Abstract
Biomolecular condensates, such as the nucleolus, stress granules/processing bodies and germ granules, are multiphase assemblages whose formation mechanisms and significance remain poorly understood. Here we identify protein constituents of the spatiotemporally ordered P, Z and M multiphase condensates in Caenorhabditis elegans germ granules using optimized TurboID-mediated proximity biotin labelling. These include 462, 41 and 86 proteins localizing to P, Z and M condensates, respectively, of which 522 were previously unknown protein constituents. Each condensate’s proteins are enriched for distinct classes of structured and intrinsically disordered domains, suggesting divergent functions and assembly mechanisms. Through a functional screen, we identify a germ granule protein, HERD-1, which prevents the mixing of P, Z and M condensates. Mixing in herd-1 mutants correlates with disorganization of germline small RNA pathways and prolonged epigenetic inheritance of RNA interference-induced gene silencing. Forced mixing of these condensate components using a nanobody with specific binding activity against green fluorescent protein also extends epigenetic inheritance. We propose that active maintenance of germ granule immiscibility helps to organize and regulate small RNA-driven transgenerational epigenetic inheritance in C. elegans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with dataset identifiers PXD046498 and PXD051885. RNA sequencing datasets have been deposited in the NCBI Sequence Read Archive under the BioProject accession number PRJNA1111473. Source data are provided with this paper. All other images supporting the findings of this study are available from figshare (https://doi.org/10.6084/m9.figshare.25955038)71.
Code availability
Custom scripts are available from GitHub (https://github.com/WANLAB2019/Multiphase-condensate-immiscibility-and-TEI).
References
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Hyman, A. A., Weber, C. A. & Julicher, F. Liquid–liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018).
Putnam, A., Cassani, M., Smith, J. & Seydoux, G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 26, 220–226 (2019).
Jain, S. et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Wan, G. et al. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679–683 (2018).
Du, Z. Z. et al. Condensate cooperativity underlies transgenerational gene silencing. Cell Rep. 42, 112859 (2023).
Sanders, D. W. et al. Competing protein–RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28 (2020).
Pitt, J. N., Schisa, J. A. & Priess, J. R. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. 219, 315–333 (2000).
Phillips, C. M., Montgomery, T. A., Breen, P. C. & Ruvkun, G. MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444 (2012).
Manage, K. I. et al. A tudor domain protein, SIMR-1, promotes siRNA production at piRNA-targeted mRNAs in C. elegans. eLife 9, e56731 (2020).
Price, I. F., Wagner, J. A., Pastore, B., Hertz, H. L. & Tang, W. C. elegans germ granules sculpt both germline and somatic RNAome. Nat. Commun. 14, 5965 (2023).
Uebel, C. J., Rajeev, S. & Phillips, C. M. Caenorhabditis elegans germ granules are present in distinct configurations and assemble in a hierarchical manner. Development 150, dev202284 (2023).
Phillips, C. M. & Updike, D. L. Germ granules and gene regulation in the Caenorhabditis elegans germline. Genetics 220, iyab195 (2022).
Uebel, C. J. et al. Distinct regions of the intrinsically disordered protein MUT-16 mediate assembly of a small RNA amplification complex and promote phase separation of Mutator foci. PLoS Genet. 14, e1007542 (2018).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Cipriani, P. G. et al. Novel LOTUS-domain proteins are organizational hubs that recruit C. elegans Vasa to germ granules. eLife 10, e60833 (2021).
Marnik, E. A. et al. The Caenorhabditis elegans TDRD5/7-like protein, LOTR-1, interacts with the helicase ZNFX-1 to balance epigenetic signals in the germline. PLoS Genet. 18, e1010245 (2022).
Price, I. F., Hertz, H. L., Pastore, B., Wagner, J. & Tang, W. Proximity labeling identifies LOTUS domain proteins that promote the formation of perinuclear germ granules in C. elegans. eLife 10, e72276 (2021).
Castel, S. E. & Martienssen, R. A. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112 (2013).
Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108 (2019).
Barucci, G. et al. Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol. 22, 235–245 (2020).
Bagijn, M. P. et al. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578 (2012).
Ashe, A. et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99 (2012).
Gu, W. et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231–244 (2009).
Tsai, H. Y. et al. A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160, 407–419 (2015).
Shukla, A. et al. poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 582, 283–288 (2020).
Buckley, B. A. et al. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451 (2012).
Guang, S. et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321, 537–541 (2008).
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998).
Batista, P. J. et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78 (2008).
Ouyang, J. P. T., Zhang, W. L. & Seydoux, G. The conserved helicase ZNFX-1 memorializes silenced RNAs in perinuclear condensates. Nat. Cell Biol. 24, 1129–1140 (2022).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Chen, W. et al. The dynamics of P granule liquid droplets are regulated by the Caenorhabditis elegans germline RNA helicase GLH-1 via its ATP hydrolysis cycle. Genetics 215, 421–434 (2020).
Wan, G. et al. ZSP-1 is a Z granule surface protein required for Z granule fluidity and germline immortality in Caenorhabditis elegans. EMBO J. 40, e105612 (2021).
Wang, J. et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018).
Hondele, M. et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573, 144–148 (2019).
Yang, P. G. et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28 (2020).
Mondal, S. et al. Multivalent interactions between molecular components involved in fast endophilin mediated endocytosis drive protein phase separation. Nat. Commun. 13, 5017 (2022).
Elbaum-Garfinkle, S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl Acad. Sci. USA 112, 7189–7194 (2015).
Wylie, C. Germ cells. Curr. Opin. Genet. Dev. 10, 410–413 (2000).
Simon, M. et al. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7, 762–773 (2014).
Sheth, U., Pitt, J., Dennis, S. & Priess, J. R. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137, 1305–1314 (2010).
Shirayama, M., Stanney, W. 3rd, Gu, W., Seth, M. & Mello, C. C. The Vasa homolog RDE-12 engages target mRNA and multiple argonaute proteins to promote RNAi in C. elegans. Curr. Biol. 24, 845–851 (2014).
Tabara, H., Hill, R. J., Mello, C. C., Priess, J. R. & Kohara, Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1–11 (1999).
Grishok, A., Tabara, H. & Mello, C. C. Genetic requirements for inheritance of RNAi in C. elegans. Science 287, 2494–2497 (2000).
Billi, A. C., Fischer, S. E. & Kim, J. K. Endogenous RNAi pathways in C. elegans. WormBook 1–49 (2014).
Ishidate, T. et al. ZNFX-1 Functions within Perinuclear Nuage to Balance Epigenetic Signals. Mol. Cell 70, 639–649.e6 (2018).
Shukla, A., Perales, R. & Kennedy, S. piRNAs coordinate poly(UG) tailing to prevent aberrant and perpetual gene silencing. Curr. Biol. 31, 4473–4485.e3 (2021).
Xu, F. et al. A cytoplasmic argonaute protein promotes the inheritance of RNAi. Cell Rep. 23, 2482–2494 (2018).
Svendsen, J. M. et al. henn-1/HEN1 promotes germline immortality in Caenorhabditis elegans. Cell Rep. 29, 3187–3199.e4 (2019).
Claycomb, J. M. et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123–134 (2009).
Reed, K. J. et al. Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 48, 1811–1827 (2020).
Huang, X. et al. Compartmentalized localization of perinuclear proteins within germ granules in C. elegans. Preprint at bioRxiv https://doi.org/10.1101/2024.03.25.586584 (2024).
Hanazawa, M., Yonetani, M. & Sugimoto, A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J. Cell Biol. 192, 929–937 (2011).
Perales, R. et al. Transgenerational epigenetic inheritance is negatively regulated by the HERI-1 chromodomain protein. Genetics 210, 1287–1299 (2018).
Lev, I. et al. MET-2-dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr. Biol. 27, 1138–1147 (2017).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Arribere, J. A. et al. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014).
Ghanta, K. S. & Mello, C. C. Melting dsDNA donor molecules greatly improves precision genome editing in Caenorhabditis elegans. Genetics 216, 643–650 (2020).
Uebel, C. J., Agbede, D., Wallis, D. C. & Phillips, C. M. Mutator foci are regulated by developmental stage, RNA, and the germline cell cycle in Caenorhabditis elegans. G3 (Bethesda) 10, 3719–3728 (2020).
Uebel, C. J., Manage, K. I. & Phillips, C. M. SIMR foci are found in the progenitor germ cells of C. elegans embryos. MicroPubl Biol. 2021 (2021).
Mair, A., Xu, S. L., Branon, T. C., Ting, A. Y. & Bergmann, D. C. Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife 8, e47864 (2019).
Zhang, X. et al. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 13, 530–550 (2018).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Zhao, C. et al. Source data for the article entitled "Multiphase condensate immiscibility contributes to transgenerational epigenetic inheritance". figshare https://figshare.com/s/df235a052c06e03f87dd (2024).
Acknowledgements
We thank members of G.W.’s laboratory for discussions. We thank S. Guang’s laboratory for sharing unpublished strains. Figs. 1a and 6i and Extended Data Fig. 10 were created with BioRender.com. This work was supported by the National Natural Science Foundation of China (grants 32070798 and 32370729), Shenzhen Medical Research Fund (B2302029) and Natural Science Foundation of Guangdong (2024A1515012650) to G.W. and by the Guangdong Science and Technology Department (2023B1212060028).
Author information
Authors and Affiliations
Contributions
C.Z. conducted the majority of the experiments with assistance from S.Y., J.H., Y.L., P.L., Z.Y. and H.J. and contributed to Figs. 1b–g, 3a,b,h,i, 4a–e, 5a–e, 6a–h and 7a–f and Extended Data Figs. 1a–h, 2a–l, 4a,b,d, 5a–r, 6a–o, 7c–g, 8a–n and 9a–f. S.C. generated a portion of the CRISPR deletions and insertions, performed immunofluorescence and HERD-1 immunoprecipitation mass spectrometry and contributed to Fig. 3c–g and Extended Data Figs. 4c and 7a. X.L., R.L. and Z.J. performed the bioinformatics analysis of PZM proteomics and HERD-1 immunoprecipitation mass spectrometry. X.L. contributed to Fig. 2c–e and Extended Data Figs. 3b,d–f and 5s–v. R.L. contributed to Fig. 6a and Extended Data Figs. 3c and 7b. R.S. and X.Z. conducted the mass spectrometry analysis and contributed to Fig. 2a and Extended Data Fig. 3a. B.D. contributed to Figs. 1a and 6i and Extended Data Fig. 10. S.K. contributed to editing the manuscript. G.W. conceived of and supervised the project, interpreted the results, contributed to Figs. 2b–d and 6a and Extended Data Fig. 3b and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
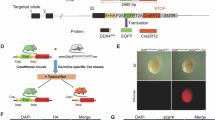
Extended Data Fig. 1 Appending tags to endogenous locus using CRISPR/Cas9 produces functional fusion proteins.
a-b, The addition of the ha::turboID epitope tag to pgl-1 (a) or znfx-1 (b) by CRISPR/Cas9 mediated homologous recombination did not affect the function of tagged proteins in dpy-11 RNAi inheritance assay. n = 4 biological replicates, data are presented as means ± s.d. Student’s t-test, two-tailed. P = 3.53 × 10−9 (pgl-1(bn101)), 0.207 (pgl-1::turboID::ha), 8.12 × 10−9 (znfx-1(gg561)), and 8.12 × 10−9 (ha::turboID::znfx-1). c, The addition of turboID::ha to the mut-16 locus did not affect MUT-16 function in pos-1 RNAi assay. n = 4 biological replicates, data are presented as means ± s.d. Student’s t-test, two-tailed. P = 3.84 × 10−12 (WT), 0.74 (mut-16(pk710)), and 0.001 (mut-16::turboID::ha). d, Brood size of animals with indicated genotype was scored at 20 °C. Animals expressing PGL-1::TurboID::HA or MUT-16::TurboID::HA exhibited normal brood size comparable to WT animals. n = 3 biological replicates, data are presented as means ± s.d. Student’s t-test, two-tailed. P = 0.896 (pgl-1::turboID::ha vs WT), 0.002 (pgl-1::turboID::ha vs pgl-1(bn101)), 0.506 (mut-16::turboID::ha vs WT), and 0.0007 (mut-16::turboID::ha vs mut-16(pk710)). e, Appending TurboID to the N terminal of SIMR-1 affected SIMR-1 function. Animals with indicated genotype were grown at 25 °C for indicated generations, and progeny per animal was scored at each generation. n = 3 biological replicates. Data are presented as mean ± s.d. Student’s t-test, two-tailed. P = 0.009 (ha::turboID::simr-1), >0.9999 (ha::simr-1), and 0.009 (simr-1(ths40)). f, Appending TurboID to N or C terminal of SIMR-1 affected SIMR-1 localization. IF with anti-HA antibody followed by Alexa Fluor 488-conjugated secondary antibody. Scale bar, 5 μm. g-h, pos-1 RNAi assay on animals with indicated genotype to confirm the functionality of 3xFLAG::GFP::HERD-1 (g) and HA::MUT-2 (h). n = 3 biological replicates. Data are presented as mean ± s.d. Student’s t-test, two-tailed. P = 0.167 (herd-1(ths351)), 0.0002 (3xflag::gfp::herd-1) in g. P = 7.92 × 10−8 (WT), 0.368 (mut-2(ne298)), and 2.86 × 10−6 (ha::mut-2) in h. ns P ≥ 0.05, ** P < 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001. Source numerical data are available in source data.
Extended Data Fig. 2 Optimization of TurboID-mediated biotinylation labeling conditions in C. elegans.
a, c-e, WT and ha::turboID::znfx-1 (a), WT and pgl-1::turboID::ha (c, d) or WT and mut-16::turboID::ha animals (e) were grown on bioB(-) bacteria with varying biotin concentrations for the indicated time. Proteins from each treatment were detected using anti-HA and anti-Tubulin antibodies with HRP-conjugated secondary antibodies or Strep-HRP. 3D, 3 days. n = 3 biological replicates. b, Embryos from either WT or PGL-1::TurboID::HA animals were cultivated on bioB(+) or bioB(-) HT115 bacteria cultured in either Luria broth medium (LB) or M9CG medium (MM). Proteins from young adult animals were analyzed by Western blot using Strep-HRP, α-HA, and α-Tubulin antibodies. n = 3 biological replicates. f, The bioB knockout strategy is illustrated, showing the bioB locus before and after knockout, repair template, and genotype primers. PCR confirmed successful bioB knockout in HT115 bacteria. g-h, WT or ha::turboID::znfx-1 animals were grown under specified conditions; progeny per animal was counted (g), and young adult animals were imaged (h), scale bar, 10 μm. Data are mean values ± s.d. i-j, Animals expressing HA::ZNFX-1 or HA::TurboID::ZNFX-1 (i) and WT animals or animals expressing PGL-1::TurboID::HA (j) were cultured on bioB(-) bacteria with biotin for 2 hours. Treated germlines were dissected, fixed, and stained with anti-HA or anti-PGL-1 antibodies (Alexa fluor 488-conjugated secondary antibody). Biotinylated proteins were visualized with Strep-Alexa fluor 568, and pachytene germ cell nuclei imaged using a Leica TCS SP8 microscope. Scale bar, 5 μm. n = 3 biological replicates. k-l, Animals expressing specified proteins were maintained on bioB(-) bacteria with 500 μM biotin for 2 hours. Dissected germlines were visualized with immunofluorescence using anti-HA or anti-PGL-1 antibodies followed by Alexa-568 conjugated secondary antibody, and biotinylated proteins were detected with Strep-Alexa fluor 488(k). Fluorescent signal colocalization was quantified (l, 27 granules: 3 animals/group, 3 cells/animal, 3 granules/cell). Scale bar, 2.5 μm. Data are presented as mean values ± s.d. Student’s t-test, two-tailed. P = 2.66 × 10−17. **** P ≤ 0.0001. Source numerical data and unprocessed blots/gels are available in source data.
Extended Data Fig. 3 System-wide analysis of PZM proteomics.
a, Principal component analysis (PCA) of top 500 significantly enriched proteins. PCA analysis showed the primary source of variation, PC1, was associated with genotype, where proteins enriched in animals expressing PGL-1::TurboID, TurboID::ZNFX-1, and MUT-16::TurboID showed the least similarity to the wild-type (WT) group. The second largest source of variation was observed along the PC2 axis, with proteins enriched from animals expressing PGL-1::TurboID and TurboID::ZNFX-1 exhibiting greater deviation from those expressed in MUT-16::TurboID. Proteins enriched from animals expressing PGL-1::TurboID and TurboID::ZNFX-1 clustered closer together than the WT group and animals expressing MUT-16::TurboID, but still formed distinct groups. This finding supports the notion that P compartment and Z compartment are distinct but closely related compartments. WT, N2 Bristol. b, DAVID analysis of proteins localized to PZM compartments. Molecular function (MF) is shown. Fisher’s exact test. c, Potential protein-protein interaction network revealed by Metascape. Note, the interaction network in the Z compartment was added manually after the interactions were confirmed in this work (see Fig.6a–i). d, Enrichment analysis of motifs in IDR regions of proteins localized to PZM compartments. Binomial Test. P = 0.0232 (top), 0.0313 (middle), and 0.0376 (bottom) for PZ compartment. P = 0.001 (top), 0.00374 (middle), and 0.0083 (bottom) for ZM compartment. P = 0.0235 for PM compartment. P = 0.0262 for PZM compartment. * P < 0.05, ** P < 0.01, and *** P ≤ 0.001. e-f, Enrichment analysis of proteins localized to P, Z, M, PZ and PM (e) or ZM and PZM (f) compartments in RNAi screens. Fisher's exact test.
Extended Data Fig. 4 Screen for genes encoding PZM compartment components required for germline immortality and/or RNAi.
a, Introducing HA or GFP to the endogenous locus of newly identified Mrt genes using CRISPR/Cas9 did not abolish the protein product function. Lineages of animals of the indicated genotypes were grown in triplicate at 25 °C. ‘F’ indicates the generation at which one replicate lineage became sterile. As different lineages of the same mutant strain often become sterile on different generations, 1 to 3 'F' numbers are provided for each strain. For those generations where a lineage became sterile, brood sizes of control (WT, N2 Bristol) and animals harboring the corresponding epitope-tagged protein lineages were scored. Note, to minimize labor, brood sizes were only scored on the generation where a mutant lineage displayed sterility. n = 3 biological replicates, error bar, ± s.d. b, Fluorescence imaging of approximately a dozen germline cells in the pachytene region of WT or zsp-1(-) animals expressing GFP::MIP-2. Scale bar, 5 μm. n > 3 animals. c, Localization of F55A12.5. F55A12.5::HA was detected with an anti-HA antibody followed by Alex Fluor 568-conjugated secondary antibody. Scale bar, 20 μm for germline, 10 μm for embryos. n > 3 animals. d, CRISPR/Cas9-generated viable deletion mutants were subjected to pos-1 RNAi, and the percentage of hatched progeny was scored. Note, F01G4.4a.1 mutants exhibited an egg-lay defect phenotype; thus, the hatching rate was scored in utero. n = 3 biological replicates. Data are presented as mean ± s.d. Student’s t-test, two-tailed. P = 7.82 × 10−5 (T26A5.2(ths364)), 3.72 × 10−6 (mip-2(ths285)), 3 × 10−5 (hpo-40(ths394)), 0.25 (herd-1(ths351)), 4.23 × 10−7 (B0001.3(ths358)), 3.07 × 10−10 (F52D2.12(ths438)), 1.48 × 10−8 (pqn-53(ths427)), 0.0001 (C36B1.11(ths356)), 0.09 (F56C9.6(ths410)), 0.12 (B0261.7(ths367)), 1.79 × 10−8(ubr-4(ths407)), 5.92 × 10−5 (gln-3(ths387)), 0.0001 (H05C05.1(ths448)), 8.39 × 10−5 (F59A3.12(ths520)), 5.12 × 10−6 (ZK1307.1(ths597)), 1.52 × 10−5 (B0001.8(ths352)), 0.051 (Y57A10A.31(ths272)), and 1.43 × 10−5 (Y37H2A.1(ths603)). ns P ≥ 0.05, and **** P ≤ 0.0001. Source numerical data are available in source data.
Extended Data Fig. 5 HERD-1 is required for germline RNAi but limits the transgenerational inheritance of RNAi.
a, WT or herd-1(-) animals were subjected to dpy-6 RNAi treatment. Young adult animals, with or without dpy-6 RNAi, were shown. n = 3 biological replicates. b, Fluorescence imaging of -3 to -1 oocytes in WT, hrde-1(-), and herd-1(-) animals expressing GFP::H2B (pkIS32) treated with gfp RNAi are shown. Scale bar, 10 μm. c, Quantification of GFP fluorescence intensity in (b). n = 10 animals. Mean values ± s.d. Student’s t-test, two-tailed. P = 0.0002 (herd-1(ths351) gfp RNAi vs WT gfp RNAi), 0.0002 (herd-1(ths351) gfp RNAi vs hrde-1(ths682) gfp RNAi), 0.00013 (herd-1(ths351) gfp RNAi vs control RNAi). d-e, Progeny of animals from (b) were grown on non-RNAi plates for the indicated generations. Fluorescence imaging (d) and quantification of animals with silenced GFP (e) are shown. Scale bar, 10 μm. n = 3 biological replicates (50 animals per replicate). Error bar, ± s.e.m. Log-rank test. P = 0.0253 (hrde-1(ths685)), and <0.0001(herd-1(ths351)). f, Fluorescence imaging of oocytes at indicated times after gfp RNAi treatment from (b) shows % of animals with silenced GFP. Scale bar, 10 μm. g, WT and herd-1(-) animals expressing single-copy mCherry::H2B in the germline were subjected to control or mCherry RNAi, and fluorescence was imaged. Scale bar, 10 μm. h, Fluorescence intensity from -1 to -3 oocytes was quantified from (g). n = 10 animals. mean values ± s.d. Student’s t-test, two-tailed. P = 8.19 × 10−8 (WT gfp RNAi vs control RNAi), 0.004 (herd-1(ths351) gfp RNAi vs control RNAi, and 0.0005 (herd-1(ths351) gfp RNAi vs WT gfp RNAi). i, Progenies from (g) were grown on non-RNAi plates. Representative imaging of oocytes from animals at indicated generations shows % of animals with silenced mCherry. Scale bar, 10 μm. j, The percentage of animals that failed to express mCherry::H2B was scored under a 63X oil objective every generation from (i). n = 3 biological replicates. Error bar, ± s.e.m. Log-rank test. P = 0.0455 (hrde-1(tm1200)), and <0.0001 (herd-1(ths351)). (k–l) The pUG PCR assay detected pUG RNA in the P0 and F1 (k), as well as the F15 and F30 (l) generations, after WT, znfx-1(-), hrde-1(-), or herd-1(-) animals expressing pie-1::gfp::h2b (mjIS31) were exposed to gfp RNAi. n = 3 biological replicates. (m–n) The pUG PCR assay detected pUG RNA in the P0 and F1 (m), as well as the F15 and F30 (n) generations, after WT, hrde-1(-), or herd-1(-) animals expressing pie-1::gfp::h2b (pkIS32) were exposed to gfp RNAi. (o–p) The pUG PCR assay detected pUG RNA in the P0 and F1 (o), as well as the F15 and F30 (p) generations, after WT, znfx-1(-), hrde-1(-), or herd-1(-) animals expressing pie-1::mcherry::h2b were exposed to mcherry RNAi. q, smFISH detected pUG RNA in animals with indicated genotype. Scale bar, 5 μm. n = 3 biological replicates. r, Fluorescence imaging of -3 to -1 oocytes in young adult animals with indicated genotype expressing a piRNA sensor. Scale bar, 10 μm. s, Classification of genes differentially targeted by endo siRNAs in herd-1(ths351) relative to WT animals. * P < 0.05, ** P < 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001. Source numerical data and unprocessed gels are available in source data.
Extended Data Fig. 6 PZM compartments are mixed in herd-1(-) animals.
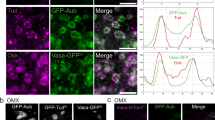
a, Fluorescence imaging of WT and herd-1(-) animals expressing PGL-1::GFP or GFP::ZNFX-1. Scale bar, 5 μm. n > 3 animals. b, Fluorescence imaging of WT and herd-1(-) animals expressing PGL-1::TagRFP and GFP::ZNFX-1. Scale bar: 5 μm for top 6 images, 2 μm for bottom 6 cell inset. c, Quantification of granule size in (b). For each group, approximately a dozen pachytene region germ cells from 3 animals were measured using ImageJ. Data are presented as mean values ± s.d. Student’s t-test, two-tailed. P = 0.0003 (GFP::ZNFX-1), and 0.0109(PGL-1::TagRFP). d, Quantification of the degree of colocalization between PGL-1 and ZNFX-1 in (b). n = 27 granules (3 animals per condition, 3 cells per animal, 3 granules per cell). Data are presented as mean values ± s.d. Student’s t-test, two-tailed. P = 1× 10−7. e-n, Fluorescence imaging of WT and herd-1(-) animals expressing PGL-1::TagRFP and MUT-16::GFP (e), PGL-1::TagRFP and GFP::WAGO-4 (g), MUT-2::mCherry and PGL-1::GFP (i), TagRFP::ZNFX-1 and MUT-16::GFP (k), HA::MUT-2 and GFP::ZNFX-1 (m). For images in (m), HA::MUT-2 and GFP::ZNFX-1 were detected using anti-HA and anti-GFP-based immunofluorescence. The degree of colocalization between PGL-1::TagRFP and MUT-16::GFP (f), PGL-1::TagRFP and GFP::WAGO-4 (h), MUT-2::mCherry and PGL-1::GFP (j), TagRFP::ZNFX-1 and MUT-16::GFP (l) and HA::MUT-2 and GFP::ZNFX-1 (n). Scale bar: 5 μm for top 6 images, 2 μm for the bottom 6 cell inset in b, g, k, and m, and 1 μm for the bottom 6 cell inset in e and i. For colocalization analysis, n = 27 granules (3 animals per condition, 3 cells per animal, 3 granules per cell). Data are presented as mean values ± s.d. Student’s t-test, two-tailed. P = 4 × 10−12 (f), 1 × 10−17 (h), 3 × 10−4 (j), 1 × 10−9 (l), and 2 × 10−19 (n). o, Western blot to detect indicated proteins in WT and herd-1(-) animals. n = 3 biological replicates. * P < 0.05, *** P ≤ 0.001, and **** P ≤ 0.001. Source numerical data and unprocessed blots are available in source data.
Extended Data Fig. 7 Identification of HERD-1 interacting proteins.
a, Silver staining of FLAG antibody immunoprecipitated proteins from WT animals (N2 Bristol) or animals expressing 3xFLAG::GFP::HERD-1. n = 2 biological replicates. b, Reactome and GO Cellular Components analysis of the top enriched HERD-1 interaction proteins (FOT>=3). c-g, Co-immunoprecipitation assays to detect interactions between the following proteins: MUT-16::HA and 3xFLAG::GFP::HERD-1 (c), HA::GLH-1 and PGL-1::GFP::3xFLAG in WT and herd-1(-) backgrounds (d), OLLAS::GFP::LOTR-1 and HA::GLH-1 in WT and herd-1(-) backgrounds (e), OLLAS::GFP::LOTR-1 and MUT-16::HA in WT and herd-1(-) backgrounds (f), DEPS-1::GFP::3xFLAG and MUT-16::HA in WT and herd-1(-) backgrounds (g). Lysate from animals expressing indicated proteins were incubated with anti-FLAG or anti-GFP antibodies. Input proteins or co-immunoprecipitated proteins were detected using Western blot with indicated antibodies. n = 2 biological replicates. Unprocessed blots/gels are available in source data.
Extended Data Fig. 8 Anti-GFP nanobody based recruitment induces mixing of PZM compartments.
a-n, Animals expressing the following proteins were detected using immunofluorescence with the specified antibodies: PGL-1::GFP; MUT-16::HA or PGL-1::GFP; MUT-16::anti-GFP annobody::HA (a), GFP::ZNFX-1; MUT-16::HA or GFP::ZNFX-1; MUT-16::anti-GFP annobody::HA (c), GFP::MIP-2; MUT-16::HA or GFP::MIP-2; MUT-16::anti-GFP annobody::HA (e), PGL-1::GFP; MUT-2::mCherry or PGL-1::GFP; MUT-16::anti-GFP annobody::HA; MUT-2::mCherry (g), GFP::ZNFX-1; MUT-2::mCherry or GFP::ZNFX-1; MUT-16::anti-GFP nanobody::HA; MUT-2::mCherry (i), PGL-1::GFP; MUT-16::HA or PGL-1::GFP; OLLAS::anti-GFP nanobody::LOTR-1; MUT-16::HA (k), PGL-1::GFP; MUT-16::HA or PGL-1::GFP; OLLAS::anti-GFP nanobody::ZNFX-1; MUT-16::HA (m). For each animal, a representative cell from the pachytene region is shown. The degree of colocalization between the following proteins was quantified using ImageJ: PGL-1 and MUT-16 (b), ZNFX-1 and MUT-16 (d), MIP-2 and MUT-16 (f), PGL-1 and MUT-2 (h), ZNFX-1 and MUT-2 (j), MUT-16 and PGL-1 (l), MUT-16 and PGL-1 (n). Scale bar, 2 μm. For each condition, n = 27 granules (3 animals for each condition, 3 cells per animal, 3 granules per cell). All Data are presented as mean values ± s.d. Student’s t-test, two-tailed. P = 2 × 10−25 (b), 2 × 10−17 (d), 6 × 10−12 (f), 7 × 10−20 (h), 2 × 10−5 (j), 1 × 10−19 (l), and 3 × 10−9 (n). **** P ≤ 0.001. Source numerical data are available in source data.
Extended Data Fig. 9 Forced mixing of PZM compartments results in RNAi defect and extended TEI.
a-b, Animals with the following genotype were subjected to control RNAi (L4440) or pos-1 RNAi:WT, mut-16(pk710), mut-16::anti-GFP nanobody::ha, gfp::znfx-1, gfp::mip-2, pgl-1::gfp, mut-16::anti-GFP nanobody::ha; gfp::znfx-1, mut-16::anti-GFP nanobody::ha; gfp::mip-2, mut-16::anti-GFP nanobody::ha; pgl-1::gfp (a) and WT, pgl-1::gfp, mut-16::anti-GFP nanobody::ha, ollas::anti-GFP nanobody::znfx-1, ollas::anti-GFP nanobody::lotr-1, pgl-1::gfp; mut-16::anti-GFP nanobody::ha, pgl-1::gfp; ollas::anti-GFP nanobody::znfx-1, pgl-1::gfp; ollas::anti-GFP nanobody::lotr-1, pgl-1::gfp; mut-16::anti-GFP nanobody::ha; ollas::anti-GFP nanobody::znfx-1, pgl-1::gfp; mut-16::anti-GFP nanobody::ha; ollas::anti-GFP nanobody::lotr-1 (b). The percentage of hatched animals was scored. n = 3 biological replicates. Data are presented as mean values ± s.d. Student’s t-test, two-tailed. P = 0.03 (mut-16::nanobody::ha; gfp::znfx-1 vs mut-16::nanobody::ha), 0.012 (mut-16::nanobody::ha; gfp::znfx-1 vs gfp::znfx-1), 0.02 (mut-16::nanobody::ha; gfp::mip-2 vs mut-16::nanobody::ha), 0.004 (mut-16::nanobody::ha; gfp::mip-2 vs gfp::mip-2), 0.002 (mut-16::nanobody::ha; pgl-1::gfp vs mut-16::nanobody::ha), 0.002 (mut-16::nanobody::ha; pgl-1::gfp vs pgl-1::gfp). c-e, Animals with the follwing genotype expressing mCherry::H2B in the germline were exposed to control RNAi (L4440) or mcherry RNAi: WT, znfx-1(ths859), mut-16::anti-GFP nanobody::ha, ollas::anti-GFP nanobody::lotr-1, pgl-1::gfp, pgl-1::gfp; mut-16::anti-GFP nanobody::ha, pgl-1::gfp; ollas::anti-GFP nanobody::lotr-1, pgl-1::gfp; mut-16::anti-GFP nanobody::ha; ollas::anti-GFP nanobody::lotr-1, herd-1(ths351) (c) or WT, znfx-1(ths859), mut-16::anti-GFP nanobody::ha, ollas::anti-GFP nanobody::znfx-1, pgl-1::gfp, pgl-1::gfp; mut-16::anti-GFP nanobody::ha, pgl-1::gfp; ollas::anti-GFP nanobody::znfx-1, pgl-1::gfp; mut-16::anti-GFP nanobody::ha; ollas::anti-GFP nanobody::znfx-1, herd-1(ths351) (d), or WT, znfx-1(ths859), gfp::znfx-1, gfp::mip-2, pgl-1::gfp, mut-16::anti-GFP nanobody::ha, gfp::mip-2; mut-16::anti-GFP nanobody::ha, gfp::znfx-1; mut-16::anti-GFP nanobody::ha, pgl-1::gfp; mut-16::anti-GFP nanobody::ha, herd-1(ths351) (e). The fluorescence intensity of -1 to -3 oocytes from these animals was measured with ImageJ. n = 10 animals. Data are presented as mean values ± s.d. Student’s t-test, two-tailed. P = 9.84 × 10−6 (herd-1(-) vs WT), 9.84 × 10−6 (mut-16::nanobody vs pm), 1.58 × 10−7 (mut-16::gfp::nanobody vs plm), 0.003 (gfp nanobody::lotr-1 vs pl), 1.58 × 10−7 (gfp nanobody::lotr-1 vs plm), 9.88 × 10−5 (pgl-1::gfp vs pm), 0.003 (pgl-1::gfp vs pl), and 1.58 × 10−7 (pgl-1::gfp vs plm) in (c). P = 1.86 × 10−5 (herd-1(-) vs WT), 0.0002 (mut-16::nanobody vs pm), 1.47 × 10−6 (mut-16::nanobody vs pzm), 6.03 × 10−7 (gfp nanobody::znfx-1 vs pz), 1.47 × 10−6 (gfp nanobody::znfx-1 vs pzm), 0.0002 (pgl-1::gfp vs pm), 6.03 × 10−7 (pgl-1::gfp vs pz), and 1.47 × 10−6 (pgl-1::gfp vs pzm) in (d). P = 3.78 × 10−12 (herd-1(-) vs WT), 9.66 × 10−10 (gfp::znfx-1 vs zm), 2.44 × 10−6 (gfp::mip-2 vs mm), 6.27 × 10−20 (pgl-1::gfp vs pm), 2.44 × 10−6 (mut-16::gfp nanobody vs mm), 9.66 × 10−10 (mut-16::gfp nanobody vs zm), and 6.27 × 10−20 (mut-16::gfp nanobody vs pm) in (e). f, Animals with the indicated genotype expressing mCherry::H2B in the germline were exposed to mcherry RNAi, and progenies were grown on non-RNAi plates. The percentage of silenced animals was scored using a 63x oil objective on a Leica DMI8 inverted microscope. n = 3 biological replicates. For each replicate, at least 50 animals were scored. Data are presented as mean values ± s.e.m. Log-rank test. P = 0.0253 (znfx-1(ths859)), >0.9999 (gfp::znfx-1, gfp::mip-2, pgl-1::gfp, and mut-16::gfp nanobody), 0.1573 (zm), and 0.0143 (pm). ns P ≥ 0.05, * P < 0.05, ** P < 0.01, *** P ≤ 0.001, and **** P ≤ 0.0001. Source numerical data are available in source data.
Extended Data Fig. 10 Working model for HERD-1 in regulating multiphase condensates immiscibility and limiting TEI.
In WT animals, siRNAs undergo amplification within the M compartment through pUG RNA-siRNA cycles. These siRNAs can degrade RNA in the cytoplasm when bound to WAGOs. Additionally, pUG RNAs can be memorized by ZNFX-1 within the Z compartment, facilitating TEI. Upon depletion of HERD-1, the PZM proteins interaction network becomes mixed, bringing previously separated small pathways into proximity. This coalescence may lead to increased incorporation of pUG RNAs or proteins into TEI and reduced distribution of siRNAs or proteins to the cytoplasm for RNAi-mediated RNA degradation. Created with BioRender.com.
Supplementary information
Source data
Source Data Figs. 1, 3–5 and 7 and Extended Data Figs. 1, 2, 4–6, 8 and 9
Statistical source data.
Source Data Figs. 1 and 6 and Extended Data Figs. 2 and 5–7
Unprocessed western blots
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, C., Cai, S., Shi, R. et al. HERD-1 mediates multiphase condensate immiscibility to regulate small RNA-driven transgenerational epigenetic inheritance. Nat Cell Biol 26, 1958–1970 (2024). https://doi.org/10.1038/s41556-024-01514-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-024-01514-8
This article is cited by
-
The dynamic and heterogeneous composition of biomolecular condensates and its functional relevance
Nature Reviews Molecular Cell Biology (2025)
-
Nucleoporins shape germ granule architecture and balance small RNA silencing pathways
Nature Communications (2025)
-
A nuclear pore-anchored condensate enables germ granule organization and transgenerational epigenetic inheritance
Nature Structural & Molecular Biology (2025)
-
Proximal proteomics reveals a landscape of human nuclear condensates
Nature Cell Biology (2025)
-
TurboID-based proximity labeling identifies novel germline proteins that maintain E granule integrity and small RNA homeostasis in C. elegans
Science China Life Sciences (2025)