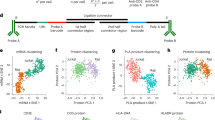
Abstract
During the process of engulfment, phosphatidylserine is exposed on the surface of dead cells as an ‘eat-me’ signal and is recognized by Protein S (ProS), a secreted factor that also binds to the Mer tyrosine kinase (MerTK) on phagocytes. Despite its robust activity, this engulfment mechanism has not been exploited for therapeutic purposes. Here we develop a synthetic protein modality called Crunch (connector for removal of unwanted cell habitat) by modifying ProS, inspired by the high engulfment capability of the ProS–MerTK pathway. In Crunch, the phosphatidylserine-binding motif of ProS is replaced with a nanobody or single-chain variable fragment that recognizes the surface proteins of targeted cells. Green fluorescent protein nanobody-conjugated Crunch eliminates green fluorescent protein-expressing melanoma cells in transplantation mouse models. In addition, CD19+B cells are eliminated by anti-CD19 single-chain variable fragment-conjugated Crunch, resulting in a therapeutic effect on systemic lupus erythematosus. Both mouse and human versions of Crunch are effective, establishing this synthetic ligand as a promising tool for the elimination of targeted cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Human acral melanoma datasets for single-cell analysis were obtained from the GEO database under accession number GSE115978. Source data are provided with this paper.
References
Soussi, T. & Wiman, K. G. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell 12, 303–312 (2007).
Rahman, A. & Isenberg, D. A. Systemic lupus erythematosus. N. Engl. J. Med. 358, 929–939 (2008).
Smolen, J. S. et al. Rheumatoid arthritis. Nat. Rev. Dis. Prim. 4, 1–23 (2018).
Huston, J. S. et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl Acad. Sci. USA 85, 5879–5883 (1988).
Brudno, J. N. & Kochenderfer, J. N. Current understanding and management of CAR T cell-associated toxicities. Nat. Rev. Clin. Oncol. 21, 501–521 (2024).
Baker, D. J., Arany, Z., Baur, J. A., Epstein, J. A. & June, C. H. CAR T therapy beyond cancer: the evolution of a living drug. Nature 619, 707–715 (2023).
Leidi, M. et al. M2 macrophages phagocytose rituximab-opsonized leukemic targets more efficiently than M1 cells in vitro. J. Immunol. 182, 4415–4422 (2009).
Shi, Y. et al. Trastuzumab triggers phagocytic killing of high HER2 cancer cells in vitro and in vivo by interaction with Fcγ receptors on macrophages. J. Immunol. 194, 4379–4386 (2015).
Iwai, Y. et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl Acad. Sci. USA 99, 12293–12297 (2002).
Bader, J. E. et al. Obesity induces PD-1 on macrophages to suppress anti-tumour immunity. Nature 630, 968–975 (2024).
Ishida, Y., Agata, Y., Shibahara, K. & Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 11, 3887–3895 (1992).
Löffler, A. et al. A recombinant bispecific single-chain antibody, CD19 × CD3, induces rapid and high lymphoma-directed cytotoxicity by unstimulated T lymphocytes. Blood 95, 2098–2103 (2000).
Dreier, T. et al. Extremely potent, rapid and costimulation-independent cytotoxic T-cell response against lymphoma cells catalyzed by a single-chain bispecific antibody. Int. J. Cancer 100, 690–697 (2002).
Hosseini, S. S. et al. Bispecific monoclonal antibodies for targeted immunotherapy of solid tumors: recent advances and clinical trials. Int. J. Biol. Macromol. 167, 1030–1047 (2021).
Kris, M. G. et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 311, 1998–2006 (2014).
Thress, K. S. et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nat. Med. 21, 560–562 (2015).
Arandjelovic, S. & Ravichandran, K. S. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 16, 907–917 (2015).
Nagata, S. & Tanaka, M. Programmed cell death and the immune system. Nat. Rev. Immunol. 17, 333–340 (2017).
Suzuki, J., Denning, D. P., Imanishi, E., Horvitz, H. R. & Nagata, S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406 (2013).
Nyberg, P., He, X., Härdig, Y., Dahlbäck, B. & García De Frutos, P. Stimulation of Sky tyrosine phosphorylation by bovine Protein S. Eur. J. Biochem. 246, 147–154 (1997).
Lai, C. & Lemke, G. An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron 6, 691–704 (1991).
Graham, D. K., Dawson, T. L., Mullaney, D. L., Snodgrass, H. R. & Earp, H. S. Cloning and mRNA expression analysis of a novel human protooncogene, c-mer. Cell Growth Differ. 5, 647–657 (1994).
Prasad, D. et al. TAM receptor function in the retinal pigment epithelium. Mol. Cell Neurosci. 33, 96–108 (2006).
Saerens, D. et al. Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies. J. Mol. Biol. 352, 597–607 (2005).
Kubala, M. H., Kovtun, O., Alexandrov, K. & Collins, B. M. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 19, 2389–2401 (2010).
Suzuki, J., Umeda, M., Sims, P. J. & Nagata, S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 (2010).
Maruoka, M. et al. Caspase cleavage releases a nuclear protein fragment that stimulates phospholipid scrambling at the plasma membrane. Mol. Cell 81, 1397–1410.e9 (2021).
Attallah, C., Etcheverrigaray, M., Kratje, R. & Oggero, M. A highly efficient modified human serum albumin signal peptide to secrete proteins in cells derived from different mammalian species. Protein Expr. Purif. 132, 27–33 (2017).
Massagué, J. Epidermal growth factor-like transforming growth factor. II. Interaction with epidermal growth factor receptors in human placenta membranes and A431 cells. J. Biol. Chem. 258, 13614–13620 (1983).
Revest, J. M., DeMoerlooze, L. & Dickson, C. Fibroblast growth factor 9 secretion is mediated by a non-cleaved amino-terminal signal sequence. J. Biol. Chem. 275, 8083–8090 (2000).
Sasada, R., Marumoto, R. & Igarashi, K. Secretion of human EGF and IgE in mammalian cells by recombinant DNA techniques; use of a IL-2 leader sequence. Cell Struct. Funct. 13, 129–141 (1988).
Kamiyama, D. et al. Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7, 11046 (2016).
Ling, L., Templeton, D. & Kung, H.-J. Identification of the major autophosphorylation sites of Nyk/Mer, an NCAM-related receptor tyrosine kinase*. J. Biol. Chem. 271, 18355–18362 (1996).
Nishi, C., Yanagihashi, Y., Segawa, K. & Nagata, S. MERTK tyrosine kinase receptor together with TIM4 phosphatidylserine receptor mediates distinct signal transduction pathways for efferocytosis and cell proliferation. J. Biol. Chem. 294, 7221–7230 (2019).
Carbone, F. R., Sterry, S. J., Butler, J., Rodda, S. & Moore, M. W. T cell receptor alpha-chain pairing determines the specificity of residue 262 within the Kb-restricted, ovalbumin257-264 determinant. Int Immunol. 4, 861–867 (1992).
Price, P. A., Fraser, J. D. & Metz-Virca, G. Molecular cloning of matrix Gla protein: implications for substrate recognition by the vitamin K-dependent gamma-carboxylase. Proc. Natl Acad. Sci. USA 84, 8335–8339 (1987).
van der Meer, J. H. M., van der Poll, T. & van ‘t Veer, C. TAM receptors, Gas6, and Protein S: roles in inflammation and hemostasis. Blood 123, 2460–2469 (2014).
Reynisson, B., Alvarez, B., Paul, S., Peters, B. & Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 48, W449–W454 (2020).
Jespersen, M. C., Peters, B., Nielsen, M. & Marcatili, P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 45, W24–W29 (2017).
Naoi, Y. et al. CD106 in tumor-specific exhausted CD8+ T cells mediates immunosuppression by inhibiting TCR signaling. Cancer Res. 84, 2109–2122 (2024).
Beck, J. D. et al. Long-lasting mRNA-encoded interleukin-2 restores CD8+ T cell neoantigen immunity in MHC class I-deficient cancers. Cancer Cell 42, 568–582.e11 (2024).
Jerby-Arnon, L. et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997.e24 (2018).
Kochenderfer, J. N., Yu, Z., Frasheri, D., Restifo, N. P. & Rosenberg, S. A. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood 116, 3875–3886 (2010).
Mitchell, K. G. et al. High-volume hybridoma sequencing on the NeuroMabSeq platform enables efficient generation of recombinant monoclonal antibodies and scFvs for neuroscience research. Sci. Rep. 13, 16200 (2023).
Schrand, B. et al. Hapten-mediated recruitment of polyclonal antibodies to tumors engenders antitumor immunity. Nat. Commun. 9, 3348 (2018).
Carmi, Y. et al. Allogeneic IgG combined with dendritic cell stimuli induce antitumour T-cell immunity. Nature 521, 99–104 (2015).
Patel, D. et al. Generation and characterization of a therapeutic human antibody to melanoma antigen TYRP1. Hum. Antibodies 16, 127–136 (2007).
Xiong, W. & Lahita, R. G. Pragmatic approaches to therapy for systemic lupus erythematosus. Nat. Rev. Rheumatol. 10, 97–107 (2014).
Mackensen, A. et al. Anti-CD19 CAR T cell therapy for refractory systemic lupus erythematosus. Nat. Med. 28, 2124–2132 (2022).
Doglio, M. et al. Regulatory T cells expressing CD19-targeted chimeric antigen receptor restore homeostasis in systemic lupus erythematosus. Nat. Commun. 15, 2542 (2024).
Edman, N. I. et al. Modulation of FGF pathway signaling and vascular differentiation using designed oligomeric assemblies. Cell 187, 3726–3740.e43 (2024).
Krishna, R. et al. Generalized biomolecular modeling and design with RoseTTAFold All-Atom. Science https://doi.org/10.1126/science.adl2528 (2024).
Roy, A. et al. De novo design of highly selective miniprotein inhibitors of integrins αvβ6 and αvβ8. Nat. Commun. 14, 5660 (2023).
Kunert, R. & Reinhart, D. Advances in recombinant antibody manufacturing. Appl. Microbiol. Biotechnol. 100, 3451–3461 (2016).
Tihanyi, B. & Nyitray, L. Recent advances in CHO cell line development for recombinant protein production. Drug Discov. Today Technol. 38, 25–34 (2020).
Xu, W.-J., Lin, Y., Mi, C.-L., Pang, J.-Y. & Wang, T.-Y. Progress in fed-batch culture for recombinant protein production in CHO cells. Appl. Microbiol. Biotechnol. 107, 1063–1075 (2023).
Thim, L. et al. Secretion and processing of insulin precursors in yeast. Proc. Natl Acad. Sci. USA 83, 6766–6770 (1986).
Mukherjee, J. et al. Prolonged prophylactic protection from botulism with a single adenovirus treatment promoting serum expression of a VHH-based antitoxin protein. PLoS ONE 9, e106422 (2014).
Mandrup, O. A. et al. Programmable half-life and anti-tumour effects of bispecific T-cell engager-albumin fusions with tuned FcRn affinity. Commun. Biol. 4, 1–11 (2021).
Pyzik, M. et al. The neonatal Fc receptor (FcRn): a misnomer? Front Immunol. 10, 1540 (2019).
Borgel, D. et al. Implication of protein S thrombin-sensitive region with membrane binding via conformational changes in the gamma-carboxyglutamic acid-rich domain. Biochem. J. 360, 499–506 (2001).
Dahlbäck, B. & Villoutreix, B. O. Regulation of blood coagulation by the Protein C anticoagulant pathway. Arter. Thromb. Vasc. Biol. 25, 1311–1320 (2005).
Griffin, J. H., Zlokovic, B. V. & Mosnier, L. O. Protein C anticoagulant and cytoprotective pathways. Int J. Hematol. 95, 333–345 (2012).
de Córdoba, S. R. et al. The gene coding for the β-chain of C4b-binding protein (C4BPB) has become a pseudogene in the mouse. Genomics 21, 501–509 (1994).
Hillarp, A., Thern, A. & Dahlbäck, B. Bovine C4b binding protein. Molecular cloning of the alpha- and beta-chains provides structural background for lack of complex formation with protein S. J. Immunol. 153, 4190–4199 (1994).
DeRyckere, D., Huelse, J. M., Earp, H. S. & Graham, D. K. TAM family kinases as therapeutic targets at the interface of cancer and immunity. Nat. Rev. Clin. Oncol. 20, 755–779 (2023).
Genetic dissection of TAM receptor–ligand interaction in retinal pigment epithelial cell phagocytosis. Neuron 76, 1123–1132 (2012).
Sandahl, M., Hunter, D. B., Strunk, K. E., Earp, H. S. & Cook, R. S. Epithelial cell-directed efferocytosis in the post-partum mammary gland is necessary for tissue homeostasis and future lactation. BMC Dev. Biol. 10, 122 (2010).
Lu, Q., Gore, M. & Zhang, Q. et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398, 723–728 (1999).
Wu, J. et al. Disturbed flow impairs MerTK-mediated efferocytosis in aortic endothelial cells during atherosclerosis. Theranostics 14, 2427–2441 (2024).
Albelda, S. M. CAR T cell therapy for patients with solid tumours: key lessons to learn and unlearn. Nat. Rev. Clin. Oncol. 21, 47–66 (2024).
Jung, H. et al. Anti-inflammatory clearance of amyloid-β by a chimeric Gas6 fusion protein. Nat. Med. 28, 1802–1812 (2022).
Morioka, S. et al. Chimeric efferocytic receptors improve apoptotic cell clearance and alleviate inflammation. Cell 185, 4887–4903.e17 (2022).
Liu, X. et al. CD47 blockade triggers T cell–mediated destruction of immunogenic tumors. Nat. Med. 21, 1209–1215 (2015).
Vonderheide, R. H. CD47 blockade as another immune checkpoint therapy for cancer. Nat. Med. 21, 1122–1123 (2015).
Niesen, F. H., Berglund, H. & Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 (2007).
Foss, S. et al. Human IgG Fc-engineering for enhanced plasma half-life, mucosal distribution and killing of cancer cells and bacteria. Nat. Commun. 15, 2007 (2024).
Tarhan, L. et al. Single Cell Portal: an interactive home for single-cell genomics data. Preprint at bioRxiv https://doi.org/10.1101/2023.07.13.548886 (2023).
Muraoka, M. et al. IK cytokine ameliorates the progression of lupus nephritis in MRL/lpr mice. Arthritis Rheumatism 54, 3591–3600 (2006).
Acknowledgements
We thank A. Fujimoto for administrative assistance and A. Wee for proof-reading. This work was supported by JST-CREST (grant no. 1199566), Kusunoki125, Sumitomo Kyoto University Innovation Promotion System (SKIPS) to J. Suzuki. Computation time was provided by the Supercomputer System, Institute for Chemical Research, Kyoto University.
Author information
Authors and Affiliations
Contributions
Y.Y. and J.S. designed the overall research and interpreted experimental results. Y.Y. conducted all experiments. Y.Y. and J.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
J.S. and Y.Y. are inventors on a patent application (2024-191258) of a protein modality to eliminate unwanted cells. This work was supported in part by the Collaborative Research Grant from Sumitomo Pharma Co Ltd.
Peer review
Peer review information
Nature Biomedical Engineering thanks Jeanette Leusen, Lun Tsou and Thomas Valerius and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Production and characterization of Crunch.
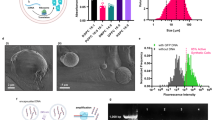
a–b, Generation of high Crunch-secreting CHO cells. CHO cells transduced with Crunch-IRES-RFP lentivirus were RFP-sorted twice (S2), reinfected, and sorted again to yield S3. Cells were seeded, switched to serum-free medium after 16 hr, and supernatant was collected at 48 hr. Crunch expression was analyzed by by immunoblotting with anti-FLAG antibody (a), and binding to GFPm⁺ BDKO cells was assessed by flow cytometry (b). c, Production kinetics. S3 cells were cultured as above, and daily supernatants were analyzed by ELISA (n = 3, independent biological samples). d–e, Crunch purification. Supernatant (540 ml, Day 6) was filtered, pH-adjusted (25 mM HEPES, pH 7.5), and incubated with Ni-NTA beads overnight. Input, flow-through, wash, and elution were assessed by immunoblotting with anti-FLAG (d). Eluates were buffer-exchanged to PBS and analyzed with BSA standards by CBB staining (e). Final concentration: 3.7 mg/ml; yield: ~9.61 mg/L. f, Glycosylation analysis. Crunch was treated with PNGaseF and analyzed by by immunoblotting with anti-FLAG. g, Binding affinity. ELISA using GFP-coated plates compared GFPNb-Crunch and GFP nanobody. Absorbance (450 nm) was used to calculate KD via nonlinear regression (R). h–i, Thermal stability. DSF analysis of 1D3 antibody and Crunch (mean of triplicates shown in h); aggregation onset temperatures (Tm) shown in i (n = 3, triplicate using the same sample). j, Vitamin K effect. GFPNb-Crunch produced with or without vitamin K was PNGaseF-treated and analyzed by by immunoblotting with anti-FLAG. All graphs show mean ± S.D.
Extended Data Fig. 2 MerTK dimerization and phosphorylation.
a–b, Split-GFP assay for MerTK-sfGFP. NIH3T3 cells expressing MerTK-sfGFP and tagRFP were co-cultured with BDKO or GFPm⁺ BDKO cells in conditioned medium containing mock or GFPNb-Crunch (from HEK293T) at 37 °C for 2 hr. After removing target cells, RFP⁺ MerTK-sfGFP⁺ NIH3T3 cells were analyzed by flow cytometry. Dot plots show % GFP⁺ cells among RFP⁺ NIH3T3 cells (a); bar plot shows % GFP⁺ cells among MerTK-sfGFP⁺ NIH3T3 cells with BDKO (b) (n = 4, independent biological samples). GFPm⁺ BDKO data in Fig. 2e. Mean ± S.D.; Student’s t test; N.S.P > 0.05 c, MerTK phosphorylation by apoptotic thymocytes and ProS. Mouse thymocytes were treated with PBS (–) or FasL (+) for 3 hr at 37 °C, incubated with 10% FBS medium for ProS loading, washed, and co-cultured with NIH3T3 or MerTK⁺ NIH3T3 cells in serum-free medium. After centrifugation (300 × g, 2 min, 37 °C) and 15 min incubation, NIH3T3 cells were washed, lysed, and analyzed by immunoblotting with anti–phospho-MerTK and anti-MerTK antibodies. d, MerTK phosphorylation by PtdSer-exposed living cells and ProS. Ba/F3 or aXkr4⁺ Ba/F3 cells were incubated with 10% FBS medium, washed, and added to MerTK⁺ NIH3T3 cells. After centrifugation and 15 min incubation, MerTK⁺ cells were lysed and analyzed by immunoblotting as in (c). e, Time-dependent MerTK phosphorylation by GFPNb-Crunch. GFPm⁺ BDKO cells were added to MerTK⁺ NIH3T3 cells with (+) or without (-) 1 µg/ml GFPNb-Crunch in serum-free medium with 5 mg/ml BSA. After centrifugation and incubation (0–30 min, 37 °C), MerTK⁺ cells were lysed and analyzed by immunoblotting as in (c).
Extended Data Fig. 3 Cell survival assay.
a, Experimental design. Ba/F3 cells were engineered to express MerTK, aXkr4 (to expose PtdSer), or both. 2.5 × 105 cells were cultured in 500 µl IL-3(–) RPMI1640 with FBS. MerTK activation supports survival and proliferation. b–e, Cell survival by PtdSer and MerTK. Cell number and viability were measured by Trypan blue exclusion every 24 hr. Line graphs show mean cell number (b) and viability (c); bar plots show both parameters at 48 and 72 hr (d, e) (n = 3, independent biological samples). f–i, Cell survival by Crunch. MerTK⁺ Ba/F3 cells were cultured with (+) or without (-) 10 µg/ml GFPNb-Crunch in IL-3(–) medium. Cell number (f) and viability (g) were measured every 24 hr. Bar plots show both at 48 and 72 hr (h, i) (n = 3, independent biological samples). Data are mean ± S.D. Statistical analysis: one-way ANOVA with Tukey-Kramer t-test (d–e, g); Student’s t test (f–i). N.S., P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Extended Data Fig. 4 ROSA26GFPm-OVA mice.
a, Schematic of the ROSA26GFPm-OVA mouse model. The CMV enhancer, chicken β-actin promoter, GFPm-OVA fusion protein (linked by GGGGS) and rabbit β-globin polyadenylation signal (pA) were inserted into the Rosa26 locus of C57BL/6 mice. b, GFP expression in ROSA26GFPm-OVA mice. GFP expression was analyzed on thymocytes, splenocytes, bone marrow (BM), and blood cells from WT or ROSA26GFPm-OVA mice using an anti-GFP antibody by flow cytometry. c, Crunch binding to cells from ROSA26GFPm-OVA mice. Thymocytes, splenocytes, bone marrow, and blood cells from WT or ROSA26GFPm-OVA mice were incubated with or without 10 µg/ml GFPNb-Crunch, and binding was detected using an anti-FLAG antibody by flow cytometry.
Extended Data Fig. 5 Pharmacokinetics and immunogenicity of Crunch.
a, Effect of vitamin K on Crunch. Engulfment assay of MerTK+ NIH3T3 cells targeting thymocytes from ROSA26GFPm-OVA mice, in the presence of 5% FBS and 10 µg/ml GFPNb-Crunch produced with or without vitamin K (Extended Data Fig. 1j). b-d, Plasma half-life of Crunch. 150 µg of GFPNb-Crunch was administered intravenously (i.v.) or intraperitoneally (i.p.) to mice (b). Plasma was collected at various time points (10 min to 120 hr), and GFPNb-Crunch levels were quantified by ELISA (c). The plasma remaining after 12 hr was used to calculate the half-life (d) (n = 4 mice). e, Antibody epitope prediction of Crunch. Mouse Protein S (mProS) and GFPNb-Crunch antibody epitopes were predicted by BepiPred-2.0. The epitope score was shown as threshold 0.5. f-g, Anti-drug antibody (ADA) assay. Mice were intravenously injected with 150 µg of GFPNb-Crunch or GFP, and plasma was collected 24 days later. Antibodies against GFP, GFPNb-Crunch, GFPNb, and Gla domain-deleted mProS (f) were measured by ELISA (Crunch n = 4, GFP n = 3). h, Macrophage stimulation. Macrophages were stimulated with 10 µg/ml GFPNb-Crunch, mouse IgG2a, 10% FBS, or 10 ng/ml LPS. RNA expression of GAPDH, IFNγ, IL-6, CCL5, IL-10, and Arg1 was measured by RT-PCR (n = 3, independent biological samples). All data are shown as mean ± S.D. Statistical analysis in g used Student’s unpaired t test; h used one-way ANOVA with Tukey-Kramer t-test. *P < 0.05 indicates significance.
Extended Data Fig. 6 Tumor suppression and effector cells of Crunch in melanoma.
a, Strategy for Crunch treatment in tumor engraftment. GFPm+ B16.F10 melanoma cells (5 ×105) were injected subcutaneously into WT mice. Saline or 100 µg GFPNb-Crunch were administered on Day 1, 8, and 15. b-c, Tumor growth following Crunch treatment. Tumor volumes were measured every three days and calculated as V = πLW²/6 (V: volume, L: long diameter, W: short diameter). The graph shows the mean tumor volume (b) and individual tumor volumes (c) for each mouse (Saline n = 6, Crunch n = 4 mice). Significant differences were observed on Day 12 (P = 0.030), Day 15 (P = 0.024), and Day 18 (P = 0.021). Data are shown as mean ± S.D. Statistical analysis was performed using Student’s unpaired t test. *P < 0.05 d-e, t-SNE analysis of non-malignant cells in the tumor microenvironment (TME) of human melanoma. Single-cell data (4857 cells, GSE115978, non-malignant cells) were clustered into various cell types: B cells, T cells, cancer-associated fibroblasts (CAF), endothelial cells, macrophages, NK cells, CD4+ T cells, and CD8+ T cells (d). MerTK expression was assessed in each cell type (e). f-h, MerTK expression in the TME of B16.F10 xenograft tumors. B16.F10 cells (5 ×105) were injected into C57BL/6 mice, and primary tumors were extracted at Day 9. Single-cell suspensions were stained for MerTK, CD45, CD11b, Ly6C, Ly6G, and F4/80 antibodies, and analyzed by flow cytometry. Singlet cells population plot was shown with or without an anti-MerTK antibody (f). MerTK expression in each cell population is shown (g). CD45 + CD11b+ cells and CD45 + CD11b+ MerTK+ cells, were analyzed by Ly6C/F4/80 or Ly6C/Ly6G expression (h). Gating strategy is shown in Supplementary Fig. 10.
Extended Data Fig. 7 scFv Crunch.
a, Binding of scCD19–Crunch to splenocytes. Splenocytes from C57BL/6 mice were incubated with or without 10 µg/ml scCD19–Crunch. Binding was analyzed by flow cytometry using an anti-FLAG antibody and anti-CD19-APC antibody. b, Engulfment of splenocytes by macrophages. Splenocytes were co-cultured with macrophages and scCD19–Crunch (10 µg/ml) in D-MEM, and analyzed as in Figs. 3g and 5d. c, Thermal stability of scGFP-Crunch. The thermal stability of scGFP-Crunch was measured by DSF, as described in Extended Data Fig. 1h. d, Binding affinity of scGFP-Crunch. Binding affinity of scGFP-Crunch to GFP was measured by ELISA, as described in Extended Data Fig. 1g. e, Binding of scGFP-Crunch. GFPm+ BDKO cells were incubated with or without 10 µg/ml GFPNb-Crunch, scGFP-Crunch, or Gla del mProS, and binding was detected by flow cytometry using an anti-FLAG antibody. f, Engulfment assay by scGFP-Crunch. Thymocytes from ROSA26GFPm-OVA mice were co-cultured with MerTK+ NIH3T3 cells in the presence or absence of 10 µg/ml GFPNb-Crunch, scCD19–Crunch, or scGFP-Crunch. Engulfment was quantified as described in Fig. 3a. The bar plot shows the percentage of engulfment+ cells among MerTK+ NIH3T3 cells (n = 3 independent biological samples). g, Epitope prediction of scGFP-Crunch. The epitope score for mProS and scGFP-Crunch was determined as in Extended Data Fig. 5e. h, ADA assay of scGFP-Crunch. Plasma antibodies against GFP, GFPNb-Crunch, GFPNb, scGFP-Crunch, and Gla del mProS were measured 24 days after injection of GFPNb-Crunch, scGFP-Crunch, or GFP (GFPNb-Crunch n = 4, scGFP-Crunch n = 3, GFP n = 3 mice). Plasma of GFPNb-Crunch- and GFP-injected mice were from Extended Data Fig. 5g. Data are mean ± S.D. Statistical analysis for f and h was performed using one-way ANOVA with Tukey-Kramer t test. Significance: N.S. P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, *****P < 0.00001.
Extended Data Fig. 8 Comparison of scCD19–Crunch and anti-CD19 antibody.
a-c, Blocking of CD19 detection by anti-CD19 antibody (1D3). Representative data of Day 3 from Fig. 5j (a). Blood cells from 1D3-treated mice were incubated with anti-rat IgG-488 antibody without (b) or with (c) anti-CD45-PE, anti-CD19-APC, and anti-B220-PE-Cy7 antibodies. The histogram shows anti-rat IgG-488 (1D3 binding) signals among DAPI-negative lymphocytes (b). Dot plots show 1D3-bound cells among CD45+ cells (c). d, AAV9 construct. Crunch and tagRFP are driven by the EF1 core promoter. e-f, Infection of AAV9 into SH-SY5Y cells. SH-SY5Y neuroblastoma cells were incubated with different titers of AAV9, and the tagRFP+ cell population was analyzed by flow cytometry (e). Plots of tagRFP+ cells and viral titer (f). g-h, AAV9 infection in mice. 1 ×1011 vg AAV9 encoding tagRFP or scCD19–Crunch were intravenously injected into mice. Two weeks later, hepatocytes and blood cells were analyzed. The histogram shows the tagRFP+ hepatocyte population (g). Dot plots show CD19+ B220+ B cells among CD45+ cells, stained with anti-mCD19, anti-B220, and anti-CD45 antibodies (h). i-j, Crunch in plasma of AAV9-injected mice. Plasma was collected from AAV9-EF1-tagRFP or scCD19–Crunch-injected mice 24 weeks after injection (Fig. 5m). Plasma was tested for Crunch binding to CD19+ BDKO cells (i), and Crunch concentration in plasma was calculated by the binding assay (j) (RFP n = 3, scCD19–Crunch n = 4 mice). Data are shown as mean ± S.D. Statistical analysis was performed using Student’s unpaired t test.
Extended Data Fig. 9 Crunch therapy in an SLE mice model.
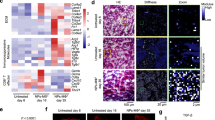
a, Strategy for Crunch therapy in MRLlpr/lpr SLE mice. MRLlpr/lpr mice were treated with saline, 150 µg scGFP-Crunch (scGFP), or scCD19–Crunch (scCD19) twice per week from 8 to 13 weeks of age. After therapy, spleen, blood, and kidneys were collected for B cell and renal analysis. MRL mice without treatment served as controls. b-d, Crunch therapy effects. The number of splenocytes (b), body weight (c), and spleen weight normalized by body weight (d) were assessed (MRL n = 1, Saline n = 7, scGFP n = 3, scCD19 n = 9 mice). e-f, Renal histology. Histopathologic scores of glomerular lesions were graded on a scale from 0-3 (e), based on H&E staining in Fig. 6k (Saline n = 7, scGFP n = 3, scCD19 n = 9 mice). IgG deposition levels in glomerular lesions were measured by Alexa647 MFI (f), as shown in Fig. 6l (Saline n = 4, scGFP, n = 3, scCD19 n = 6 mice). Saline (e P = 0.00000081, f P = 0.0019), scGFP (e P = 0.00025, f P = 0.0041), compared with scCD19. Data are mean ± S.D. Statistical analyses were performed using one-way ANOVA with Tukey-Kramer t test. Significance is indicated by N.S. P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary information
Supplementary Information
Supplementary Figs. 1–18 and Tables 1 and 2.
Supplementary Data
Statistical source data.
Source data
Source Data Fig. 1
Unprocessed western blots and gels data and statistical source data.
Source Data Fig. 2
Unprocessed western blots data and statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western Blots data and statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Table. 1
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yamato, Y., Suzuki, J. Phagocytic clearance of targeted cells with a synthetic ligand. Nat. Biomed. Eng (2025). https://doi.org/10.1038/s41551-025-01483-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41551-025-01483-9