clades in fecal metagenomes from Westernized and non-Westernized countries. Each dot denotes the prevalence of the Segatella clade in sample from each country. Error bars showing the standard errors. Categorization of the populations in (C) and (D) were provided by curatedMetagenomicData (cMD)22. D Probability distribution of the numbers of Segatella clades present in samples from Westernized or non-Westernized countries. The y-axis denotes the percentage of samples from each country. Error bars showing the standard errors. E A bar plot showing the percentages of each/all MAG genes identified in each isolate genome according to clade or all clades. F A bar plot showing the percentages of genes in each/all clade(s) that can be found in the MAGs.
Given that these clades were derived from a single individual living in the Fiji Islands, we sought to explore how representative these clades were of strains found globally. We examined both core and clade-specific genes in publicly available fecal metagenomic datasets from diverse geographical regions1,12. As expected, all six clades have higher prevalence in non-Western countries than Western countries (Fig. 1C, Supplementary Fig. 2A). Whereas a substantial number of gut microbiomes from non-Western populations comprise members from all six clades, the majority of gut microbiomes from Western countries contain only one clade, Clade IV (Fig. 1D). The prevalence of most clades decreases gradually with increasing income levels with the notable exceptions of Clade I (S. sanihominis) and Clade V (S. brunsvicensis), which have considerable prevalence in some populations with upper middle income (Supplementary Fig. 2B). We also checked whether the presence of specific clades was enriched in 17 specific diseases22, but no obvious associations were observed (see “Methods”).
Limitations of metagenomic assembly in distinguishing clades
Metagenomic assembly methods are not often benchmarked against isolate genomes derived from the same microbiome, but rather synthetic microbiomes or assembly statistics. Leveraging the unique opportunity of having both metagenomes and numerous isolates and the extensive use of S. copri metagenomic assembled genomes (MAGs), we assessed whether standard pipelines accurately capture genomic content. We applied four established metagenomic assembly pipelines on the fecal sample from which we obtained the Segatella isolates: (1) assembly with MEGAHIT23, followed by contig-binning by MetaBAT224; (2) assembly with MEGAHIT23, followed by multiple contig binning tools (MetaBAT225, CONCOCT26 and MaxBin 2.027) and bin refinement using DAS Tool28; (3) the same as pipeline (2) but using metaSPAdes for assembly29,30; and (4) assembly with metaSPAdes and IDBA-UD, then binning with MetaBAT230,31,32. These yielded few if any MAGs (1, 5, 3, and 0, respectively) annotated as Segatella, and, despite removing contaminating DNA using MAGpurify28, none of these methods produced MAGs meeting field standards (>90% completeness, <5% contamination, as determined by CheckM33) (Supplementary Fig. 2C). We were not able to determine whether co-assembly produces higher fidelity genomes, yet most of these methods utilize the initial assembly steps above as starting points.
Although the average genome size was similar between the isolate genomes and MAGs, there was greater variability in MAG size and gene content (Supplementary Fig. 2D, E), likely indicating incomplete assembly or contamination. The MAGs overall had poor recovery of the Segatella genomes: on average, 51.7% of the core genes and 20% of all 11,885 Segatella genes were absent from any MAG (Fig. 1E, F). The pangenome analysis incorporating both MAGs and isolate genomes showed that the majority of MAGs did not cluster with the isolate clades (Supplementary Fig. 2E). Rather, they formed into a distinct cluster, closer to Clade IV, the most abundant clade in this metagenome. Examination of genes identified in both isolate genomes and MAGs revealed that, on average, the isolate gene pool exhibited higher coverage of MAG genes than vice versa. Particularly, bin 8 displayed notably low coverage for all the isolate clades regardless of its high completeness and low contamination compared to other MAGs (Fig. 1E, F). These findings underscore the importance of obtaining whole genome sequencing for identifying novel clades of Segatella and obtaining a more complete depiction of the Segatella pangenome.
Segatella clades exhibit different cell morphologies
When cultured under the same conditions, the clades exhibit distinct cell morphologies, as observed by scanning electron microscopy (SEM), and varied in size (Fig. 2A, Supplementary Fig. 3A). While most of the selected isolates grew as rod shapes at stationary phase in Schaedler Broth, substantial filamentation repeatedly occurred in Clade VI (C6-F5) cells (Fig. 2A, Supplementary Fig. 3B). Filamentation of bacterial cells can be considered as a stress response and has been reported in several intestinal Bacteroides species, albeit not in Segatella34,35. Filamentation was observed exclusively in Clade VI (C6-F5) cells, but not in other isolates, highlighting another instance of clade-specific responses, even within identical environmental conditions.
A Scanning electron microscopy (SEM) images of select Segatella isolates, cultured in Schaedler broth until early stationary phase. Isolates names and corresponding clades are labeled on the top. B Quantification of biofilm formation by select Segatella isolates using crystal violet staining. Optical density at 590 nm was measured. Isolates are colored by the clades they belong to (n = 6 for each isolate, error bars showing the standard deviations). Dark and light gray bars denote the genomes that with or without T9SS present, respectively. C Crystal violet staining readings (OD590) of isolates with and without T9SS in the genomes. Statistical significance was calculated by two-tailed Mann-Whitney test (****p ≤ 0.0001, n = 78 for both groups). D Minimum Inhibitory Concentration (MIC, μg/mL) ranges of antibiotics on select Segatella isolates belong to different clades, as indicated by the color bars. The yellow dots denote the predicted antibiotic resistances as shown in (C). The > symbols indicate that no growth inhibition was observed within the concentration range we tested for the antibiotic.
Surprisingly, Clade II (S6-D2), even after going through a series of fixation and dehydration steps, retained a considerable amount of extracellular substance observable with (Fig. 2A). This observation aligns with the findings of a crystal violet assay, which supports the idea that S6-D2 produces significantly higher amounts of biofilm than the other isolates tested (Fig. 2B). Biofilm formation often involves self-aggregation in a secreted extracellular polymeric substance that provide metabolic advantages, in addition to antibiotic resistance36,37. While oral Segatella species have been documented to produce biofilms38, such behavior is not observed in gut variants.
The genetic mechanisms regulating biofilm formation in the Bacteroidota phylum, including Segatella species, are poorly understood. Type IX secretion systems (T9SS) were previously reported to be involved in biofilm formation of the oral species Prevotella intermedia39. Using TXSscan, we found T9SSs in isolate genomes belonging to Clade III, IV, V, and VI40. Isolates detected with T9SS had significant higher readings after crystal violet staining, further suggesting involvement of T9SS in regulating biofilm formation in S. copri even though it may not be the sole contributing factor as isolate S6-D2, which does not harbor T9SS, exhibited high readings compared with other tested isolates (Fig. 2C).
Segatella clades differ in their intrinsic antibiotic susceptibilities
Biofilm formation is often considered one tactic bacteria use to counter the effects of antibiotics. Upon inspection, overall, the Segatella genomes we isolated harbor few annotated antibiotic resistance genes. Fourteen of the isolates, the ones belonging to Clade III, IV, V, and VI, harbor the cfxA6 beta-lactamase gene and the type strain S. copri DSM18205 carries a tetQ gene providing resistance to tetracycline, both of which were confirmed by higher minimum inhibitory concentrations (MICs) to ampicillin and tetracycline, respectively (Fig. 2D, Supplementary Fig. 3C). Yet, most of the tested isolates showed low susceptibilities to multiple antibiotics, including gentamicin, chloramphenicol, ampicillin, and vancomycin. In addition, none of the isolates tested was susceptible to kanamycin. Interestingly, a few Segatella isolates proved susceptible to gentamicin, which is supposed to be ineffective against anaerobic bacteria due to its oxygen-dependent mechanism of cell membrane penetration41. These unexpected results suggest that Segatella may possess additional uncharacterized mechanisms of antibiotic resistance. Furthermore, in some cases, isolates from the same clade, even those with high ANI, exhibited different multi-drug resistance profiles, implicating mutations in metabolic genes that can alter antibiotic susceptibilities and/or acquired antimicrobial resistances from the environment or other bacteria species (Fig. 2D).
Antimicrobial resistance can be mediated through the acquisition of mobile genetic elements (MGEs). Our isolates harbored various types of MGEs, including transposons, conjugative elements, phage-like elements (lacking phage structural genes), integrons, and mobility islands. Transposable elements were found across all clades, although significantly less in Clade I isolate genomes. Other MGE types were restricted to Clade II, III, IV, and V (Supplementary Fig. 3D). MGEs play pivotal roles in bacterial evolution by enabling bacteria to acquire fitness advantages from their environment, although many of the genes within MGEs are poorly annotated. Being equipped with various MGEs likely facilitates the rapid and divergent evolution of Segatella clades within the extensive gene pool maintained by intestinal bacteria.
Clades harbor distinct sets of carbohydrate utilization machineries
As diet has been cited as a main contributor in the colonization of S. copri2,13,42,43, we directed our focus towards the utilization of dietary fibers as these are a key metabolic feature of members of the Bacteroidota phylum. Polysaccharide utilization loci (PULs), involved in the sensing, transportation, and digestion of available polysaccharides in the environment44, have been reported to be differentially distributed among S. copri isolates, enabling their digestion of different sets of polysaccharides10. We predicted PULs from our isolate genomes by examining loci containing both susC and susD genes, known to be involved in carbohydrate transfer, and at least one known gene belonging to any carbohydrate active enzyme (CAZyme)45 family. The FijiCOMP isolates were rich in PULs, averaging 16 PULs per genome, albeit lower than what was reported in Bacteroides species but similar to previous reports in S. copri10,44 (Supplementary Fig. 4A). This amounted to a large number of CAZymes identified within the isolate genomes (Fig. 3A), with surprisingly few consistencies across all Segatella isolates; only three PUL-associated CAZymes were present in all isolates (GH2, GH3, and GH10). Some PULs are predicted to hydrolyze specific polysaccharides, for instance, the α-mannan-cleaving GH99 exclusively presents in Clade V, which may underlie colonization niches in the gut46. Others have enzymes capable of hydrolyzing animal-derived polysaccharides, such as those from sialidase family GH33, which can cleave N-glycolylneuraminic acid (Neu5Gc), a polysaccharide rich in red meat47,48.
A PUL-associated CAZymes identified in Segatella isolate genomes. Predicted substrates are depicted at the top. The red dots denote the isolates selected to study the polysaccharides utilization and fermentation products (from top to bottom: Clade I (F2-D6), Clade VI (C6-F5), Clade V (F2-C11), Clade II (S6-D2), S. copri DSM18205, Clade III (F2-H8), and Clade IV (S6-C12)). B The growth of select Segatella isolates with different carbon sources and the concentration of acids in the spent media at stationary phase measured by HPLC. GMC denote the 1:1:1 mixture of glucose, maltose, and cellobiose, with a same total concentration as the polysaccharides. Side bars showing the growth of bacteria, measured by optical density (OD600), using the given polysaccharides or mono-/di-saccharide cocktail as the only carbon source. Shaded cells are values below the calibration range (0–50 mM), indicating possible consumption of acids in the medium.
To test carbohydrate preferences empirically, we performed growth experiments using different polysaccharides as the sole carbon source. We specifically chose six plant-derived polysaccharides, including starch, xylan, and inulin, which are commonly found in the typical Fijian diet, such as taro. Additionally, substrates identified through genomic analysis of PULs, such as xyloglucan, were also included (Fig. 3B). To broaden the comparison, we also added a group of mixed mono-/di-saccharides and one animal-derived polysaccharide, heparin. Overall, Segatella isolates exhibited more robust growth in media containing the plant-derived polysaccharides compared to the two animal-derived polysaccharides, in accordance with previously reported diet correlations2,43.
Segatella isolates showed distinct preferences and abilities in utilizing different polysaccharides (Fig. 3B), consistent with the variation observed in PUL content. Five subfamilies of GH5, which is thought to facilitate the degradation of xyloglucan10,49, were detected from Segatella isolate genomes, whose presence correlated with the growth on xyloglucan (Fig. 3B, Supplementary Fig. 4B). Clades achieving moderate to high biomass all had more than three of the five subfamilies including at least one of GH5_2 and GH5_7 subfamilies (Supplementary Fig. 4B, C). Multiple subfamilies of CAZyme family GH13, classified as pullulan degrading CAZymes50,51, were detected from isolate genomes, and, specifically, subfamily GH13_7, annotated as α-amylase, was present in all isolates capable of utilizing pullulan (final OD600 > 0.1) and was absent from those unable to grow on pullulan. Additionally, Clade IV (S6-C12), which had the highest number of detected CAZymes, was able to grow on all plant-derived polysaccharides, albeit with varying maximum OD600. The highest OD600, observed with xyloglucan as the carbon source, was 5.5 times higher than the lowest value, observed with pullulan. On the contrary, despite a high number of PULs, Clade III (F2-H8) exhibited mild growth on all of the selected polysaccharides, with the exception of pectin. Surprisingly, none of the isolates preferred the mono/disaccharides mixture (GMC) over the plant-derived polysaccharide options, despite cellobiose and glucose being the major carbon sources in M10, the rich medium for the cultivation of S. copri complex.
Segatella isolates produce short-chain fatty acids as a result of polysaccharide degradation
Dietary fibers are degraded by gut bacteria into different types of short-chain fatty acids (SCFAs), which compose up to 10% of the host’s energy budget and confer numerous health benefits, including regulation of host metabolism, immunity, and anti-inflammatory responses48,52,53,54. Bacteroides species predominantly produce propionate and acetate55,56,57, whereas intestinal Firmicutes are the main producers of butyrate58. Other important fermentation products also include lactate and succinate, the latter of which is reported to benefit the host glucose metabolism by activating intestinal gluconeogenesis54. To identify the fermentation products of Segatella isolates from different clades, we conducted high-performance liquid chromatography (HPLC) analysis on the spent media of seven diverse isolates inoculated with different polysaccharides.
Segatella isolates showed the ability to produce a variety of SCFAs including formate, lactate, succinate, propionate, and acetate. Butyrate and valerate were also detected from some samples even though at low concentrations. Despite differences in growth of the isolates in different carbon sources, isolates produced similar ratios of SCFAs (Fig. 3B), with some notable differences. Clade I (F2-D6), Clade II (S6-D2), and Clade IV (S6-C12) had similar SCFA profiles. Although Clade III (F2-H8) was unable to grow to a large extent in any of the supplied carbon sources, this isolate was able to produce a considerable amount of propionic acid (18.92 mM) from pectin utilization, a feat unmatched by the other isolates, despite their higher growth. Interestingly, although S6-C12 and S. copri DSM18205, two strains within Clade IV, are genetically very similar (Fig. 1A), they exhibited markedly different carbohydrate preferences and SCFA production (Fig. 3B). This suggests that the metabolism of SCFAs may be governed by specific sets of enzymes whose presences do not necessarily align with genome clustering.
Only some Segatella isolates induced strong transcriptional responses from intestinal epithelial cells
Given the challenge in colonizing murine models with Segatella, especially in light of their highly variable carbohydrate preferences, we chose to analyze host transcriptional responses to Segatella clades using cultured human Caco-2 cells, an intestinal carcinoma cell line. Two-hour incubation with the selected Segatella isolates did not induce significant mortality of Caco-2 cells (Supplementary Fig. 5A). There were a large number of differentially expressed genes (DEGs) that were clade-specific. However, Caco-2 responses to Segatella isolates’ transcriptomes clustered into two groups, with one largely reminiscent of the untreated cells (“hypo-stimulating”), with fewer DEGs compared to the second group (“hyper-stimulating”) (Fig. 4A, B). Interestingly, the type strain S. copri DSM18205 resulted in few differentially expressed genes (DEGs), despite being reported as pro-inflammatory in previous studies7,59.
A The gene expression profiles of Caco-2 cells treated with Segatella isolates calculated from the normalized read counts. A Z-score normalization was performed across groups for each gene. Smaller heatmap on the right shows the gene expression files of the Caco-2 cells in Hypo and Hyper groups. The select clade-representative isolates are Clade I (F2-D6), Clade II (S6-D2), Clade III (F2-H8), Clade IV (S6-C12), Clade V (F2-C11), and Clade VI (C6-F5). The bar plot on the bottom shows the number of total and group-unique DEGs found in the indicated treatment conditions compare to the Blank group. B Volcano plot visualization the differential expression of genes between Hypo and Hyper groups. Horizontal dash line denoting the p-value cutoff of 0.05. Log2(fold change) is used to quantify the differential expression of genes in the Hyper group compared to the Hypo group. C The top 20 up- and down-regulated Gene Ontologies (GOs) in Hyper group compared to Hypo group, ranking by p-values. Gene counts are the numbers of DEGs assigned to each GO term.
Among the DEGs with the largest effect sizes between the Hypo and Hyper groups were genes with known association with gastro-intestinal disease. For instance, G protein-coupled receptor 55 (GPR55), a gene associated with intestinal inflammation, was found among the DEGs upregulated in the Hyper group60. Regulated expression of many long non-coding RNAs (lncRNAs) was also observed including those correlated to gastric cancer and colorectal cancer. For example, DLGAP1 and pcsk2-2:1 (or RPS27AP2) showed highly increased expression in the Hyper group as well61,62. Curiously, among the genes with increased expression in the Hypo group compared to Hyper group (Fig. 4B) was SEMA3E, a gene whose expression is significantly reduced in ulcerative colitis patients63. The regulation of these genes leads to the hypothesis that isolates classified into the Hyper and Hypo groups could potentially be disease-promoting or -preventing, respectively. However, further study will be required to understand the full regulatory networks of these genes in vivo.
Functional enrichment analyses on the DEGs between the Hyper and Hypo groups identified nearly half of the 19 significantly up-regulated genes in the Hyper group were associated with actin production, implying potential changes in cell morphology, including cell migration or internalization of bacterial cells. (Fig. 4C). Hypothesizing that this may contribute to barrier defects, we tested whether exposure to Segatella isolates causes hyperpermeability of Caco-2 cell layers. Fluorescein isothiocyanate-labeled dextran (FITC-dextran) of various sizes is used to measure permeability. Yet, no significant changes in cell layer permeability were observed, except for a slight increase in permeability seen for one of the Clade IV isolates (S6-C12) (Supplementary Fig. 5B).
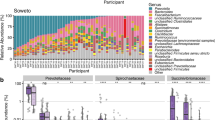
Among all microbiome-derived Segatella clades, Clade II and V exhibited the most different gene expression profiles (Supplementary Fig. 5C, Fig. 4A). Among the 2972 DEGs identified between Clade II- and Clade V-treated groups, 6 of the top 20 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with significantly reduced expression in the Clade II-treated group were associated with immune signaling pathways, including IL-17, NF-kB, Nod-like receptor, C-type lectin receptors, mTOR, and TNF signaling pathway (Supplementary Fig. 5D). These findings highlight significant differences of Clade II in modulating the host immune responses. We further annotated the DEGs according to their associations with disease. We observed that, in comparison to the non-treated group, only the treatment with Clade VI bacteria led to elevated expression of numerous genes that are correlated with intestinal/colorectal cancers, amounting to a total of 59 genes (Supplementary Fig. 6A).
Interactions between different isolates led to the changes in relative abundances in co-culture communities
Despite the realization that humans harbor closely related strains with dynamic strain replacement happening occasionally, there is a surprising lack of understanding regarding how closely-related consortia persist14,64,65. Within diverse communities, complex competitive interactions are thought to provide stability66. Acknowledging the distinct metabolisms and nutritional preferences of different Segatella clades, we performed co-culture experiments to decipher pairwise interactions. To identify individual isolates, we designed primers to the marker gene, rplN, which spanned a region of sufficient genetic diversity to distinguish between isolates. Employing amplicon sequencing, we were able to track the relative abundances of isolates in the co-culture community (Fig. 5). In the time frame, most of the pairwise cocultures reached relative abundance ratios different from the starting point (0.5:0.5). Some co-cultures resulted in reduced growth overall, including a strain cocktail involving five isolates, suggesting competitive interactions (Supplementary Fig. 6B). Clade IV isolate S6-C12 dominated in all combinations, often reaching higher relative abundances than any other co-cultured isolate (Fig. 5 upper left), whereas Clade V isolate F2-C11 was uncompetitive with all the other isolates. These results are in accordance with the estimated clade relative abundances in the fecal metagenomes, of which Clade IV and V presented the highest and lowest abundances, respectively. (Note that Clade VI was not included in the coculture experiment, although its abundance in its source metagenome is below 5%, Fig. 1B).
Changes in relative abundances of each isolate, represending different clades, in the cocultures over a 25-hour period. The top plot shows the relative abundances of isolates in the cocktail of the five isolates, which are in Clade I (F2-D6), Clade II (S6-D2), Clade III (F2-H8), Clade IV (S6-C12), Clade V (F2-C11).
We performed a second assay to explore whether any metabolites or proteins produced by one organism could promote or inhibit the growth of another, to further probe the interactions between the isolates. Segatella isolates cultured in the spent medium of other isolates showed growth inhibition to different extents. The results are consistent with what was observed in the coculture experiment, with Clade IV isolate S6-C12 spent medium showing stronger inhibition of all other isolates. Clade I isolate F2-D6 was inhibited by all other isolates (Supplementary Fig. 6C). The results indicated that the interactions between cocultured Segatella isolates are, at least partially, achieved by secreted small molecule metabolites or proteins during bacteria growth.
Discussion
S. copri was first isolated in 2007, and its genome was made available in 200967. Whereas the study of Bacteroides species has been facilitated by the ease at which they can be cultured, since the first S. copri isolates were obtained, only one S. copri genome has been made available, that from the gut microbiome of a healthy Indian male68, and until recently, only one strain was publicly available through commercial strain catalogs. Therefore, most of our knowledge about the role of S. copri in the gut microbiome has come from correlative data from case-control metagenomic studies. The diversity within the gut commensal S. copri complex has gained attention in recent years. Tett et al. assembled thousands of MAGs from fecal metagenomes in 2019, clustering them into four distinct lineages1. Most recently, Blanco-Míguez and colleagues further expanded the S. copri complex into 13 distinct species3.
We performed whole genome sequencing on 63 of our Fijian S. copri complex isolates clustered into six clades based on their gene content, which were classified into six of the new species identified by Blanco-Míguez et al.3. The detected differences in functional gene content are likely reflective of deeper evolutionary relationships rather than recent gene transfer events, as gene clustering largely matches the isolates’ phylogeny. The data presented here, regarding the nutritional preferences, production of metabolites, and interactions with host cells, supports the notion that genomic diversity-driven variations in metabolism and phenotypes provide a possible explanation to the conflicting correlations between S. copri complex and host health. As S. copri is being considered to serve as a diagnostic indicator69,70 or even a putative therapeutic target71, it is critical to understand the potential effects driven by the genetic diversity between S. copri and closely related Segatella species within the gut microbiome.
The two prominent genera within the Bacteroidota phylum, Bacteroides and Segatella, are thought to play a major role in carbohydrate degradation in the gut. Bacteroides and Segatella species harbor numerous diverse PULs and consistently make up a large portion of individuals’ microbiome composition72,73. Segatella species are pervasive in developing countries worldwide, where Bacteroides dominate in Westernized countries, and it is suspected that diets higher in fiber drive this difference4,19. There are some major similarities, including the production of propionate. Bacteroides-produced propionate has been reported to play a beneficial role in intestinal immunity and homeostasis56,57. Mapping of the central carbon metabolism of S. copri DSM18205 (Clade IV) indicated that it is equipped to produce succinate, formate, and acetate with glucose9,67,74. Our results using more complex carbohydrates reveal low production of SCFAs by this isolate. On the contrary, our in vitro experiments reveal that Segatella clades isolated from the Fijian individual broadly produce propionate as a result of degrading plant-derived polysaccharides, among other SCFAs. Our analysis of the PUL-associated CAZymes which have known or predicted substrates, correlates to large extent with the growth observed in each fiber. However, the majority of the CAZymes identified are poorly characterized. Although Segatella isolates have universal abilities to produce propionate, preliminary searching of known enzymes responsible of propionate synthesis in Bacteroides yielded no homologous genes, suggesting unique, or sufficiently diverged, mechanisms in Segatella.
In addition to SCFAs, we previously reported that members of these Segatella clades produce diverse and novel sphingolipids, which serve both as important structural components of the cell membrane as well as signaling molecules11,75. Due to the structural conservation between bacteria and mammalian sphingolipids, microbiota-produced sphingolipids were reported to be involved in host metabolism and immune homeostasis75,76,77,78,79. The production of sphingolipids and SCFAs may underlie some of the transcriptional differences we observed within the Segatella-Caco-2 cell coculture experiment presented here. Progress in genetic engineering and/or colonization of Segatella in mice models is required to further investigate their roles in host metabolism and immunity. The observed multidrug resistance in Segatella clades warrants attention, given the opportunistic pathogenicity of S. copri, which has been implicated in bloodstream infections as reported in a prior case study80.
Members of the genus Segatella have been associated with inflammation in respiratory mucosa and the oral cavity, as well as the vaginal tract7. Only recently has their colonization within the gut been associated with gut-associated inflammation and chronic inflammation7,81. Given that Segatella are highly prevalent worldwide1,82, understanding whether or not all clades of S. copri complex contribute equally to inflammation is of high importance. Confusingly, case-control studies have both implicated and absolved S. copri in inflammation-associated metabolic disorders. S. copri has been associated with insulin resistance8, but not type 2 diabetes83,84,85; with both obesity86,87,88 and leanness89; and yet is found elevated in patients with rheumatoid arthritis59, hypertension90, non-alcoholic fatty liver disease (NAFLD)87,91 and inflammatory bowel disease92. However, these observations may be confounded by previously unrecognized diversity within the S. copri complex. Our study did not find any strong correlation between S. copri presence and disease occurrence at the strain-level. Mouse models are often used as a way to examine host-microbe interactions, providing some advantages that cell culture does not such as the effects of long-term colonization. A few studies have incorporated mouse models in studying the impacts of S. copri on the host, however, those were limited to the type strain S. copri DSM18205, which has also been shown to induce pro-inflammatory cytokines IL-6 and IL-23 in vitro, thereby promoting Th17-mediated immune responses recapitulating responses seen in rheumatoid arthritis7,93. The colonization of various clades remains challenging as it requires the supplement of plant-derived polysaccharides. Our analysis of nutritional preferences provides more guidance on the practice of facilitating colonization of Segatella in murine models by polysaccharides supplementation that can further understanding of the role of Segatella in human populations. We expect that the growing availability of strains and associated genomes will further research into the roles of individual strains in disease and multi-strain consortia that may co-exist in individual’s microbiomes.
Methods
Human subject and stool sample collection
Human stool samples used in this study were collected as part of the Fiji Community Microbiome Project (FijiCOMP)19. This study was initially approved by the Institutional Review Boards at Columbia University, the Massachusetts Institute of Technology, and the Broad Institute and ethics approvals were received from the Research Ethics Review Committees at the Fiji National University and the Ministry of Health in the Fiji Islands. The Cornell University Institute Review Board additionally approved this study (#1608006528). Human subjects were consented prior to participation in the study. Stool samples were collected into PBS with 20% glycerol within 30 min of voiding, preserved in RNALater (QIAGEN), and stored at −80 °C prior to metagenomic library preparation. The prepared library was sequenced on the Illumina HiSeq2000 platform, 2 × 250 bp paired end reads19. The quality of the metagenomic sequencing reads were assessed by FastQC94.
Segatella genomes included in this study
The type strain used in this study, Segatella copri DSM18205, was purchased from DSMZ. Its genome was downloaded from NCBI RefSeq (GCF_000157935.1). The 63 S. copri isolates used in this study were obtained as described in the METHOD DETAILS section. Apart from our own isolate genomes, we included in part of our analysis the isolates from previous publications available at the time this study was conducted. This includes 83 genomes from Tett et al., 11 genomes from Li et al. and 32 from Blanco-Míguez et al1,3,12. The reference genomes of all other Segatella species (named as Prevotella by NCBI) used in phylogenetic analysis were downloaded from NCBI RefSeq database.
Bacteria isolation and cultivation
The formula of Modified Medium 10 (M10) agar was modified from a previous study and is prepared as described in Supplementary Note 1, degassed overnight, and used to obtain Segatella isolates from human gut microbiomes67,95. Stool samples were diluted with PBS (10−1 to 10−8), plated onto M10 agar, and incubated in the anaerobic chamber (3%H2, 20% CO2, remainder N2. Coy Lab Products.) for up to 48 h. To further identify Segatella isolates, we performed Polymerase chain reactions (PCR) using previously developed Segatella 16S rRNA-specific primers96 (primers: g-Prevo-F, g-Prevo-R. Supplementary Table 2). Colonies yielded bands with correct sizes were streaked and individual isolates were further verified by Sanger sequencing of the full-length 16S rRNA gene (primers: 27 F, 1042 R. Supplementary Table 2).
When needed, the frozen stocks were inoculated and cultured for 24 h at 37 °C in the anaerobic chamber. In order to get the best revival, the glycerol stocks were first inoculated onto degassed M10 agar plates and then subculture to either BBL™ Schaedler Broth (BD Biosciences or HiMedia) or M10 plates, depending on the requirements of the following experiments. Solid and liquid media were freshly made and stored in the anaerobic chamber overnight to degas before use.
The liquid medium was prepared by dissolving Schaedler Broth powder with water inside anaerobic chamber, adding in 0.05% resazurin, aliquoting desired amount into sample tubes or serum bottles, sealing and autoclaving at 121 °C for 15 min. Schaedler Broth from two different manufacturer were used throughout the study as indicated due to the discontinuation of BBL™ Schaedler Broth by BD Biosciences.
Whole-genome sequencing
S. copri DSM18205 and 63 Segatella isolates obtained from previous steps were anaerobically cultured on M10 plates for 24 h and then resuspended in PBS. Genomic DNA was isolated using E.Z.N.A.® Bacterial DNA Kit (Omega). Libraries were prepared using the NEBNext® Ultra™ II DNA Library Prep Kit (Illumina). Libraries were sequenced on the Illumina MiSeq 2 × 250 bp platform.
Computational methods
Fecal metagenome processing and analysis
The Fijian fecal sample metagenome was processed using three different pipelines used in previous studies and one pipeline used in the lab to get the MAGs for following analysis.
(1) First method was adopted from the study of Pasolli et al24. Contigs shorter than 1000 nucleotides were filtered out after assembling with MEGAHIT22. The reads were mapped using bowtie2 (--very-sensitive-local), followed by binning with MetaBAT2 (-m 1500)25,97.
(2) In the pipeline from Nayfach et al.28, the metagenome was assembled using MEGAHIT followed by contig binning with MaxBin, MetaBAT2 and CONCOCT23,25,26,27. The obtained bins were then refined using DAS Tool and cleaned using MAGpurify28,98.
(3) The third method is a pipeline established in the lab combining several previously used and proved pipelines29. Briefly, the reads were assembled with metaSPAdes30 followed by contig binning MaxBin27, MetaBAT225 and CONCOCT26 binning and bin refinement using DAS Tool30.
(4) The last method was used by Chen et al.31. BBTools was used to remove the adapter sequences, contamination from PhiX, and other illumine trace contaminants from the raw reads (bbduk.sh ktrim = r k = 23 mink = 11 hdist = 1 tbo = t)99. Low-quality bases and reads were removed using Sickle100. The filtered reads were then assembled using metaSPAdes30 and IDBA-UD32, followed by read mapping using bowtie2 with default parameters97. Scaffolds longer than 2.5 kb went through binning by MetaBAT2 with default parameters25. The original pipeline used in Chen et al.’s research includes manual cleaning and curation of the MAGs obtained from steps above, which we skipped in order to compare the performance of pipelines without manual processing.
The qualities of the bins were assessed with and the taxonomic classifications of the MAGs were acquired from CheckM33. The composition of the fecal metagenome was profiled by MetaPhlAn220.
Isolate genome assembly and annotation
The paired-end raw reads were trimmed by Trimmomatic and assembled into genomes using SPAdes v3.10.1101,102. Any contigs that are less than 500 bp in length were filtered out. The completeness and quality of assembled genomes were checked with QUAST v4.0 and CheckM v1.0.11 with a contamination cutoff of 5% and completeness cutoff of 95%33,103. Open reading frames were predicted by running Prodigal v2.6.3 on obtained genomes104. Proteins were annotated from the KEGG (Kyoto Encyclopedia of Genes and Genomes) prokaryotic protein database using DIAMOND v0.9.21 blastx105. Sequences were then filtered based on e-value and percent identity. Hits with e-values higher than 1e-5 or less than 30% identity to the reference sequences were removed.
Phylogenic analysis
16S rRNA gene
The 16S rRNA sequences were identified from the genome of each Segatella isolate using rnammer v1.2106. For the twelve isolates from which we failed to get 16S genes by rnammer, amplification and sequencing of full-length 16S rRNA genes were performed with primers 27 F and 1492 R (Supplementary Table 2) and Phusion® Hot Start Flex DNA Polymerase (NEB). Cycles were performed as: 98 °C for 3 min, then 30 cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s. The PCR products were cleaned using Agencourt AMPure XP Beads (Beckman Coulter) before Sanger sequencing.
The ANI values were calculated using FastANI between each two isolates including the type strain S. copri DSM18205, of which the genome was downloaded from NCBI RefSeq107. Genome based-phylogenomic trees of all Segatella isolate genomes and reference genomes of other Segatella species were constructed by PhyloPhlAn3 using the default library containing more than 400 marker genes with following options108: --diversity low, --tree raxml. The tree was annotated and visualized using GraPhlAn109.
Presence of Segatella clades in cMD
Genes were called from Segatella isolate genomes using prodigal 2.6.3104 and blasted against Uniref90 database (September 2023 release) using DIAMOND 2.1.8105,110. Genes mapped to the same Uniref90 ID with identity ≥90% were clustered together. If a cluster had genes from ≥95% of genomes of particular clade and has no genes from the other clades, the centroid gene, the gene with longest sequence, was considered as a marker gene for that particular clade1. This resulted in 1393 genes for Clade I, 622 genes for Clade II, 437 genes for Clade III, 580 genes for Clade IV, 1206 genes for Clade V, and 490 genes for Clade VI.
cMD22 were filtered for fecal metagenomes and only countries with more than 5 samples and samples with ≥20 M reads, resulting in 10,400 samples. Read files were adapter trimmed using following parameters with BBTools99: ktrim = r k = 23 mink = 11 hdist = 1 tpe tbo. To estimate presence of Segatella clades in cMD, the database of clade-specific genes was indexed using default parameters in KMA111 and concatenated paired-end clean reads from cMD were mapped to it with default parameters and filtered for genes with ≥90% identity and ≥95% coverage. A clade was considered present in the metagenome if ≥75% of the clade marker genes were present. The relative abundance of Segatella clades in each sample was calculated as:
Western and non-western classification was used as provided in cMD and countries were assigned different income-class based on World Bank’s classification downloaded in March 2024 (https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups). To assess correlation of disease with prevalence and abundance of Segatella clades, we only considered studies with both control and disease samples and performed Fisher’s exact test and Mann-Whitney test respectively.
Estimation of clade relative abundance
The relative abundance of each Segatella clade in the fecal metagenome was estimated using DiTASiC with the default parameters112. One isolate genome was picked from each clade as the reference genome. Due to the lower similarity of F2-F6 to other isolates in Clade III, two genomes were picked from this clade as reference genomes (C6-B8 and F2-F6). The proportion of clade in isolate collection was calculated by dividing the total number of isolates by the number of isolates belong to each clade.
Antibiotic resistance prediction and test
Antimicrobial resistance genes were predicted in the Segatella isolate genomes using ABRicate113 combining following databases: CARD114, EcOH115, ARG-ANNOT116, Ecoli_VF, VFDB117, MEGARES 2.00118, Resfinder119, PlasmidFinder120.
Resistance against various antibiotics were tested using Minimum Inhibitory Concentration (MIC) assays with broth microdilution method121. To ensure sufficient growth of Segatella isolates, Schaedler broth was used in the MIC tests. Briefly, select Segatella isolates were inoculated onto M10 agar plates and cultured for 24 h. Colonies were collected and resuspended in sterile PBS to 5 × 105 CFU/mL. Schaedler broth with concentration gradients of select antibiotics were prepared in polystyrene 96 well plates (Costar) and degassed overnight. In the anaerobic chamber, 10 μL of bacteria cell suspension was inoculated into 200 μL of medium per well. Microplates were incubated for 48 h anaerobically and the optical density at 600 nm was read on Biotek Cytation 5 multimode reader with necessary dilutions. The interpretation of the results referred to the Reading guide for broth microdilution and the Breakpoint tables for interpretation of MICs and zone diameters and from the European Committee on Antimicrobial Susceptibility Testing (EUCAST)122.
Identification of MGEs from isolate genomes
The MGEs were identified from the isolate genomes following the method provided by by Khedkar et al.123. Briefly, HMM profiles were built for recombinases using known protein sequences and additional HMMs from Pfam124. The recombinases within the isolate genomes were annotated using these HMMs and were mapped to the accessory gene regions to identify recombinase islands. Then the annotated phage structural genes from EggNOG and genes involved in conjugation from TXSscan were mapped to the recombinase islands to assign potential MGEs40,125.
SEM and TEM imaging
For SEM imaging, the bacteria cells were fixed with 2% Glutaraldehyde and 1% OsO4 and transferred to a filter paper after dehydration with serial gradients of ethanol. After overnight critical point drying, the filter papers were sputter-coated with approximately 10 nm gold-palladium particles (ratio 60:40) for 60 s at 30 mA of current and the images were acquired from the Zeiss Sigma 500 SEM. For S. copri DSM18205 and isolate F2-D6, the samples were imaged at 0.5 kv with a working distance of 2.0 mm and 1.7 mm, respectively. For the rest of the samples, imaging was performed at 1.0 kv with 5.0 mm working distance. Images were acquired with a secondary electron signal using a side angle Everhart-Thornley detector. For TEM imaging, after the same fixation procedures as used for SEM, the pellets were resuspended in 1.5% uranyl acetate and incubated in dark for 1 h. After dehydration with serial gradients of ethanol, samples were infiltrated and embedded with Quetol 651 for overnight. The samples were viewed on a JEM-1400 transmission electron microscope (JEOL, USA, Inc., Peabody, MA) operated at 100 kV and images were captured on a Veleta 2 K × 2 K CCD camera (EM-SIS, Germany).
Biofilm quantification
The biofilm formation by Segatella isolates was quantified using crystal violet staining assay126. Select Segatella isolates were cultured anaerobically in 200 μL of Schaedler Broth on a polystyrene 96-well plate (Costar) for 48 h. Wells with broth incubated at the same time were used as blanks. The liquid cultures were then aspirated, and plates were dried at 60 °C for one hour. Each well was added with 150 μL 0.1% crystal violet solution and stained at room temperature for 15 min. After washing three times with water to remove excess staining, the residual liquid was removed and the plates were dried at 60 °C for 10 min. Finally, the biofilm was solubilized and destained with 150 μL 33% acetic acid per well. Absorbance at 590 nm was read on Biotek Cytation 5 multimode reader to quantify biofilm formation. Dilutions were made when necessary.
Polysaccharides utilization
CAZyme and PUL predictions
The CAZyme genes in each Segatella isolate were predicted and annotated from dbCAN-HMMdb-V1145 database using hmmscan (version 3.3) filtered with recommended cutoffs (e-value < 1e-18, coverage > 0.35)127. The detected CAZymes were categorized based on the correlated polysaccharide substrates referring to the information in previous studies128,129. PULs were predicted from the isolate genomes using PULpy130.
Bacteria cultivation and sample preparation for HPLC
Segatella Defined Medium (SDM) was adopted from Defined Minimal Medium Glucose (DMMG) and optimized for the growth of different Segatella clades (Supplementary Note 2)131,132. Select Segatella isolates were first inoculated onto M10 plates and cultured anaerobically at 37 °C for 24 h. Colonies were then collected and resuspended in sterile PBS. After adjusting the OD600 to 1.0, 200 uL of the bacteria cell suspension was inoculated into 5 mL SDM with 0.5% (w/v) different carbon sources and cultured anaerobically at 37 °C for 48 h. Blank cultures were inoculated with same amount of PBS. The final optical density at 600 nm (OD600) of liquid cultures was measured on Biotek Cytation 5 and was calculated by subtracting the blank readings of medium with corresponding carbon sources.
The rest of the liquid cultures was centrifuged at 5000 g, 4 °C for 15 min to pellet the bacteria cells. The supernatants were collected and filtered through 0.45 um filters. To prepare samples for HPLC, 1 mL of filtered supernatant was transferred to an autosampler vial and then mixed well with 100 uL of concentrated HCl.
HPLC settings
The HPLC measurement protocol was modified from a previous study133. The Shimadzu HPLC–UV system used consisted of the following modules: a LC-20AD pump, a LC-10AD-VP pump, a DGU-14A degasser, a CBM-20A controlling module, a SIL-20A Auto-sampler, a CTO-20AC oven, an SPD-10A UV detector, and a RF-10A Fluorescence detector. Chromatographic separation was performed using the Hypersil GOLD aQ C18 column (150 mm × 4.6 mm i.d, particle size = 3 µm. ThermoFisher.). The column was thermostatized at 30 °C while running. Two mobile phases were used for the optimal separation of different organic acids: mobile phase A was 20 mM NaH2PO4 with pH adjusted to 2.2 using phosphoric acid and filtered with a 0.2 µm filter. Mobile phase B was mobile phase A mixed with acetonitrile (6:4, v/v). The washing buffer was acetonitrile in HPLC water (6:4, v/v). The program of the bi-gradient elution performed is shown in Supplementary Table 3. Ten microliters of the prepared samples were injected into HPLC and the UV detector read at a wavelength of 210 nm. The heights of peaks and baselines were acquired from the chromatography and the concentrations of each acid in the samples were calculated from the standard curves described below.
Calibration and standard curve acquisition
HPLC on calibrator solutions
Stock solutions (SS) of select SCFAs were prepared in either HPLC water or 1:1 mix (v/v) of water and HPLC-grade methanol with the concentrations indicated in Supplementary Table 3 For the acquisition of calibration curves, blank SDM was filter sterilized using a 0.45 µm filter and was used to prepare calibrator solutions containing following concentrations of all SCFAs: 50 mM, 25 mM, 10 mM, 5 mM, 2.5 mM, 1 mM, 0.5 mM, 0.25 mM, 0.1 mM, 0 mM. Acids with similar elution times were assigned to two different calibration groups (A and B) to better separate the peaks. To run calibrators on HPLC, 1 mL of each calibrator solution was added into an autosampler vial with 100 uL concentrated HCl and vortexed for 15 s to fully mix. Ten microliters of the prepared solution were injected and each calibrator solution was run for three times as biological replicates.
Calculation of limits of detection and quantification
Linear regression was performed on the peak heights of each SCFA which were acquired as described in the HPLC data analysis section below. The Limit of Detection (LOD) and Limit and Quantification (LOQ) were calculated as suggested by the International Conference on Harmonisation (ICH) as follows where σ is the standard deviation of the response and S is the slope of the calibration curve134:
Bacteria-human cells co-culture experiment
Co-culture experiment was performed with human Caco-2 cell line at passage 9. Dulbecco’s Modified Eagle Medium (DMEM, Corning) supplemented with 10% Fetal Bovine Serum (FBS) was used as the culture medium. Caco-2 cells were cultured in 6-well Transwell inserts (Corning, 0.4 uM pore size, Polycarbon membrane) for 4 weeks for differentiation before experiment. On the day of experiment, medium was changed from DMEM supplemented with 10% FBS to pure DMEM.
Seven Segatella isolates including the type strain S. copri DSM18205 were selected to represent different clades. Bacteria cells were collected from overnight-cultured M10 plates and washed with sterile PBS twice. Bacteria pellets were then resuspended in sterile DMEM medium with OD600 adjusted to 1.0. The bacteria suspensions were diluted 1: 50 with sterile DMEM and added into the Transwell inserts at an MOI of 1: 25. Plates were cultured at 37°C aerobically with 5% CO2 for two hours. Based on our test, all Segatella isolates can maintain good viability after two-hour aerobic incubation in DMEM and the viability dropped dramatically afterwards (Supplementary Fig. 6D).
Total RNA extraction and sample quality control
After two-hour incubation, medium was removed from the plates and 1 mL of TRIzol reagent (Invitrogen) was added into each well immediately to collect cells. The total RNAs of Caco-2 cells was extracted by using TRIzol method. Before library preparation, the concentrations of obtained RNA samples were measured using Qubit Fluorometer and the sample integrities and purities were examined using Agilent RNA Sample Quality Control Analysis and agarose gel electrophoresis. Samples with an RNA integrity number (RIN) higher than 8.0 were qualified for library construction.
Library construction and RNA sequencing
Messenger RNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first strand cDNA was synthesized using random hexamer primers, followed by the second strand cDNA synthesis using dTTP for non-directional library. After end repair, A-tailing, adapter ligation, size selection, amplification, and purification, libraries were ready for sequencing. The library was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on Illumina NovaSeq 150 bp Paired-end platform, according to effective library concentration and data amount.
RNA-sequencing data analysis
Part of the RNA-sequencing data analysis was performed by Novogene.
Quality control
The raw data in fastq format was first processed using the fastp software. This step involved extracting clean data (clean reads) by filtering out reads containing adapters, ploy-N sequences, and low-quality reads from the raw dataset. Concurrently, metrics such as Q20, Q30, and GC content were calculated for the clean data. Subsequent analyses were conducted exclusively using this cleaned high-quality dataset.
Read mapping
Reference genome and gene model annotation files were downloaded from genome website directly. Index of the reference genome was built and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.5135. Hisat2 was selected as the mapping tool for that it can generate a database of splice junctions based on the gene model annotation file, leading to more accurate mapping results compared to other tools that do not account for splice junctions.
Gene expression level quantification
featureCounts v1.5.0-p3 was used to count the numbers of reads mapped to each gene136. Then we calculated the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) for each gene based on the gene length and mapped read counts.
Differential gene expression analysis
Pairwise differential expression analysis of the treatment conditions (three biological replicates per condition for the eight treatment conditions) was performed using the DESeq2 R package (1.20.0)137. The same analysis was also performed between the hyper and hypo group using all samples clustered into each group. The resulting p-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted p-value <= 0.05 identified by DESeq2 were assigned as differentially expressed.
Functional enrichment analysis
We performed enrichment analyses of Gene Ontology (GO), KEGG pathways, DO (Disease Ontology), and disease related genes with DisGeNET database of differentially expressed genes using the clusterProfiler R package138, in which gene length bias was corrected. Functional terms with corrected p-value <= 0.05 were considered significantly enriched by differential expressed genes.
Cell permeability assay
Caco-2 cells (Passage 13) were cultured in 6-well Transwell inserts (Corning, 0.4 uM pore size, Polycarbon membrane) for four weeks to differentiate. The same Bacteria-Human cells co-culture experiment was performed following the protocol above. After two hours incubation, the medium was removed from both the inserts and the basolateral compartments. The cells were washed with PBS and 1.5 mL FITC-Dextran solutions (1 mg/mL) were added to the apical side of the cell layers. The wells were refilled with DMEM and cultured at 37 °C with 5% CO2. At 1 h, 2 h, and 4 h, 1 mL of samples were collected from the basolateral compartments and refilled with 1 mL fresh DMEM. The concentration of FITC-Dextran was measured on Biotek Cytation 5 (excitation: 490 nm, emission: 520 nm, Bandwidth: 10).
Primer design and testing for the co-culture interaction experiment
Five isolates from different phylogenetic clades and S. copri DSM18205 were selected for the co-culture interaction experiment. Marker genes were identified using AMPHORA2 (e-value cutoff = 1e-7) from each isolate genome139. A region of rplN gene was found to provided sufficient variation to distinguish between each two isolates while a single pair of primers can be used to amplify this region from all select isolates (Supplementary Table 2).
To test our method, qPCR was performed on QuantStudio3 Real-Time PCR System using Luna® Universal qPCR Master Mix (NEB) with the following program and different amount of input genomic DNAs were tested: 95 °C for 1 min, then 40 cycles of 95 °C for 15 s and 60 °C for 30 s. The qPCR results confirmed equal amplification of targeting regions from the select isolates, and the differences in input genomic DNAs can be maintained during amplification. Based on the amplification curves, 1 μg of sample DNA was used in library preparation.
Co-culture interaction experiment
Selected Segatella isolates were first inoculated onto M10 plates and cultured anaerobically at 37 °C for 24 h. Colonies were resuspended in sterile PBS and adjusted to have an optical density at 600 nm (OD600) of ~1.0. For each pair of organisms, 250 μL suspension of each isolate was mixed and inoculated into 15 mL of warm degassed Schaedler broth. In parallel, 500 μL cultures were set up for each individual organism. Liquid medium inoculated with the same volume of sterile PBS was used as a negative control. For the isolate cocktail group, equal amount of each isolate suspension except the type strain was mix together and 1.5 mL of the cocktail was inoculated into 50 mL freshly made prewarmed Schaedler broth in serum bottle. One mL of liquid culture was drawn at each time point (0 h, 5 h, 7 h, 9 h, 11 h, 13 h, 25 h). Cells were collected by centrifuging at 10,000 g for 5 min. After removing the supernatant, cell pellets were flash-frozen and stored at −80 °C.
Co-culture interaction library preparation and data processing
Genomic DNA extraction
1 mL of co-culture samples were spun down by centrifuging at 10,000 g for 5 min, and the cell pellets were used for genomic DNA extraction using Mag-Bind® Bacterial DNA 96 Kit (Omega, Bio-Tek). The DNA extraction was performed following the protocol and automated by epMotion 5075vtc robot (Eppendorf). The concentration of yielded DNAs was measured using Quant-iT™ PicoGreen™ dsDNA Assay Kit (Invitrogen) in 96-well plates.
Library preparation
The obtained genomic DNAs were prepared into sequencing libraries and sequenced on Illumina MiSeq 2 × 250 bp platform. The amplicon region of the rplN gene was first amplified with a pair of primers containing adapter sequences (rplN-adp-fw, rplN-adp-rev, Supplementary Table 2) using the following cycle: 98 °C for 30 s, then 22 cycles of 98 °C for 10 s, 63 °C for 30 s, and 72 °C for 10 s, followed by final extension 72 °C for 5 min. The PCR products was purified using AMPure XP beads. After clean-up, the DNA concentration was measured using PicoGreen assay as described above, and the yield DNA was diluted to 0.2 ug/ul. In the second PCR, amplicons from different samples were indexed with a set of unique barcodes designed by Diebold et al.140. One microgram of DNA yielded from the first PCR was input as the template and the cycles were performed as follows: 98 °C for 30 min, then 8 cycles of 98 °C for 10 s, 64 °C for 30 s, and 72 °C for 10 s, followed by final extension 72 °C for 5 min. Both PCRs used Phusion® Hot Start Flex DNA Polymerase in a 30 μL reaction. Each sample was run as two 15 μL reactions in the second PCR and were pooled together afterwards. The products of the second PCR were again cleaned up by AMPure XP beads purification and the concentrations were determined using PicoGreen assays. Two nanomolar of each sample were pooled together, and the sample purity and fragment size were checked with Fragment Analyzer. The pooled sample was then sequenced on Illumina MiSeq 2 × 250 bp platform. Set-up of PCR reactions, AMPure beads purification, sample dilution and pooling steps were performed on the Eppendorf epMotion 5075vtc robot. PCR reactions were performed on Eppendorf Mastercycler® nexus.
Cleaning and merging of sequencing reads
After barcode trimming, the paired-end reads were merged and filtered using USEARCH v11.0.667141. The sequencing reads were first merged with the following parameters: length range = expected length of the amplicon ± 20 bp, maximum differences allowed = 22 bases, percent identity ≥ 85%. The merged reads were then passed through the filter with the maximum error threshold of 1.0. Unique sequences were then detected from the filtered reads and the count of each unique sequence was calculated.
Relative abundances calculation
The unique sequences from each sample were aligned to the rplN genes extracted from isolate genomes to determine the corresponding source strain of each unique sequence. The percentage of unaligned reads or reads from unexpected source strain was calculated to test the quality and purity of the samples. Three samples were found with either inadequate merged reads or overabundant reads from unexpected source strains and therefore were removed from the following analyses. Then the counts of the unique sequences provided by USEARCH were used to calculate the relative abundances of the two isolates in each co-culture sample.
Isolate supernatants inhibitory assay
Supernatant collection
To collect culture supernatants, S. copri DSM18205 strain and eight select isolates representing different clades were first inoculated onto fresh-made M10 plates and cultured anaerobically at 37 °C for 24 h. Colonies were collected from the plates and resuspended in sterile PBS with OD600 readings adjusted to ~1.0. Three milliliter of cell suspension of each isolate was inoculated into 100 mL warm degassed Schaedler broth in a serum bottle and anaerobically cultured for 18 h until they have reached the early stationary phase. Medium inoculated with sterile PBS was processed and used as the negative control in the experiments. The OD600 readings of samples at the time of collection were measured using a SpectraMax M3 microplate reader. Liquid cultures were centrifuged at 7000 g for 10 min to spin down the bacteria cells. The supernatants were collected and passed through 0.22 μm filter cups to sterilize. The supernatant filtrates were flash-frozen and stored at −80 °C for future use.
Inhibitory assay
Before inoculation of the Segatella isolates, 5 mL of each sterile supernatant was combined with 5 mL of autoclaved Schaedler broth in a sample tube and degassed in the anaerobic chamber overnight. The nine select isolates were streaked onto fresh-made M10 plates and cultured anaerobically at 37 °C for 24 h. Colonies were collected from the plates and resuspended in sterile PBS with OD600 readings adjusted to ~1.0. Three hundred microliters of cells suspension of each isolate was inoculated into culture tubes containing different supernatants and fresh medium and cultured anaerobically at 37 °C. The OD600 values were measured 24 h after inoculation using the SpectraMax M3 microplate reader.
Statistical analysis
Statistical analyses of Figs. 2C and 5C were performed in GraphPad Prism. The differences of two groups were compared using two-tailed Mann-Whitney test and multiple t-test with Bonferroni correction, respectively. The methods, sample sizes, and significance levels are indicated in the figure legends.
Data availability
The Whole-genome sequencing data of Segatella isolates passed filtering were deposited to NCBI BioProject PRJNA217052. The metagenome data of Fijian sample Fiji_W2.48.ST was obtained in a previous work and is available under the same BioProject. The accession numbers of all corresponding SRAs are listed in Supplementary Table 4.
Materials availability
Segatella fecal isolates acquired in this study can be provided upon request.
References
Tett, A. et al. The prevotella copri complex comprises four distinct clades underrepresented in Westernized populations. Cell Host Microbe 26, 666–679.e7 (2019).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl Acad. Sci. 107, 14691–14696 (2010).
Blanco-Míguez, A. et al. Extension of the Segatella copri complex to 13 species with distinct large extrachromosomal elements and associations with host conditions. Cell Host Microbe 31, 1804–1819 (2023).
Hitch, T. C. A. et al. A taxonomic note on the genus Prevotella: description of four novel genera and emended description of the genera Hallella and Xylanibacter. Syst. Appl. Microbiol. 45, 126354 (2022).
Alpizar-Rodriguez, D. et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 78, 590–593 (2019).
Kovatcheva-Datchary, P. et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 22, 971–982 (2015).
Larsen, J. M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 151, 363–374 (2017).
Pedersen, H. K. et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381 (2016).
Dillon, S. M. et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 9, 24–37 (2016).
Fehlner-Peach, H. et al. Distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe 26, 680–690.e5 (2019).
Xiao, X. et al. Prevotella copri variants among a single host diverge in sphingolipid production. mBio 0, e02409–e02423 (2024).
Li, J. et al. A versatile genetic toolbox for Prevotella copri enables studying polysaccharide utilization systems. EMBO J. 40, e108287 (2021).
Gellman, R. H. et al. Hadza Prevotella require diet-derived microbiota accessible carbohydrates to persist in mice. http://biorxiv.org/lookup/doi/10.1101/2023.03.08.531063 (2023) https://doi.org/10.1101/2023.03.08.531063.
Smillie, C. S. et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 23, 229–240.e5 (2018).
Faith, J. J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Brito, I. L. et al. Transmission of human-associated microbiota along family and social networks. Nat. Microbiol. 4, 964–971 (2019).
Valles-Colomer, M. et al. The person-to-person transmission landscape of the gut and oral microbiomes. Nature 614, 125–135 (2023).
Lo Presti, A. et al. Phylogenetic analysis of Prevotella copri from fecal and mucosal microbiota of IBS and IBD patients. Ther. Adv. Gastroenterol. 16, 17562848221136328 (2023).
Brito, I. L. et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature 535, 435–439 (2016).
Truong, D. T. et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Pasolli, E. et al. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods 14, 1023–1024 (2017).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019).
Alneberg, J. et al. Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014).
Wu, Y.-W., Simmons, B. A. & Singer, S. W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016).
Nayfach, S., Shi, Z. J., Seshadri, R., Pollard, K. S. & Kyrpides, N. C. New insights from uncultivated genomes of the global human gut microbiome. Nature 568, 505–510 (2019).
Kent, A. G., Vill, A. C., Shi, Q., Satlin, M. J. & Brito, I. L. Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 11, 4379 (2020).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017).
Chen, L.-X., Anantharaman, K., Shaiber, A., Eren, A. M. & Banfield, J. F. Accurate and complete genomes from metagenomes. Genome Res. 30, 315–333 (2020).
Peng, Y., Leung, H. C. M., Yiu, S. M. & Chin, F. Y. L. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 (2012).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Eley, A., Greenwood, D. F. & O’Grady, F. Comparative growth of bacteroides species in various anaerobic culture media. J. Med. Microbiol. 19, 195–201 (1985).
Justice, S. S., Hunstad, D. A., Cegelski, L. & Hultgren, S. J. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6, 162–168 (2008).
Béchon, N. & Ghigo, J.-M. Gut biofilms: Bacteroides as model symbionts to study biofilm formation by intestinal anaerobes. FEMS Microbiol. Rev. 46, fuab054 (2022).
Vert, M. et al. Terminology for biorelated polymers and applications (IUPAC Recommendations 2012). Pure Appl. Chem. 84, 377–410 (2012).
Albaghdadi, S. Z. et al. In vitro characterization of biofilm formation in Prevotella species. Front. Oral Health 2, 724194 (2021).
Naito, M., Shoji, M., Sato, K. & Nakayama, K. Insertional inactivation and gene complementation of Prevotella intermedia Type IX secretion system reveals its indispensable roles in black pigmentation, hemagglutination, protease activity of interpain A, and biofilm formation. J. Bacteriol. 204, e0020322 (2022).
Abby, S. S. et al. Identification of protein secretion systems in bacterial genomes. Sci. Rep. 6, 23080 (2016).
Al-Majdoub, Z. M., Owoseni, A., Gaskell, S. J. & Barber, J. Effects of gentamicin on the proteomes of aerobic and oxygen-limited Escherichia coli. J. Med. Chem. 56, 2904–2910 (2013).
Jiang, L. et al. A high-fiber diet synergizes with Prevotella copri and exacerbates rheumatoid arthritis. Cell. Mol. Immunol. 19, 1414–1424 (2022).
Precup, G. & Vodnar, D.-C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: a comprehensive literature review. Br. J. Nutr. 122, 131–140 (2019).
Grondin, J. M., Tamura, K., Déjean, G., Abbott, D. W. & Brumer, H. Polysaccharide utilization loci: fueling microbial communities. J. Bacteriol. 199, e00860–16 (2017).
Zhang, H. et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018).
Sobala, L. F. et al. An epoxide intermediate in glycosidase catalysis. ACS Cent. Sci. 6, 760–770 (2020).
Zaramela, L. S. et al. Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates. Nat. Microbiol. 4, 2082–2089 (2019).
Wardman, J. F., Bains, R. K., Rahfeld, P. & Withers, S. G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 20, 542–556 (2022).
Junghare, M. et al. Biochemical and structural characterisation of a family GH5 cellulase from endosymbiont of shipworm P. megotara. Biotechnol. Biofuels Bioprod. 16, 61 (2023).
Kahar, U. M., Latif, N. A., Amran, S. I., Liew, K. J. & Goh, K. M. A bibliometric analysis and review of pullulan-degrading enzymes—past and current trends. Catalysts 12, 143 (2022).
Vester-Christensen, M. B. et al. Exploration of the transglycosylation activity of barley limit dextrinase for production of novel glycoconjugates. Molecules 28, 4111 (2023).
Parada Venegas, D. et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10, 277 (2019).
den Besten, G. et al. Short-chain fatty acids protect against high-fat diet-induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes 64, 2398–2408 (2015).
De Vadder, F. et al. Microbiota-produced succinate improves glucose homeostasis via intestinal gluconeogenesis. Cell Metab. 24, 151–157 (2016).
Louis, P. & Flint, H. J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41 (2017).
Price, C. E. et al. Intestinal Bacteroides modulates inflammation, systemic cytokines, and microbial ecology via propionate in a mouse model of cystic fibrosis. mBio 0, e03144–23 (2024).
Wang, X. et al. Bacteroides methylmalonyl-CoA mutase produces propionate that promotes intestinal goblet cell differentiation and homeostasis. Cell Host Microbe 32, 63–78.e7 (2024).
Rios-Covian, D. et al. Shaping the metabolism of intestinal Bacteroides population through diet to improve human health. Front. Microbiol. 8, 376 (2017).
Scher, J. U. et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013).
Schicho, R. & Storr, M. A potential role for GPR55 in gastrointestinal functions. Curr. Opin. Pharmacol. 12, 653–658 (2012).
Cai, C. et al. Serum exosomal long noncoding RNA pcsk2-2:1 as a potential novel diagnostic biomarker for gastric cancer. OncoTargets Ther. 12, 10035–10041 (2019).
Wang, X. et al. Long noncoding RNA DLGAP1-AS2 promotes tumorigenesis and metastasis by regulating the Trim21/ELOA/LHPP axis in colorectal cancer. Mol. Cancer 21, 210 (2022).
Eissa, N. et al. Semaphorin 3E regulates apoptosis in the intestinal epithelium during the development of colitis. Biochem. Pharmacol. 166, 264–273 (2019).
Costea, P. I. et al. Subspecies in the global human gut microbiome. Mol. Syst. Biol. 13, 960 (2017).
Zhao, S. et al. Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe 25, 656–667.e8 (2019).
Coyte, K. Z., Schluter, J. & Foster, K. R. The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666 (2015).
Hayashi, H., Shibata, K., Sakamoto, M., Tomita, S. & Benno, Y. Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 57, 941–946 (2007).
Bag, S., Ghosh, T. S. & Das, B. Draft genome sequence of Prevotella copri Isolated from the Gut of a Healthy Indian Adult. Genome Announc 5, e00834–17 (2017).
Scher, J. U. et al. Prevotella copri and enhanced susceptibility to arthritis. U.S. Patent Application No. 14/532,586.
Krajmalnik-Brown, R. et al. Microbiome markers and therapies for autism spectrum disorders. U.S. Patent No. 9,719,144. 1 Aug. 2017.
Björck, I. et al. Treatment of obesity, the metabolic syndrome, type 2 diabetes, cardiovascular diseases, dementia, alzheimer's disease and inflammatory bowel disease by using at least one bacterial strain from prevotella. U.S. Patent Application No. 16/175,299.
Accetto, T. & Avguštin, G. Polysaccharide utilization locus and CAZYme genome repertoires reveal diverse ecological adaptation of Prevotella species. Syst. Appl. Microbiol. 38, 453–461 (2015).
Desai, M. S. et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016).
Franke, T. & Deppenmeier, U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol. Microbiol. 109, 528–540 (2018).
Heaver, S. L., Johnson, E. L. & Ley, R. E. Sphingolipids in host–microbial interactions. Curr. Opin. Microbiol. 43, 92–99 (2018).
Brown, L. C. W. et al. Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLOS Biol. 11, e1001610 (2013).
An, D. et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133 (2014).
Oh, S. F. et al. Host immunomodulatory lipids created by symbionts from dietary amino acids. Nature 600, 302–307 (2021).
Le, H. H., Lee, M.-T., Besler, K. R. & Johnson, E. L. Host hepatic metabolism is modulated by gut microbiota-derived sphingolipids. Cell Host Microbe 30, 798–808.e7 (2022).
Posteraro, P. et al. First bloodstream infection caused by Prevotella copri in a heart failure elderly patient with Prevotella-dominated gut microbiota: a case report. Gut Pathog. 11, 44 (2019).
Iljazovic, A. et al. Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol. 14, 113–124 (2021).
Yeoh, Y. K. et al. Prevotella species in the human gut is primarily comprised of Prevotella copri, Prevotella stercorea and related lineages. Sci. Rep. 12, 9055 (2022).
Forslund, K. et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Karlsson, F. H. et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013).
Moreno-Indias, I. et al. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. Am. J. Transl. Res. 8, 5672–5684 (2016).
Zhu, L. et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatol. Baltim. Md 57, 601–609 (2013).
Hu, H.-J. et al. Obesity alters the microbial community profile in Korean adolescents. PLoS ONE 10, e0134333 (2015).
Vangay, P. et al. US immigration Westernizes the human gut microbiome. Cell 175, 962–972.e10 (2018).
Li, J. et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5, 14 (2017).
Michail, S. et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol. Ecol. 91, 1–9 (2015).
Elinav, E. et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757 (2011).
Maeda, Y. et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 68, 2646–2661 (2016).
Andrew, S. FastQC: a quality control tool for high throughput sequence data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Caldwell, D. R. & Bryant, M. P. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl. Microbiol. 14, 794–801 (1966).
Matsuki, T. et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68, 5445–5451 (2002).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018).
Bushnell, B. BBMap: a fast, accurate, splice-aware aligner. https://www.osti.gov/biblio/1241166 (2014).
Joshi, N. A. & Fass, J. N. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software]. Available at https://github.com/najoshi/sickle (2011).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455 (2012).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 11, 119 (2010).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Lagesen, K. et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108 (2007).
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T. & Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114 (2018).
Asnicar, F. et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 11, 2500 (2020).
Asnicar, F., Weingart, G., Tickle, T. L., Huttenhower, C. & Segata, N. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3, e1029 (2015).
The UniProt Consortium. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res 51, D523–D531 (2023).
Clausen, P. T. L. C., Aarestrup, F. M. & Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinforma. 19, 307 (2018).
Fischer, M., Strauch, B. & Renard, B. Y. Abundance estimation and differential testing on strain level in metagenomics data. Bioinforma. Oxf. Engl. 33, i124–i132 (2017).
Torsten Seemann. ABRicate, Github https://github.com/tseemann/abricate.
Jia, B. et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573 (2017).
Ingle, D. J. et al. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genomics 2, e000064 (2016).
Gupta, S. K. et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58, 212–220 (2014).
Chen, L., Zheng, D., Liu, B., Yang, J. & Jin, Q. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 44, D694–D697 (2016).
Doster, E. et al. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 48, D561–D569 (2020).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Kowalska-Krochmal, B. & Dudek-Wicher, R. The minimum inhibitory concentration of antibiotics: methods, interpretation, clinical relevance. Pathogens 10, 165 (2021).
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 14.0, 2024. http://www.eucast.org.
Khedkar, S. et al. Landscape of mobile genetic elements and their antibiotic resistance cargo in prokaryotic genomes. Nucleic Acids Res. 50, 3155–3168 (2022).
Mistry, J. et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419 (2021).
Huerta-Cepas, J. et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019).
Peeters, E., Nelis, H. J. & Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 72, 157–165 (2008).
Eddy, S. R. Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011).
Berlemont, R. & Martiny, A. C. Glycoside hydrolases across environmental microbial communities. PLoS Comput. Biol. 12, e1005300 (2016).
Nguyen, S. T. C., Freund, H. L., Kasanjian, J. & Berlemont, R. Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy. Appl. Microbiol. Biotechnol. 102, 1629–1637 (2018).
Stewart, R. D., Auffret, M. D., Roehe, R. & Watson, M. Open prediction of polysaccharide utilisation loci (PUL) in 5414 public Bacteroidetes genomes using PULpy. 421024 Preprint at https://doi.org/10.1101/421024 (2018).
Sperry, J. F., Appleman, M. D. & Wilkins, T. D. Requirement of heme for growth of Bacteroides fragilis. Appl. Environ. Microbiol. 34, 386–390 (1977).
Esteve, A. N., Huang, F. & Sardari, R. Development of a defined minimal culture medium for Prevotella copri DSM 18205. (Lund University, Master’s Thesis, 2020).
De Baere, S. et al. Development of a HPLC–UV method for the quantitative determination of four short-chain fatty acids and lactic acid produced by intestinal bacteria during in vitro fermentation. J. Pharm. Biomed. Anal. 80, 107–115 (2013).
Abraham, J. International conference on harmonisation of technical requirements for registration of pharmaceuticals for human use. in Handbook of Transnational Economic Governance Regimes (eds. Tietje, C. & Brouder, A.) 1041–1053 (Brill | Nijhoff, 2010). https://doi.org/10.1163/ej.9789004163300.i-1081.897.
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The innovation 2.3 (2021).
Wu, M. & Scott, A. J. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28, 1033–1034 (2012).
Diebold, P. J., New, F. N., Hovan, M., Satlin, M. J. & Brito, I. L. Linking plasmid-based beta-lactamases to their bacterial hosts using single-cell fusion PCR. eLife 10, e66834 (2021).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinforma. Oxf. Engl. 26, 2460–2461 (2010).
Acknowledgements
This work was funded by grants from the Packard Foundation (to I.L.B.), Pew Charitable Trusts (to I.L.B.) and the National Health Institutes (1DP2HL141007 to I.LB.). We thank Cornell Center for Materials Research (CCMR) for performing SEM imaging and Weill Cornell Medicine Microscopy and Image Analysis Core Facility for performing TEM imaging. The whole-genome sequencing of S. copri isolates was performed by the Biotechnology Resource Center (BRC) at Cornell Institute of Biotechnology.
Author information
Authors and Affiliations
Contributions
I.L.B., and X.X. contributed to the conceptualization of the work. X.X., A.G., and A.S. built the methodology and performed the research. X.X. and A.S. contributed to the analysis and visualization of the results. X.X. and I.L.B. wrote the original draft and all authors contributed to the review and editing. The work was under the supervision of I.L.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xiao, X., Singh, A., Giometto, A. et al. Segatella clades adopt distinct roles within a single individual’s gut. npj Biofilms Microbiomes 10, 114 (2024). https://doi.org/10.1038/s41522-024-00590-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-024-00590-w