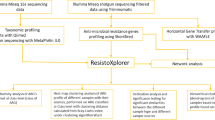
Abstract
Migratory birds are key vectors of pathogens and antibiotic-resistance genes (ARGs), yet intrapopulation variation and its microbiome-mediated basis remain poorly understood. Here, we characterized the gut microbiome of 70 individuals from a migratory Anser serrirostris population using full-length 16S rDNA sequencing, followed by metagenomic analysis of 25 representative samples. Both approaches consistently identified two distinct groups (E1 and E2). Network analysis revealed impaired microbial interactions in E1 compared to E2. E1 exhibited higher abundances of opportunistic pathogens (e.g., Pseudomonas, Erwinia) and enriched functions related to pathogenicity and ARGs, predominantly driven by these taxa. Conversely, E2 showed function enrichment in short-chain fatty acid biosynthesis and plant metabolite degradation, mediated mainly by Bradyrhizobium and Ligilactobacillus. Genome-centric analysis identified several pathogenic genomes (e.g., Salmonella, Vibrio parahaemolyticus) harboring critical virulence factors and ARGs predominantly in E1. These results provide valuable insights into microbiome-driven variation in pathogen/ARG loads within migratory bird populations.
Similar content being viewed by others
Introduction
The gut microbiome has been demonstrated to significantly influence host growth, health, and fitness at both single-individual and population scales through various mechanisms, including food digestion, nutrient absorption, modulation of the immune system, determination of consequences of antibiotic treatment, and transmission of certain important strains and functional genes such as those conferring antibiotic resistance1,2,3,4. Given the importance of the gut microbiome, there has been a significant increase in the number of studies aimed at understanding the structure and function of the gut microbiome as well as how the composition of the gut microbiome is influenced by intrinsic and extrinsic factors in humans and several animal species5,6. Previous studies of the microbiome composition of humans7 and several animal species, such as wild mice8, pandas9, and broilers10, have demonstrated that gut microbiome samples from the same host species could naturally form clusters. The formation of such clusters has been attributed to several factors, including diet and variation in the host’s genetic background11,12. However, recent studies have demonstrated that individuals in the same broiler population with identical genetic backgrounds and farming conditions exhibited distinct gut microbiome compositions, resulting in the separation of these individuals into different clusters10,13. Although the causal factors of variation in the gut microbiome within the same population remain elusive, pivotal bacterial taxa or bacterial guilds that exhibited distinct differences in relative abundance among clusters have been identified through taxonomic decomposition analysis7,14. The identification of such pivotal bacterial taxa or guilds has provided valuable insight into the roles of the identified differentially abundant taxa, ranging from diagnosis and correlation to causation, in relation to the specific traits or diseases of the host. For example, the role of Prevotella in chronic inflammatory diseases in humans has been identified, as well as the influence of Ochrobactrum and Rhodococcus on the growth and meat quality of broilers10,15.
Although significant progress has been made toward the characterization of the gut microbiome of humans and several agriculturally important animals, such as pigs16, cows17, and chickens18, those of wild animals, particularly migratory birds, remain largely unexplored19,20. Migratory birds have been demonstrated to play an important role in the transmission of pathogens and antibiotic-resistance genes (ARGs) during their migration19,21,22, and individuals of the same species frequently carry distinct abundances of pathogens and ARGs21,23. However, our understanding of the microbiome basis of such variation remains limited, possibly due to the small sampling sizes for a given species from the same population and the consequent interference of several other factors, including diet19,24,25.
Anser serrirostris is a migratory bird species that breeds across the Russian Arctic tundra, ranging from northeastern Siberia to Kamchatka, and winters primarily in China, Korea, and Japan, with China being the major wintering destination (accounting for >71.5% of the total estimated wintering population in East Asia)26,27. The wintering population of A. serrirostris in China was estimated to exceed 229,000 individuals in 2020, and the population continues to grow27. However, the structure, function, and associated pathogen and ARG contents of the A. serrirostris gut microbiome have not yet been investigated. The migrations of many bird species, including A. serrirostris, are performed by large social populations, where individuals within the same population have access to nearly identical food sources27,28. Consequently, this uniformity in diet can minimize the effect of dietary variation on the gut microbiome25. Hence, conducting a comprehensive sampling of a specific population of migratory bird species, such as A. serrirostris, and thoroughly investigating the structure, function, and associated pathogens and ARG contents in the gut microbiome will enhance our understanding of the extent of variation and the underlying microbiome basis of this variation. Such insights will inform mitigation strategies-for example, targeted probiotic interventions to suppress ARG-harboring taxa and pathogens in wild bird populations29. To this end, we conducted comprehensive sampling and performed full-length 16S rDNA amplicon and metagenomic sequencing analyses of an A. serrirostris population.
Fecal samples are widely used as a proxy for gut microbiome composition analysis because they can be collected non-invasively and are particularly suitable for studying species with strong migratory tendencies. However, fecal sample–based analyses provide a less comprehensive profile of the gut microbiome compared with intestinal content–based analyses, which, although more informative, require euthanasia and are therefore not feasible for protected animal species30. Here, we collected fresh fecal samples of various individuals in an A. serrirostris population from Jiangsu Yancheng Wetland National Nature Reserve, Rare Birds, an important stopover wintering site for A. serrirostris located in lower Yangtze River floodplain27 (Supplementary Figs. 1 and 2), for characterizing and comparing their gut microbiomes as well as estimating the pathogen and ARG contents using full-length 16S rDNA amplicon and deep metagenomic sequencing.
Results
The A. serrirostris population was stratified into two groups through microbiome profiling analysis
Seventy fresh fecal samples were collected from an A. serrirostris population and subjected to full-length 16S rRNA gene sequencing (PacBio ccs). Read processing, OTU clustering at 97% similarity, and taxonomic annotation against SILVA v132.99 were performed as described in Methods. Twenty-two bacterial phyla were identified in the gut microbiota of these 70 individuals, with Bacillota, Pseudomonadota, Actinomycetota, and Bacteroidota being the predominant phyla (average relative abundance >1%) (Supplementary Fig. 3). Ward linkage hierarchical clustering analysis based on genus-level profiles partitioned these 70 individuals into two distinct groups, namely group 1 (E1), containing 27 samples, and E2, containing 43 samples (Fig. 1A). Alpha-diversity metrics showed no significant difference in Shannon index between the two groups, whereas E1 exhibited a significantly lower number of observed genera (richness) but markedly higher evenness (Fig. 1B and Supplementary Fig. 6). Beta-diversity analysis indicated that the overall bacterial community composition of E1 differed significantly from that of E2 (PERMANOVA, F = 15.99, P < 0.001) (Supplementary Fig. 5). Compared with E2, The E1 group was found to harbor a significantly higher relative abundance of Pseudomonadota and a significantly lower relative abundance of Bacillota, Bacteroidota, and Actinomycetota (all P < 0.001, Mann–Whitney U-test, two-tailed) (Fig. 1C and Supplementary Fig. 4). When compared at the genus level, 77 genera were identified to exhibit significantly differential relative abundance between the two groups (DESeq2, False Discovery Rate, FDR < 0.05) (Supplementary Data 1). Among them, 19 genera were substantially differentially abundant (intergroup relative abundance difference >1%) between the two groups, with Erwinia (8.99% vs. 0.49%), Pseudomonas (11.89% vs 3.21%), Enterobacter (6.86% vs 0.14%) and four others exhibiting significantly higher relative abundances in E1 and Lactobacillus (6.04% vs. 39.54%) and eleven others showing a significantly higher relative abundance in E2 (Fig. 1D).
A The clustering result of 70 samples based on the bacterial community composition data determined at the genus level using STAMP. B Alpha-diversity (Shannon index) differences between E1 and E2 groups revealed by 16S-based analysis. C Relative abundance comparison between E1 and E2 at phylum levels revealed by the 16S rDNA amplicon data (two-tailed Mann–Whitney U-test). D The extended error bar plot of genera differentially abundant between E1 and E2 (>1% difference in abundance) revealed by the 16S rDNA amplicon data (DESeq2, FDR < 0.05). *denotes <0.05, **denotes <0.01, ***denotes <0.001, and n.s. denotes not significant. The Numerical p-values are shown in Supplementary Data 15.
To elucidate the functional repertoire underlying the E1/E2 stratification, 25 representative samples (11 E1, 14 E2) were selected for metagenomic sequencing. The clean reads were co-assembled, and metagenes were subsequently predicted from the resulting contigs. By filtering out genes belonging to eukaryotes (except fungi), we built an integrated gut microbial gene catalog containing 3,121,626 nonredundant genes, of which 1,917,880 (61.43%) were complete genes with an average length of 632 bp. The rarefaction curve clearly suggested that the gene catalog comprehensively represented the gut microbiome of the sampled A. serrirostris population (Fig. 2A). Metagene abundance-based analysis of gut microbiome composition yielded consistent grouping results for E1 and E2 (with 11 samples forming E1 and 14 samples forming E2) (Fig. 2B), and permutational multivariate analysis of variance (PERMANOVA) confirmed that group membership explained a significant portion of variance in microbiome structure (F = 40.64, P = 0.001). The observed differences in microbiome composition between the two groups were unlikely attributable to genomic background variation among the 25 individuals, as the samples were collected under a rigorous protocol (see “Methods”) and their mitochondrial sequences were highly conserved and showed no differentiation between the groups (Supplementary Fig. 7). The metagenome-based analysis revealed distinct variations in term of microbial community diversity and complexity between the groups at the whole microbiome level (Fig. 2C). In detail, E2 exhibited significantly higher Shannon index and evenness than E1 (Mann–Whitney U-test, two-tailed, both P < 0.001) (Fig. 2C and Supplementary Fig. 6). Furthermore, molecular ecological networks (MEN) analysis revealed that the network topological properties of E2 were significantly different from those of E1, in that the E2 network exhibited significantly higher values of average clustering coefficient (Avgcc), connectedness (Con), density (D), and geodesic efficiency (E), and significantly lower values of average path distance (GD) (Student’s t-test, all P < 0.0001) (Fig. 2D). These indices suggest that the microbial members in E2 were closely connected with their neighbors and formed a more complex bacterial community network compared with E1. Notably, the microbiome of E2 was suggested to form a modular structure (modularity score 0.682), and the microbiome structure of E1 was nonmodular (modularity score 0.625) based on the modularity score threshold of 0.634, as described by Newman31.
A Rarefaction curve of detected genes in A. serrirostris microbiomes based on 100-fold permuted sampling orders, and the ratios of complete and partial genes in the gene catalog are shown. B PCoA plot exhibiting that the 25 samples were separated into two distinct groups. C Alpha-diversity (Shannon index) differences between E1 and E2 groups. D Network structure and property differences between E1 and E2. Relative abundance comparison between E1 and E2 at the phylum (E) and genus (F) levels. Mann–Whitney U-test was used for statistical analysis in (C, E), and DESeq2 was used for statistical analysis in (F). * denotes <0.05, ** denotes <0.01, *** denotes <0.001, and n.s. denotes not significant. The Numerical p-values are shown in Supplementary Data 15.
Consistent with the full-length 16S rDNA amplicon-based analysis, the metagenome-based analysis also identified a significantly higher relative abundance of Pseudomonadota and a significantly lower relative abundance of Bacillota and Bacteroidota in E1 compared with E2 (all P < 0.05, Mann–Whitney U-test, two-tailed) (Fig. 2E and Supplementary Fig. 4). Further comparative analysis at the genus level identified 1201 differentially abundant genera between the two groups (DESeq2, FDR < 0.05), including 23 genera exhibiting substantially differential abundance (Fig. 2F and Supplementary Data 2). Among them, Pseudomonas (35.13% in E1 and 1.08% in E2), Erwinia (16.79% vs. 0.18%), Enterobacter (3.23% vs. 0.24%), and several others exhibiting significantly higher relative abundances in E1, as well as Bradyrhizobium (1.05% vs. 13.09%), Asinibacterium (1.98% vs. 14.06%), and two genera classified under the Lactobacillus group in the 16S rDNA Silva database, Levilactobacillus (0.11% vs. 9.45%) and Ligilactobacillus (0.64% vs. 13.28%), and several others showing a significantly higher relative abundance in E2. Notably, significantly higher abundance of Pseudomonas, Erwinia, and Enterobacter in E1, and significant enrichment of Lactobacillus in E2 were also identified through full-length 16S rDNA amplicon analysis (Fig. 1D).
The beneficial, pathogenicity-associated genes and ARGs differed between the two groups
KEGG orthology (KO) annotations were assigned to the A. serrirostris gut microbial gene catalog based on the eggNOG database, and 33.28% of the nonredundant genes obtained KO annotations with 10,458 KOs identified. We identified significantly more predominant KOs (i.e., KOs with abundance >1 TPM [transcripts per million]) in E1 (6,182.9 ± 226.7 KOs, mean ± SD, n = 11) than in E2 (5,150.8 ± 540.2 KOs, n = 14) (Student’s t-test, P = 3.7e–7), indicating that E1 harbored a relatively diverse functional microbiome. The phyletic distribution patterns of these predominant KOs separated the samples belonging to E1 group from those belonging to group E2 (Fig. 3A). Although 3,862 KOs were shared between the two groups, E1 was found to harbor 1,819 group-unique KOs, and this number for E2 was 405 (Fig. 3B). Through DESeq2 analysis, we identified 3,985 differentially abundant KOs, including 2,201 higher abundant KOs in E1 (E1HKs) and 1,784 higher abundant KOs in E2 (E2HKs) (False Discovery Rate, FDR < 0.01) (Supplementary Data 3). The E1HKs were mainly enriched in pathways involved in microbe-microbe and microbe-host interactions, while E2HKs were mainly enriched in metabolism-associated pathways, respectively (Fisher’s exact test, P < 0.05) (Table 1).
A Phyletic distribution of KOs among individuals. B Venn plot depicting the number of core E1 and E2 KOs based on their presence rate (>75%). The abundance difference of (C) plant-derived secondary metabolites (PSMs) degradation-, E short-chain fatty acids (SCFAs) synthesis-, and G pathogenicity-associated functions between E1 and E2, and D, F, H, the top contributors of these functions at the genus level. Mann–Whitney U-test was used for statistical analysis. *denotes <0.05, **denotes <0.01, ***denotes <0.001, and n.s. denotes not significant. The Numerical p-values are shown in Supplementary Data 15.
Multiple genes involved in microbe–microbe competition were identified in E1HKs. For example, the complete T6SS machinery system (hcp, lip, icmF, dotJ, clpV, impA, fhaL, and pppA) and several T6SS effector-encoding genes, such as pld1_2 and vgrG, were identified in the E1HKs (Fig. 4A). In addition, several ABC transporter-encoding genes dedicated to the uptake of essential but gut-limiting metals—iron, zinc, and manganese—including troBCD, sitABCD, mtsAB, and znuBC, were identified in the E1HKs (Fig. 4A). Furthermore, the pathway “biosynthesis of siderophore group nonribosomal peptides” was enriched in the E1HKs (Table 1), and almost all of the genes involved in the synthesis of two important siderophores, enterochelin (entABCDEF), and pyochelin (pchADFG), were identified in E1HKs (Fig. 4A). Pseudomonas and Erwinia, which were highly abundant in E1 but exhibited very low relative abundance in E2 (Fig. 2F), were the main contributors to inter-microbial competition-associated genes in the E1 group (Fig. 4B–D).
A The abundance difference of microbe–microbe competition associated functions between E1 and E2. B-D, the top contributors of (B) T6SS- (C) Siderophore-, and (D) metal uptake-associated functions at the genus level. Mann–Whitney U-test was used for statistical analysis. *denotes <0.05, **denotes <0.01, ***denotes <0.001, and n.s. denotes not significant. The Numerical p-values are shown in Supplementary Data 15.
Interestingly, the E1 group harbored a significantly higher relative abundance of pathogenicity-associated genes, and Pseudomonas and Erwinia were the main genera harboring these genes in the E1 group (Fig. 3G, H, and Supplementary Fig. 6). In detail, GacA-GacS, which is recognized as a global regulatory two-component system (TCS) of virulence-associated traits in many bacterial pathogens, including Pseudomonas spp. and several pathogenic Enterobacteriaceae species such as Erwinia32, was identified in E1HKs, with 64.64- and 73.25-fold higher average relative abundances in E1 than in E2 for gacA and gacS, respectively (Supplementary Fig. 8). Multiple KOs associated with inflammatory excitation and pathogenicity, such as those involved in lipopolysaccharide (LPS) synthesis (e.g., waaBEOPQ and lpxMT), the T3SS machinery system and associated effectors (e.g., bopN, sspH2, ipaH9.8, pipB3, and exoU), cationic antimicrobial peptide (CAMP) resistance (e.g., phoPQ and pagP), and RTX toxins (e.g., cyaC, ptxA, rtxA, and exlA), were identified in E1HKs (Fig. 3G).
Several pathways involved in the degradation of phenolic compounds, such as “benzoate degradation”, “aminobenzoate degradation”, “fluorobenzoate degradation”, “xylene degradation”, and “styrene degradation”, were enriched in E2HKs (Table 1), and Bradyrhizobium was the main contributor of the functional properties involved in phenolic compound degradation in the E2 group, accounting for 62.12 ± 11.68% (mean ± SD) of the normalized total relative contribution (Fig. 3C, D). However, in addition to the relative decrease in the functional properties involved in phenolic compound degradation, the contribution of Bradyrhizobium to these functions was also significantly decreased in E1 (21.71 ± 17.01%) compared with E2 (62.12 ± 11.68%) (P = 0.0014, Mann–Whitney U-test, two-tailed). The pathways “propanoate metabolism” and “butanoate metabolism” were also enriched in E2HKs (Fig. 3E and Table 1). Propanoate and butanoate are two major short-chain fatty acids (SCFAs) produced by the gut microbiome that play essential roles in maintaining intestinal health33. Multiple KOs involved in SCFAs synthesis were identified in E2HKs (Fig. 3E). Escherichia, Bradyrhizobium, Asinibacterium, Levilactobacillus, and Ligilactobacillus were the main contributors to the KOs in E2 (Fig. 3F). However, the contribution of these genera to SCFAs biosynthesis-related functions was decreased in the E1 group.
Intriguingly, the pathway “beta-lactam resistance” was found to be overrepresented in the E1HKs (Table 1), with beta-lactamase-encoding genes, such as blaCTX-M, blaZ, blaCARB-1, blaADC, blaPDC, and blaOXA-213, exhibiting a significantly higher relative abundance in the E1 group than in E2. We then analyzed the overall ARG prevalence in the A. serrirostris gut microbiome, and 1,505 of the 3,121,626 nonredundant genes were identified as ARGs. These 1,505 ARGs were further grouped into 224 antibiotic resistance ontologies (AROs) that conferred resistance to various drug classes (Fig. 5A and Supplementary Data 4). Notably, the E1 group harbored a significantly higher abundance of ARGs (on average 32.44 folds higher) compared with that of E2 (Mann–Whitney U-test, two-tailed, P < 0.001) (Fig. 5B). Erwinia, Enterobacter, Leclercia, and Pseudomonas were the main contributors to ARGs in the E1 group (Fig. 5C). Of the 1,505 ARGs, 270 were predicted to be located in mobile genetic elements (MGEs) associated contigs, including 254 plasmid- and 16 phage-derived contigs (Supplementary Data 4), and these ARGs accounted for an average of 19.82% (ranging from 9.10 to 39.38%) of the total abundance of ARGs, with significantly higher relative abundance in the E1 group compared with E2 (Fig. 5B).
A Distribution of the identified 1505 ARGs. The resistance mechanisms and number of ARGs are shown. B, C Abundance difference of ARGs between E1 and E2 and the top contributors. The abundance difference of plasmid- and phage-derived ARGs is shown inside (B). Mann–Whitney U-test was used for statistical analysis. *denotes <0.05, **denotes <0.01, ***denotes <0.001, and n.s. denotes not significant. The Numerical p-values are shown in Supplementary Data 15.
Genome-centric analysis identified several bacterial pathogens with different prevalence levels
Through metagenome-assembled genomes (MAGs) extraction analysis using the metaWRAP pipeline, 91 MAGs (>70% completeness and <10% contamination) were recovered from the gut microbiome of A. serrirostris (Supplementary Data 5). Among these MAGs, we selected several prevalent potential pathogens carrying VFs and ARGs for further in-depth investigation. For example, a high-quality MAG affiliated with Pseudomonas designated bin58 (completeness 82.63% and contamination rate 3.51%) was identified. GacAS (the key global regulatory TCS required for virulence, also identified in E1HKs) and several other pathogenicity-associated genes, such as type IV pili and flagellum synthesis-associated genes (responsible for adhering to the host cells), the alg gene cluster (responsible for forming biofilms and overcoming host defense), and exlA (critical for cytotoxicity), and several ARGs, were identified in the bin58 genome (Supplementary Data 6 and 7). Bin58 exhibited a significantly higher abundance in E1 (average genome coverage 45.25 folds) than in E2 (0.10 folds) (Fig. 6A). Of note, compared with the available genomes in the NCBI RefSeq database, bin58 exhibited relatively high similarity (ANIm > 97%) with two strains isolated from the Lake Erie region, North America, Pseudomonas sp. 02C 26 (isolated from a water sample) and Pseudomonas sp. S07E 245 (isolated from a soil sample), and exhibited low relatedness (ANIm < 85%) to other available Pseudomonas genomes.
The phylogenetic trees were constructed using CVTree and visualized using iTOL. For Pseudomonas sp. bin58 (A), all RefSeq genomes with ANI > 80% were included in the phylogenetic analysis. For Salmonella bin89 (B) and Acinetobacter baumannii bin59 (D), the 100 RefSeq genomes with the highest ANI to each MAG were selected. For Vibrio parahaemolyticus bin35 (C), the top 100 genomes were selected, then near-identical (clonal) top hits with identical metadata (isolation source, location, date, and submitter) were dereplicated by retaining a single representative per clonal cluster prior to tree building.
Notably, we identified several high-quality MAGs as important pathogens that can infect humans and cause severe diseases of the infected, such as Vibrio parahaemolyticus34 (bin35, completeness 95.69% and contamination rate 1.80%), Salmonella enterica35 (bin89, completeness 95.81% and contamination rate 5.15%), and Acinetobacter baumannii36 (bin59, completeness 95.06% and contamination rate 3.97%). Bin89 was assigned to S. enterica by GTDB-Tk37 using 120 conserved marker genes. Comparison against a collection of >10,000 high-quality Salmonella genomes from NCBI RefSeq showed that the nearest RefSeq matches were affiliated with serotype Saintpaul (Supplementary Data 8). However, the phylogenetic tree in Fig. 6B indicates that bin89 exhibits a relatively low degree of genome similarity to those Saintpaul genomes. To further investigate its taxonomic status, we applied SeqSero2 for in silico serotyping38; SeqSero2 predicted an antigenic formula of 4:g,m:1,2 for bin89, a result that does not clearly match common reference serovars. In the phylogenetic tree, a strain affiliated with serotype Enteritidis (S. Enteritidis NCCP 16206; GCF_009884355) was included and, after midpoint rooting, formed a separate clade that effectively acted as an outgroup. Taken together, the GTDB assignment, RefSeq comparisons, and SeqSero2 serotyping support classifying bin89 as a Salmonella genome that is genomically divergent from well-represented reference strains, potentially reflecting an under-sampled wild-animal lineage. We identified a total of 300 virulence factors (VFs), including many well-known pathogenicity genes, such as those involved in host cell adhesion and invasion (T3SS machinery and associated effectors), hemolytic activity (slyA), and cytotoxin (spvB) (Supplementary Data 9). Moreover, 52 ARGs were identified in bin89, including several ARGs conferring multidrug resistance (Supplementary Data 10). Bin89 was detected in 10 of the 11 samples in the E1 group and 4 of the 14 samples in the E2 group, and it exhibited significantly higher abundance in E1 than in E2 (Mann–Whitney U-test, two-tailed, P < 0.01) (Fig. 6B).
Vibrio parahaemolyticus bin35 exhibited very high genome similarity (ANIm > 99.9%) with several human patient-originated V. parahaemolyticus genomes in the NCBI Refseq database (Supplementary Data 11), and harbored thermostable direct hemolysin (tdh), which is a key VF for V. parahaemolyticus that invades the human body, as well as several other genes and genomic islands that are critical for pathogenicity39 (Supplementary Fig. 9). Notably, the phylogenetic relationship and distribution of VFs further suggested that bin35 was more closely related to human patient-originated strains than to those isolated from environmental samples and animals (Fig. 6C and Supplementary Fig. 9). Bin35 was highly abundant in sample S12 affiliated with the E1 group (genome coverage 7.8 folds) and was detected in three more samples with relatively low abundance (genome coverage <0.2-fold) (Fig. 6C).
Acinetobacter baumannii is one of the most intractable pathogens in the clinic, mainly because of its resistance to the majority of the antibiotics currently used, besides its virulence, thus leading to the prevalence of infections and outbreaks36. Bin59 exhibited high genome similarity (ANIm > 99.8%) to more than 100 A. baumannii strains available in the NCBI RefSeq database, and 98 strains out of the 100 top-hit strains were isolated from human patients with missing isolation source information for the other 2 strains (Supplementary Data 12). We also identified several important genes that were critical for host infection and virulence, such as omp33, bfmR/S TSC, and trimeric autotransporter adhesin encoding gene ata, in bin59 (Supplementary Data 13). Furthermore, several ARGs that are resistant to widely used antibiotics in the clinic, including β-lactam antibiotics (OXA-23, OXA-66, and ADC-73), chloramphenicol (craA), fosfomycin (abaF), colistin (emrABEKY), aminoglycoside (aac and armA), as well as efflux pumps (adeABCFGHIJKLNRS), were identified in bin59 (Supplementary Data 14). Bin59 was highly prevalent in the A. serrirostris population and was detected in 24 of the 25 samples with the highest genome coverage of 2.3-fold (Fig. 6D). Similar as S. enterica bin89, underrepresentation of wild animal-derived A. baumannii strains in the public genome database was also suggested by the phylogenic analysis (Fig. 6D).
Discussion
In this study, we investigated the structure and function of the gut microbiome of A. serrirostris by conducting full-length 16S rDNA amplicon analysis on 70 individuals, followed by deep metagenomic sequencing of 25 representative individuals from a population wintering in the Yancheng National Rare Birds Nature Reserve, China. Based on these analyses, we constructed a comprehensive gene catalog that accurately represents the A. serrirostris gut microbiome (Fig. 2A). Although the structure and function of the gut microbiome have been investigated in several wild bird species19,21,22, this study provides, to the best of our knowledge, the first gene catalog comprehensively representing the gut microbiome of a migratory wild bird species. Only 56.70 and 33.28% of the gene catalog obtained taxonomy and function annotation, supporting the notion that our understanding of wild animal microbiomes, including that of A. serrirostris, is at an early stage, with there being little knowledge of the composition of the microbiome. The predominance of bacterial annotations (98.85% of annotated genes) indicates that bacteria likely contribute a broader repertoire of functions within the gut microbiome of A. serrirostris than the less-represented fungi, archaea, and viruses. However, because public reference databases are heavily biased toward bacterial genomes, the representation and inferred functional contributions of fungal, archaeal, and viral genes may be systematically underestimated40. Future studies using expanded domain-specific databases may help elucidate the actual abundance and functional roles of these underrepresented microbial domains. The order of abundance of the phyla Pseudomonadota, Bacillota, Bacteroidota, and Actinomycetota was similar to that observed in a previous study on the gut microbiome of five sympatric flycatchers (Ficedula strophiata, Phoenicurus auroreus, Niltava sundara, Phoenicurus frontalis, and Tarsiger rufilatus)41, but was different from another study with 17 species of migratory birds in which Bacillota was most abundant (47.8%), followed by Pseudomonadota, Fusobacteriota, and Bacteroidota42. These differences may be related to the bird species and their dietary habits25.
One of the most interesting findings in the present study is the definition of two groups among the individuals within the same population, according to the abundance-based gut microbiome composition obtained by both 16S rDNA amplicon analysis and metagenome-based analysis (Figs. 1A, 2B, and Supplementary Figs. 4–6). In both wild and domestic birds, the host diet, genetics, age, sex, social contact, behavioral habits, and environmental factors play major roles in shaping the gut microbiome25, and even different bird species that live in the same geographic environments and consume shared food sources tend to have similar gut microbiome structures and functions6,21. In the present study, the division of two A. serrirostris groups could not be explained by the effects of diet or host genetic background, since all of the involved individuals had the same feeding environments and shared nearly identical genetic backgrounds (Supplementary Fig. 7). However, important metadata—including age, sex, and health status—were unavailable in this study due to the logistical challenges of capturing and assessing these parameters in large populations of this protected migratory species. These missing variables likely play a role in explaining the observed variations in microbiome structure. Future research should prioritize developing non-invasive metadata collection methods that comply with international migratory bird protection agreements, which may help identify the key drivers of group differentiation within the same bird population.
A high number of functional genes identified in the gut microbiome of A. serrirostris were involved in the degradation of plant-derived secondary metabolites (PSMs) (Fig. 3B and Table 1), which may be related to the herbivorous behavior of this bird species. Secondary metabolites are important compounds produced by plants to protect themselves from being foraged by herbivores through several means, including direct toxicity and digestion impairment. However, certain herbivores have evolved strategies to mitigate the negative effects of plant secondary metabolites and even benefit from these compounds, mainly with the help of the gut microbiome43,44. Bradyrhizobium was identified as the main contributor to PSM degradation, and this genus was also predominant and overrepresented in E2 (Figs. 2F and 3D). Bradyrhizobium is well known as a symbiotic nitrogen-fixing bacteria associated with diverse legume species45; meanwhile, both diazotrophic and non-diazotrophic Bradyrhizobium spp. colonize the root endophytic compartment of a wide variety of plant species, including the model species Arabidopsis46 and rice47. In association with both the symbiotic and endophytic traits, Bradyrhizobium serves as the mutualistic taxon in plant microbiomes and is capable of metabolizing a large number of phenolic acids derived from plant hosts48,49. Bradyrhizobium is also predominant in the gut microbiome of several migratory bird species that frequently consume plant material (Supplementary Fig. 10). We inspected the full-length 16S OTU table and identified one OTU annotated as Bradyrhizobium. BLASTn of the OTU representative sequence against the NCBI rRNA/ITS database returned Bradyrhizobium murdochii CNPSO:4020—a plant-derived strain50—as the top hit, with 98.23% identity over the full-length 16S alignment. Therefore, it is possible that Bradyrhizobium is recruited from the plant microbiomes by herbivorous birds, as it is in the case of Pseudomonas, as the dominant endophyte group in arrow bamboo (Fargesia nitida) and in the microbiome of arrow bamboo-feeding pandas51,52, to alleviate the negative effects of PSMs during their evolution. These results suggest how the gut microbiome contributes to the avian hosts’ fresh plant material feeding cycle. Moreover, the functional genes in the A. serrirostris gut microbiome could serve as an important source for the discovery of novel enzymes, such as those involved in plant material utilization.
The formation of a complex and modular bacterial community network with relatively high alpha-diversity (Shannon index) in the microbiome of E2 (Fig. 2C, D) could increase the robustness of the microbiome against perturbations, and in turn, benefit the host by several means, such as enhancing the normal physiological functions and the ability to resist external interference of the host53,54. Several highly abundant genera, including Levilactobacillus, Ligilactobacillus, Escherichia, Bradyrhizobium, and Asinibacterium, exhibited in E2 (Fig. 2F), are known as SCFA producers in the gut microbiome55,56,57,58, and these genera were indeed found to be the main contributors of the SCFA biosynthesis-associated genes identified in this study (Fig. 3F). SCFAs produced by the microbiome can benefit the host by modulating the immune system and physiology, as well as by other means33. Compared with E2, the microbiome of E1 exhibited a significantly decreased α-diversity with significantly attenuated positive inter-microbial connections, collectively resulting in a nonmodular structure (Fig. 2D). Several opportunistic pathogens that could cause human infections, such as Pseudomonas and Erwinia59,60, exhibited significantly higher relative abundance in the E1 group than in E2, and these genera were also found to be the main contributors to the abundant inter-microbial competition-associated functions, such as T6SS as well as the ABC transporters and siderophores responsible for absorbing essential metals into the bacterial cells (Figs. 3G and 4A). T6SS plays an important role in the inter-bacterial competition in the gut microbiome61. Several metal elements, including iron, zinc, and manganese, are critical for the growth and metabolism of bacteria that colonize the intestinal environment62. However, the availability of these metals in the gut is limited. Sequestration of essential metal nutrients in the immediate vicinity, which is primarily mediated by the siderophores and associated ABC transporters, is an important strategy adopted by certain microbial members to repress the growth of competitive microbes63. The relative abundance of genera involved in PSM degradation and SCFA production, including Ligilactobacillus and several other genera, was significantly lower in E1. In contrast, the opportunistic pathogens that were the major contributors to the inter-microbial competition-associated functions, such as Pseudomonas and Erwinia, were also the main contributors to the E1-abundant pathogenicity-associated genes (Figs. 3H and 4B–D). Thus, the high abundance of potential pathogenic genera in the E1 group probably resulted in the enrichment of pathogenicity-associated genes, including those involved in LPS synthesis, T3SS effectors, and RTX toxin production, in the gut microbiome (Fig. 3G). LPS is a component of the cell wall of Gram-negative bacteria and induces inflammatory responses in the host64. The T3SS effectors play important roles in triggering host inflammatory responses32. RTX toxins are present in a broad range of Gram-negative mammalian pathogens and function to block phagocytosis and the bactericidal production of reactive oxygen species in host cells65. Furthermore, among the E1HKs affiliated with “CAMP resistance” pathway, we identified the TCS phoPQ and the downstream pagP that endow the bacteria with resistance against the CAMPs produced by host neutrophils by increasing the acylation level of lipid A66. It is important to note that the detection of functions such as GacS/GacA, T6SS, and LPS biosynthesis, which are mainly contributed by several potential pathogenic genera (e.g., Pseudomonas and Erwinia), should not be taken as definitive evidence of pathogenicity. These loci are present in both pathogenic and non-pathogenic lineages within a given genus and can mediate either mutually beneficial interactions or inter-microbial competition, depending on the genetic context67,68,69. Therefore, species- or even strain-level annotations are required to draw conclusive inferences. In this study, however, only genus-level annotations could be reliably assigned using the metagene-centric approach, which nevertheless enabled us to quantify the contributions of different taxa to specific pathways or functions (i.e., contributional profiling)70,71. Thus, although the higher abundance of these taxa and functions in E1 suggests a greater pathogenic potential in that group, confirmation will require high-throughput cultivation, genome sequencing, and experimental infection or functional assays in future work to determine the species- and strain-level attribution of virulence traits.
Multiple ARGs that can confer resistance for several important classes of clinical antibiotics, including beta-lactam antibiotics, were identified in the A. serrirostris microbiome (Fig. 5A). Notably, although the relative abundance of ARGs in E2 was very low and even reached a level below the detection ability, we identified highly abundant ARGs in E1. The predominant pathogenic bacterial taxa identified in E1 were also the main contributors to the ARG contents (Fig. 5C). Furthermore, given that a large fraction of these ARGs were located in MGE regions that could facilitate inter-microbial transmission of the ARGs, these ARGs could pose severe threats to public health. Previous studies have demonstrated that the gut microbiomes of wild birds are reservoirs for ARGs, and that the abundance of ARGs varies among bird species21,22. Our results further demonstrated that individuals belonging to the same species and living in the same habitat could harbor distinct abundances of ARGs. These results suggest that extensive and comprehensive sampling of individuals in a migratory bird population is required in such surveys to obtain solid conclusions, as the abundances of pathogens and ARGs may be undetectable in certain individuals, whereas some members may harbor a high abundance of pathogens and ARGs (Figs. 3H, 4B, and 5C).
Several high-quality MAGs of known pathogenic bacteria that could cause human infections were recovered from the A. serrirostris microbiome. We further analyzed four representative MAGs with distinct prevalence patterns in the A. serrirostris population (Fig. 6). A. baumannii bin59 was widely distributed in the studied A. serrirostris population and was closely related to those patient-originated strains (Fig. 6D). Despite the frequent identification of A. baumannii in clinical settings and the significant attention it has garnered, the natural reservoir of A. baumannii remains undefined72. The high prevalence of bin59 in the A. serrirostris population suggests that this migratory bird species could serve as a reservoir for A. baumannii, posing a risk to public health. S. enterica bin89 exhibited a significantly higher abundance in E1 compared to E2 (Fig. 6B), which was closely related to several genomes affiliated with serotype Saintpaul (Supplementary Data 8). Many members affiliated with Salmonella, including S. Saintpaul, are zoonotic pathogens35, and the impacts of bin89 on the fitness of A. serrirostris need to be further determined. Of note, S. Saintpaul was recently identified frequently in animals and human patients in China73. Compared to the highly prevalent bin89 and bin59, V. parahaemolyticus bin35 was relatively less prevalent in the A. serrirostris population (Fig. 6C), suggesting that it was only occasionally acquired by certain individuals from environments containing water. However, considering that bin35 was closely related to V. parahaemolyticus strains originating from human patients and also harbored critical pathogenicity genes and islands for human infection, there is a potential risk for this strain to contaminate water and other environments impacted by A. serrirostris, and the risk to public health should not be overlooked. Furthermore, Pseudomonas sp. bin58 was suggested to be distinctly related to currently known genomes, but here the bacterium harbored several pathogenicity-associated genes and exhibited relatively high abundance in the E1 group. Thus, the potential risk of this strain also needs to be monitored.
This study has provided a comprehensive analysis of the composition and variation of gut microbiome structure and function, as well as the pathogen and ARG contents of an important migratory wild bird species, A. serrirostris. Although the E1 group was enriched for genera that include opportunistic pathogens (e.g., Salmonella, Pseudomonas), the consequences for individual- and population-level bird health remain uncertain. These taxa have been isolated from wild and migratory birds and have been linked to clinical disease in some avian hosts74,75; however, detection in fecal metagenomes alone does not demonstrate active infection, which is also shaped by host condition, environmental stressors, and exposure dose. Recent advances in animal tracking and identification technologies, such as GPS biologgers and drone imagery with automated re-identification, now enable repeated health monitoring and sampling of individual animals across stopover sites. These approaches are both feasible and ethically sound; integrating them with environmental sampling (e.g., water, plants, soil) along migratory flyways in future studies could provide valuable insights into the impacts of pathogens and ARGs on hosts as well as their environmental sources. Given that A. serrirostris frequently grazes grass crops and defecates in farmed agricultural land, including winter wheat, barley, and cabbage at stopover sites76, the notorious pathogens and ARGs carried by A. serrirostris could be transmitted to local animals and humans through direct contact, bioaerosols, food chains, and other means77,78, and thus pose a risk to public health. The factors contributing to the observed differentiation in the gut microbiome of the A. serrirostris population remain unclear. Further studies are needed to develop non-invasive, field-deployable methods for protected wildlife that can capture individual-level metadata (e.g., age, sex, and health status) to uncover the drivers of intrapopulation microbiome divergence. A better understanding of the microbiome basis of pathogen and ARG contents in migratory birds will contribute to the management of migratory bird-mediated pathogens and ARG transmission.
Methods
Sampling collection
In this study, we selected an A. serrirostris population that contained several hundred individuals in the Jiangsu Yancheng Wetland National Nature Reserve, Rare Birds, for sampling and analysis. This reserve is located in the lower Yangtze River floodplain and covers an area of 247,260 hectares, ranging from 119°53'45” to 121°18’ 12” E and 32°48'47” to 34°29'28” N, and is recognized as an important stopover or wintering site for A. serrirostris as well as several other migratory bird species such as red-crowned cranes (Supplementary Fig. 1). This A. serrirostris population had arrived to the reserve more than 2 weeks before the sampling date as confirmed by our continuous monitoring.
Fecal sampling was performed during the morning of 9 December 2020 in a barley seedling field (approximately 800 × 400 m) that was exclusively occupied by a foraging A. serrirostris flock (>300 individuals; see Supplementary Fig. 2). Ambient temperature during sampling remained stable at 7 °C. Prior to collection, binocular monitoring confirmed the absence of non-target avian species within a 500-m radius over a continuous 60-min observation period. Sampling was focused on a 100 × 100-m quadrat exhibiting the highest population density, as determined through the binocular monitoring. Fresh fecal specimens were identified using species-specific morphological markers, including cylindrical morphology (length: 8–10 cm; diameter: ~0.5 cm) and greenish-brown pigmentation correlating with chlorophyll-rich dietary intake. These diagnostic criteria were cross-verified by two independent ornithologists during the sample collection. To avoid environmental contamination during sampling, an aseptic stick was inserted into the central inner layer of the feces, rotated, and withdrawn to ensure that samples were obtained from inside the feces to avoid soil-derived contamination. Notably, we collected samples from various sites at least 1 m away from each other to avoid the collection of samples from the same individual. To prevent microbial DNA degradation, fresh fecal samples were immediately transported to the laboratory on dry ice and stored at −80 °C until DNA extraction.
DNA extraction and full-length 16S rDNA amplicon sequencing and analysis
Total microbial genomic DNA was extracted from fecal samples using the QIAamp® PowerFecal® DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The DNA concentration and purity of all samples were measured by NanoDrop (ThermoFisher Scientific, Waltham, MA, USA), and their quality was verified on a 1.0% agarose gel electrophoresis. Subsequently, the DNA concentrations were determined using Qubit 4.0 (ThermoFisher Scientific) to ensure satisfactory library preparation. A total of 70 samples were sequenced. Full-length 16S rRNA genes were amplified using the primer set 27 F and 1492 R with barcodes79. The PCR products were sequenced using a PacBio HiFi sequencing platform (Guangzhou, China), and high-quality circular consensus sequence (CCS) reads were generated.
In this study, an operational taxonomic unit (OTU)-based approach was selected for processing CCS reads. This choice was made due to the recognized advantages of OTU-based methods over amplicon sequence variant-based pipelines when analyzing gut microbiome data characterized by high inter-individual variability, such as that of wild birds25,80. Following primer removal and length/quality filtering (retaining CCS reads between 1.0 and 1.6 kb), the remaining high-quality reads were processed using the CLC Microbial Genomics Module (CLC Genomics Workbench v21, QIAGEN, Hilden, Germany). OTU clustering was performed using the module’s reference-based OTU clustering workflow with a 97% sequence identity threshold against the SILVA 132.99 16S rRNA database implemented in the module. Representative OTU sequences were taxonomically annotated by mapping to the reference database; where multiple equally good matches existed, the taxonomy was assigned to the lowest common ancestor.
Metagenomic sequencing and bioinformatics analysis
A subset of 25 fecal samples was subjected to metagenome sequencing by Shanghai BIOZERON Co., Ltd. using an Illumina NovaSeq platform, and 2 × 150 bp paired reads were generated. Raw reads from metagenome sequencing were filtered, trimmed, and quality-controlled to generate clean reads using Sickle (ver. 1.33) with parameters pe, -t sanger, and -l 8081. In total, 339.8 G bp of 150 bp high-quality reads were generated with an average sequencing depth of 13.6 G bp per sample. The mitochondrial genomes of two related bird species, including Anser albifrons frontalis (NC_039888.1) and A. albifrons (AF363031.1), were downloaded from the NCBI database; then, the mitochondria-originated reads were identified from the metagenomic reads of each sample by aligning the reads to the mitochondrial genomes of the aforementioned species using bowtie282 (ver. 2.2.5) with parameters --no-unal and -t. The mapped reads in each sample were assembled into mitochondrial sequences using mitoZ with parameters all, --clade Chordata, --genetic_code 2, --assembler mitoassemble, and --requiring_taxa Chordata83, and the phylogenetic relationships between each sample and relevant reference genomes were determined using pyani v0.2.12 with default settings84.
Clean reads, after removing mitochondrial sequences, were co-assembled using MEGAHIT (ver. 1.2.9) with the “meta-large” parameter85. Taxonomic annotations were assigned to the assembled contigs using MMseqs2 easy-taxonomy module with uniref90 database as ref. 86, and the plant- and aves-derived contigs were identified and then discarded using SeqKit (ver. 0.15.0) accordingly87. Then, the contigs originating from the bird host and plants (i.e., undigested contents in the gut) were identified and removed, generating an assembly containing 1,593,935 contigs with a total length of 3.56 G bp. Subsequently, the remaining contigs were used for gene prediction using Prodigal (ver. 2.6.3) with parameters -p meta and -q88. The predicted genes were clustered using CD-HIT-est (ver. 4.8.1)89 with an identity cutoff of 0.95 (-c 0.95), and the taxonomic annotations were assigned to these genes using MMseqs2 easy-taxonomy module as mentioned above. The genes belonging to eukaryotes (except fungi) were removed using Seqkit (ver. 0.15.0)87, and the final nonredundant gene set was generated. The reads from each sample were aligned to nonredundant genes using bowtie2 (ver. 2.2.5) with parameters --no-unal and -t82, and read count tables were generated using Samtools (ver. 1.11) idxstats command90. Rarefaction curve of detected genes was generated based on a 100-fold permuted sampling order, center value represents the median of detected genes.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) annotations were derived based on the eggNOG database by eggnog-mapper (ver. 5.0.2) with parameters --itype proteins and -m diamond with the implemented eggNOG orthology database (v5.0.2) as ref. 91. The ARGs were annotated based on the Comprehensive Antibiotic Resistance Database (CARD) using Resistance Gene Identifier (RGI, ver. 5.1.0)92 main module with parameters --include_loose, --local, --clean, and sequence identity and coverage thresholds 80%. We identified gene-encoding VFs by aligning with the VF Database (VFDB) using blastn (v2.5.0+) with parameters -evalue 1e-5 and sequence identity and coverage thresholds 60%93. Only the best hits were retained for aligned VFs. The bacteriophage- and plasmid-derived ARG-carrying contigs were identified using MobileOG-db software (v1.6) with parameters -k 15, -e 1e-5, -p 80, and -q 8094.
Metagenomic binning
We constructed metagenomic bins (or MAGs) using metaWRAP (ver. 1.3.2)95 Binning module with parameters --metabat2, --maxbin2, and --concoct. We performed the refinement of MAGs using the bin-refinement module in metaWRAP with default settings. The completeness and contamination of the final MAGs were estimated using CheckM (ver. 1.1.3) with default parameters96. Representative MAGs (>70% completeness and <10% potential contamination) were classified using the Genome Taxonomy Database Toolkit (GTDB-tk) classify_wf module with default parameters37. The selected bins were manually curated using medusa97 and gapfiller98 as described by Zhang et al.99. The abundance of each MAG in each sample was determined using CoverM with parameters --min-read-percent-identity 95 and -m trimmed_mean100. MAGs were annotated using PROKKA101 with default parameters, and VFs and ARGs were identified using the previously described methods. Genomes closely related to the MAGs were retrieved from the NCBI database using the datasets software102 based on the taxonomic identifiers associated with each MAG. Genome-wide average nucleotide identity (ANI) between the MAGs and the downloaded genomes was calculated using FastANI103. The top-hit genomes based on ANI values were selected for subsequent phylogenetic analysis. All downloaded genomes were uniformly annotated using PROKKA with the same parameters applied to the MAGs. The phylogenetic tree (Neighbor-Joining tree) was constructed using CVtree104, and the Prokka-generated faa files were used as inputs in the analysis. The phylogenetic tree was rooted using the midpoint method and visualized using iTOL (https://itol.embl.de/)105.
Statistical analysis
The 70 samples were clustered using the ward-linkage algorithm in Statistical Analysis of Metagenomic Profiles (STAMP) software (v2.1.3)106, based on genus-level read count profiles derived from full-length 16S rDNA amplicon analysis. Differentially abundant genera were identified using the DESeq2107 method applied to the read count matrix. Prior to analysis, genera detected in fewer than 10% of samples (i.e., 7 samples for 16S rDNA data and 2 samples for metagenomic data) were excluded. The filtered count matrix was then analyzed using DESeq2 with a significance threshold of |log2(fold change)|>1 and an FDR < 0.05. For inter-group comparisons at the phylum level, the relative abundance-based analyses were performed using a two-tailed Mann–Whitney U-test (P < 0.05). Principal Co-ordinates analysis plot was generated based on the Bray–Curtis distance metrics calculated using the genus-level read count matrix derived from 16S rDNA and metagenome data as inputs, and permutational multivariate analysis of variance (PERMANOVA) was performed to analyze the effect of stratification on the microbial composition via the adonis2 function in the vegan package108. The differentially abundant KOs were identified using the DESeq2107 method with the following thresholds: |log2(fold change)|>1, FDR < 0.01, and average relative intragroup abundance >1 TPM (transcripts per million). Pathway enrichment among the differentially abundant KOs was evaluated using a two-tailed Fisher’s exact test (P < 0.05). Comparisons of taxonomic origins of selected KOs between E1 and E2 were performed using a two-tailed Mann–Whitney U-test (P < 0.05).
MENs were constructed based on the random matrix theory (RMT) model using the iNAP pipeline109. The correlation matrices were constructed based on RMT with Spearman correlations of the non-log-transformed genera read count matrix, and only genera present in >50% of the samples were retained in the matrixes. We applied a series of cutoff thresholds from 0.01 to 1.0 with 0.01 intervals to the matrix and used an appropriate threshold value of 0.94 to construct the networks based on the RMT χ² result. For each generated empirical network, the corresponding random network was generated by rewiring all nodes and links 100 times using the greedy modularity optimization method implemented in the iNAP. We compared the network topological properties, including the average clustering coefficient (avgCC), average path distance (GD), geodesic efficiency (E), density (D), connectedness (Con), and modularity, between MENs using Student’s t-test as suggested by the iNAP pipeline with the property values from the empirical networks and standard deviation values from the random network as inputs.
Data availability
The full-length 16S rDNA sequencing and metagenomic data have been deposited in the China National GenBank database (CNGBdb) under project ID CNP0003865. The 91 MAGs and metagenes have been deposited in Figshare (https://figshare.com/account/home#/projects/185836).
References
Peixoto, R. S., Harkins, D. M. & Nelson, K. E. Advances in microbiome research for animal health. Annu. Rev. Anim. Biosci. 9, 289–311 (2021).
Raymond, F. et al. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J. 10, 707–720 (2016).
Lee, K. et al. Population-level impacts of antibiotic usage on the human gut microbiome. Nat. Commun. 14, 1191 (2023).
Tu, V. et al. A quantitative approach to measure and predict microbiome response to antibiotics. MSphere 9, e00488–00424 (2024).
Alessandri, G., Rizzo, S. M., Ossiprandi, M. C., van Sinderen, D. & Ventura, M. Creating an atlas to visualize the biodiversity of the mammalian gut microbiota. Curr. Opin. Biotechnol. 73, 28–33 (2022).
Matheen, M. I. A., Gillings, M. R. & Dudaniec, R. Y. Dominant factors shaping the gut microbiota of wild birds. Emu-Austral Ornithol. 122, 255–268 (2022).
Costea, P. I. et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 3, 8–16 (2018).
Wang, J. et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc. Nat. Acad. Sci. USA 111, E2703–E2710 (2014).
Huang, G. et al. PandaGUT provides new insights into bacterial diversity, function, and resistome landscapes with implications for conservation. Microbiome 11, 221 (2023).
Yuan, Z. et al. Enterotype identification and its influence on regulating the duodenum metabolism in chickens. Poult. Sci. 99, 1515–1527 (2020).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Maritan, E., Quagliariello, A., Frago, E., Patarnello, T. & Martino, M. E. The role of animal hosts in shaping gut microbiome variation. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 379, 20230071 (2024).
Fan, Y., Ju, T., Bhardwaj, T., Korver, D. R. & Willing, B. P. Week-old chicks with high Bacteroides abundance have increased short-chain fatty acids and reduced markers of gut inflammation. Microbiol. Spectr. 11, e0361622 (2023).
Frioux, C. et al. Enterosignatures define common bacterial guilds in the human gut microbiome. Cell Host Microbe 31, 1111–1125.e1116 (2023).
Larsen, J. M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 151, 363–374 (2017).
Xiao, L. et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 1, 1–6 (2016).
Kim, M., Park, T. & Yu, Z. Metagenomic investigation of gastrointestinal microbiome in cattle. Asian-Australas. J. Anim. Sci. 30, 1515 (2017).
Feng, Y. et al. Metagenome-assembled genomes and gene catalog from the chicken gut microbiome aid in deciphering antibiotic resistomes. Commun. Biol. 4, 1305 (2021).
Chua, P. Y. S. & Rasmussen, J. A. Taking metagenomics under the wings. Nat. Rev. Microbiol. 20, 447–447 (2022).
Levin, D. et al. Diversity and functional landscapes in the microbiota of animals in the wild. Science 372, eabb5352 (2021).
Cao, J. et al. Metagenomic analysis reveals the microbiome and resistome in migratory birds. Microbiome 8, 1–18 (2020).
Miller, E. A., Ponder, J. B., Willette, M., Johnson, T. J. & VanderWaal, K. L. Merging metagenomics and spatial epidemiology to understand the distribution of antimicrobial resistance genes from Enterobacteriaceae in wild owls. Appl. Environ. Microbiol. 86, e00571–00520 (2020).
Lin, Y. et al. Migratory birds-one major source of environmental antibiotic resistance around Qinghai Lake, China. Sci. Total. Environ. 739, 139758 (2020).
Bodawatta, K. H., Hird, S. M., Grond, K., Poulsen, M. & Jønsson, K. A. Avian gut microbiomes taking flight. Trends Microbiol. 30, 268–280 (2022).
Sun, F., Chen, J., Liu, K., Tang, M. & Yang, Y. The avian gut microbiota: diversity, influencing factors, and future directions. Front. Microbiol. 13, 934272 (2022).
Rogacheva, E. V. & Frölich, D. The birds of central Siberia. (Husum-Druck-und Verlagsgesellschaft, 1992).
Li, C. et al. Population trends and migration routes of the East Asian Bean Goose Anser fabalis middendorffii and A. f. serrirostris. Wildfowl 6, 124–156 (2020).
Salewski, V. & Bruderer, B. The evolution of bird migration-a synthesis. Naturwissenschaften 94, 268–279 (2007).
Garcias-Bonet, N. et al. Horizon scanning the application of probiotics for wildlife. Trends Microbiol. 32, 252–269 (2024).
Ericsson, A. C. et al. The influence of caging, bedding, and diet on the composition of the microbiota in different regions of the mouse gut. Sci. Rep. 8, 4065 (2018).
Newman, M. E. Modularity and community structure in networks. Proc. Nat. Acad. Sci. USA 103, 8577–8582 (2006).
Cao, H., Baldini, R. L. & Rahme, L. G. Common mechanisms for pathogens of plants and animals. Annu Rev. Phytopathol. 39, 259–284 (2001).
Martin-Gallausiaux, C., Marinelli, L., Blottière, H. M., Larraufie, P. & Lapaque, N. SCFA: mechanisms and functional importance in the gut. Prog. Biophys. Mol. Biol. 80, 37–49 (2021).
Letchumanan, V. et al. Vibrio parahaemolyticus: The protagonist of foodborne diseases. Prog. Biophys. Mol. Biol. 2, a0000029 (2019).
Sánchez-Vargas, F. M., Abu-El-Haija, M. A. & Gómez-Duarte, O. G. Salmonella infections: an update on epidemiology, management, and prevention. Travel Med. Infect. Dis. 9, 263–277 (2011).
Lee, C.-R. et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 7, 55 (2017).
Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38, 5315–5316 (2022).
Zhang, S. et al. SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl. Environ. Microbiol. 85, e01746–01719 (2019).
Boyd, E. F. et al. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8, 110 (2008).
Yan, M. & Yu, Z. Viruses contribute to microbial diversification in the rumen ecosystem and are associated with certain animal production traits. Microbiome 12, 82 (2024).
Dong, Z. et al. Comparative analysis of the intestinal tract microbiota and feeding habits of five sympatric flycatchers. Avian Res. 13, 100050 (2022).
Wang, B. et al. Diet drives the gut microbiome composition and assembly processes in winter migratory birds in the Poyang Lake wetland, China. Front. Microbiol. 13, 973469 (2022).
Dearing, M. D., Foley, W. J. & McLean, S. The influence of plant secondary metabolites on the nutritional ecology of herbivorous terrestrial vertebrates. Annu. Rev. Ecol. Evol. Syst. 36, 169–189 (2005).
Trabelcy, B., Shteindel, N., Lalzar, M., Izhaki, I. & Gerchman, Y. Bacterial detoxification of plant defence secondary metabolites mediates the interaction between a shrub and frugivorous birds. Nat. Commun. 14, 1821 (2023).
Ormeño-Orrillo, E. & Martínez-Romero, E. A genomotaxonomy view of the Bradyrhizobium genus. Front. Microbiol. 10, 1334 (2019).
Schneijderberg, M. et al. A genetically and functionally diverse group of non-diazotrophic Bradyrhizobium spp. colonizes the root endophytic compartment of Arabidopsis thaliana. BMC Plant Biol. 18, 1–9 (2018).
Greetatorn, T. et al. Mechanisms of rice endophytic bradyrhizobial cell differentiation and its role in nitrogen fixation. Microbes Environ. 35, ME20049 (2020).
Cheynier, V., Comte, G., Davies, K. M., Lattanzio, V. & Martens, S. Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiol. Biochem. 72, 1–20 (2013).
Zhang, Y., Trivedi, P., Xu, J., Roper, M. C. & Wang, N. The citrus microbiome: from structure and function to microbiome engineering and beyond. Phytobiomes J. 5, 249–262 (2021).
Helene, L. C. F., Klepa, M. S., O’Hara, G. & Hungria, M. Bradyrhizobium archetypum sp. nov., Bradyrhizobium australiense sp. nov. and Bradyrhizobium murdochi sp. nov., isolated from nodules of legumes indigenous to Western Australia. Int. J. Syst. Evol. Micr. 70, 4623–4636 (2020).
Yao, R. et al. Fly-over phylogeny across invertebrate to vertebrate: the giant panda and insects share a highly similar gut microbiota. Comput. Struct. Biotechnol. J. 19, 4676–4683 (2021).
Zhang, N. et al. Quantification and diversity of cultivated bacteria in root endosphere and rhizosphere of bamboo species Fargesia nitida in association with the tree succession. Microbiol. Res. J. Int. 33, 1–16 (2023).
Fargione, J. E. & Tilman, D. Diversity decreases invasion via both sampling and complementarity effects. Ecol. Lett. 8, 604–611 (2005).
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A. & Afkhami, M. E. Environmental stress destabilizes microbial networks. ISME J. 15, 1722–1734 (2021).
Nakkarach, A. et al. Anti-cancer and anti-inflammatory effects elicited by short chain fatty acids produced by Escherichia coli isolated from healthy human gut microbiota. Microb. Cell Fact. 20, 1–17 (2021).
Magalhaes, R. et al. Oxidative status and intestinal health of gilthead sea bream (Sparus aurata) juveniles fed diets with different ARA/EPA/DHA ratios. Sci. Rep. 10, 13824 (2020).
Vital, M., Howe, A. C. & Tiedje, J. M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio 5, e00889 (2014).
Sun, X. et al. Dietary fiber ameliorates lipopolysaccharide-induced intestinal barrier function damage in piglets by modulation of intestinal microbiome. Msystems 6, https://doi.org/10.1128/msystems.01374-01320 (2021).
Sanz-García, F. et al. Coming from the wild: multidrug resistant opportunistic pathogens presenting a primary, not human-linked, environmental habitat. Int. J. Mol. Sci. 22, 8080 (2021).
Vouga, M. & Greub, G. Emerging bacterial pathogens: the past and beyond. Clin. Microbiol. Infect. 22, 12–21 (2016).
Sana, T. G., Lugo, K. A. & Monack, D. M. T6SS: the bacterial” fight club” in the host gut. PLoS Pathog. 13, e1006325 (2017).
Palmer, L. D. & Skaar, E. P. Transition metals and virulence in bacteria. Annu. Rev. Genet. 50, 67–91 (2016).
García-Bayona, L. & Comstock, L. E. Bacterial antagonism in host-associated microbial communities. Science 361, eaat2456 (2018).
Di Lorenzo, F., De Castro, C., Silipo, A. & Molinaro, A. Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol. Rev. 43, 257–272 (2019).
Ahmad, J. N. & Sebo, P. Bacterial RTX toxins and host immunity. Curr. Opin. Infect. Dis. 34, 187–196 (2021).
Peschel, A. How do bacteria resist human antimicrobial peptides?. Trends Microbiol 10, 179–186 (2002).
Li, E. et al. Rapid evolution of bacterial mutualism in the plant rhizosphere. Nat. Commun. 12, 3829 (2021).
Coulthurst, S. J. The Type VI secretion system–a widespread and versatile cell targeting system. Res. Microbiol. 164, 640–654 (2013).
Bertani, B. & Ruiz, N. Function and biogenesis of lipopolysaccharides. Ecosal Plus 8, https://doi.org/10.1128/ecosalplus.ESP-0001-2018 (2018).
Tamames, J., Cobo-Simón, M. & Puente-Sánchez, F. Assessing the performance of different approaches for functional and taxonomic annotation of metagenomes. BMC Genom. 20, 960 (2019).
Lu, J., Breitwieser, F. P., Thielen, P. & Salzberg, S. L. Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3, e104 (2017).
Eveillard, M., Kempf, M., Belmonte, O., Pailhories, H. & Joly-Guillou, M.-L. Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int. J. Infect. Dis. 17, e802–e805 (2013).
Li, L. et al. Genetic context of blaCTX–M–55 and qnrS1 genes in a foodborne Salmonella enterica serotype Saintpaul isolate from China. Front. Microbiol. 13, 899062 (2022).
Wigley, P. Salmonella and salmonellosis in wild birds. Animals 14, 3533 (2024).
Abd El-Ghany, W. A. Pseudomonas aeruginosa infection of avian origin: zoonosis and one health implications. Vet. World 14, 2155 (2021).
Si, Y. et al. Spring migration patterns, habitat use, and stopover site protection status for two declining waterfowl species wintering in China as revealed by satellite tracking. Ecol. Evol. 8, 6280–6289 (2018).
Chen, P., Yu, K. & He, Y. The dynamics and transmission of antibiotic resistance associated with plant microbiomes. Environ. Int. 176, 107986 (2023).
Wang, J. et al. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 38, 55 (2017).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1–e1 (2013).
Boukerb, A. M. et al. Comparative analysis of fecal microbiomes from wild waterbirds to poultry, cattle, pigs, and wastewater treatment plants for a microbial source tracking approach. Front. Microbiol. 12, 697553 (2021).
Joshi, N. & Fass, J. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33)[Software] (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Meng, G., Li, Y., Yang, C. & Liu, S. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47, e63–e63 (2019).
Pritchard, L., Glover, R. H., Humphris, S., Elphinstone, J. G. & Toth, I. K. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8, 12–24 (2016).
Li, D. et al. MEGAHIT v1. 0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11 (2016).
Mirdita, M., Steinegger, M., Breitwieser, F., Söding, J. & Levy Karin, E. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics 37, 3029–3031 (2021).
Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PloS ONE 11, e0163962 (2016).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 1–11 (2010).
Huang, Y., Niu, B., Gao, Y., Fu, L. & Li, W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829 (2021).
Alcock, B. P. et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525 (2020).
Liu, B., Zheng, D., Zhou, S., Chen, L. & Yang, J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917 (2022).
Brown, C. L. et al. mobileOG-db: a manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl. Environ. Microbiol. 88, e00991–00922 (2022).
Uritskiy, G. V., DiRuggiero, J. & Taylor, J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 1–13 (2018).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Bosi, E. et al. MeDuSa: a multi-draft based scaffolder. Bioinformatics 31, 2443–2451 (2015).
Nadalin, F., Vezzi, F. & Policriti, A. GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinform. 13, 1–16 (2012).
Zhang, Y., Xu, J., Wang, E. & Wang, N. Mechanisms underlying the rhizosphere-to-rhizoplane enrichment of cellvibrio unveiled by genome-centric metagenomics and metatranscriptomics. Microorganisms 8, 583 (2020).
Aroney, S. T. et al. CoverM: Read alignment statistics for metagenomics. Bioinformatics 41, btaf147 (2025).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
O’Leary, N. A. et al. Exploring and retrieving sequence and metadata for species across the tree of life with NCBI Datasets. Sci. Data 11, 732 (2024).
Hernández-Salmerón, J. E. & Moreno-Hagelsieb, G. FastANI, Mash and Dashing equally differentiate between Klebsiella species. PeerJ 10, e13784 (2022).
Qi, J., Luo, H. & Hao, B. CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 32, W45–W47 (2004).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82 (2024).
Parks, D. H., Tyson, G. W., Hugenholtz, P. & Beiko, R. G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Hu, Y.-J. & Satten, G. A. Testing hypotheses about the microbiome using the linear decomposition model (LDM). Bioinformatics 36, 4106–4115 (2020).
Feng, K. et al. iNAP: an integrated network analysis pipeline for microbiome studies. iMeta 1, e13 (2022).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2024YFC2310300, 2022YFC2604200) and the Yangzhou University Interdisciplinary Research Foundation for Veterinary Medicine Discipline of Targeted Support (grant number yzuxk202003).
Author information
Authors and Affiliations
Contributions
Y.Z.Z. conceived and designed the experiments, and reviewed and edited the paper. Y.W. analyzed the data and wrote the original draft. J.X. analyzed part of the data. E.T.W. reviewed and edited the paper. Y.W., Y.Z.Z., L.S., J.W., and Y.Q.Z. collected the samples. H.C. and W.H.C. monitored the migratory bird population and provided guidelines for sample collection. Y.W., G.X.L., S.L.L., and M.C.H. performed part of the experiments. X.A.J. and J.L.H. contributed to funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Y., Xu, J., Liang, G. et al. Gut microbiome profiling of a migratory Anser serrirostris population reveals two groups with distinct pathogen and ARG contents. npj Biofilms Microbiomes 12, 22 (2026). https://doi.org/10.1038/s41522-025-00888-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-025-00888-3