Abstract
Histone methyltransferases regulate chromatin organization and are frequently mutated in human diseases, including cancer. One such often mutated methyltransferase, SETD2, associates with transcribing RNA polymerase II and catalyses H3K36me3—a modification that contributes to gene transcription, splicing and DNA repair. Although its catalytic function is well-characterized, its non-catalytic roles remain unclear. Here we reveal a catalysis-independent function of SETD2 in nuclear lamina stability and genome integrity. Through its intrinsically disordered amino terminus, SETD2 associates with lamina-associated proteins, including lamin A/C, lamin B1 and emerin. Loss of SETD2 or its N terminus leads to severe nuclear morphology defects and genome instability, mirroring lamina dysfunction. Mechanistically, the N terminus of SETD2 serves as a scaffold for the mitotic kinase CDK1 and lamins, facilitating lamin phosphorylation and depolymerization during mitosis. Restoration of the N-terminal regions required for interaction with CDK1 and lamins rescues nuclear morphology and suppresses tumorigenic growth in a clear cell renal cell carcinoma model with SETD2 haploinsufficiency. These findings reveal a previously unrecognized role of SETD2 in nuclear lamina organization and genome maintenance that probably extends to its role as a tumour suppressor.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD040771. Source data are provided with this paper.
References
Venkatesh, S. & Workman, J. L. Set2 mediated H3 lysine 36 methylation: regulation of transcription elongation and implications in organismal development. Wiley Interdiscip. Rev. Dev. Biol. 2, 685–700 (2013).
Markert, J. W., Soffers, J. H. & Farnung, L. Structural basis of H3K36 trimethylation by SETD2 during chromatin transcription. Science 387, 528–533 (2024).
Sharda, A. & Humphrey, T. C. The role of histone H3K36me3 writers, readers and erasers in maintaining genome stability. DNA Repair 119, 103407 (2022).
Carvalho, S. et al. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 41, 2881–2893 (2013).
Carvalho, S. et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife 3, e02482 (2014).
de Almeida, S. F. et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 18, 977–983 (2011).
McDaniel, S. L. & Strahl, B. D. Shaping the cellular landscape with Set2/SETD2 methylation. Cell. Mol. Life Sci. 74, 3317–3334 (2017).
Fahey, C. C. & Davis, I. J. SETting the stage for cancer development: SETD2 and the consequences of lost methylation. Cold Spring Harb. Perspect. Med. 7, a026468 (2017).
Husmann, D. & Gozani, O. Histone lysine methyltransferases in biology and disease. Nat. Struct. Mol. Biol. 26, 880–889 (2019).
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Zbar, B., Brauch, H., Talmadge, C. & Linehan, M. Loss of alleles of loci on the short arm of chromosome 3 in renal cell carcinoma. Nature 327, 721–724 (1987).
Jonasch, E., Walker, C. L. & Rathmell, W. K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 17, 245–261 (2021).
The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013).
Sato, Y. et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 45, 860–867 (2013).
Hsieh, J. J. et al. Chromosome 3p loss-orchestrated VHL, HIF, and epigenetic deregulation in clear cell renal cell carcinoma. J. Clin. Oncol. 36, JCO2018792549 (2018).
Chiang, Y. C. et al. SETD2 haploinsufficiency for microtubule methylation is an early driver of genomic instability in renal cell carcinoma. Cancer Res. 78, 3135–3146 (2018).
Yuan, W. et al. Heterogeneous nuclear ribonucleoprotein L is a subunit of human KMT3a/Set2 complex required for H3 Lys-36 trimethylation activity in vivo. J. Biol. Chem. 284, 15701–15707 (2009).
Bhattacharya, S. & Workman, J. L. Regulation of SETD2 stability is important for the fidelity of H3K36me3 deposition. Epigenetics Chromatin 13, 40 (2020).
Bhattacharya, S. et al. The disordered regions of the methyltransferase SETD2 govern its function by regulating its proteolysis and phase separation. J. Biol. Chem. 297, 101075 (2021).
Bhattacharya, S. et al. The methyltransferase SETD2 couples transcription and splicing by engaging mRNA processing factors through its SHI domain. Nat. Commun. 12, 1443 (2021).
Hung, V. et al. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 11, 456–475 (2016).
van Steensel, B. & Belmont, A. S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017).
Geyer, P. K., Vitalini, M. W. & Wallrath, L. L. Nuclear organization: taking a position on gene expression. Curr. Opin. Cell Biol. 23, 354–359 (2011).
Dittmer, T. A. & Misteli, T. The lamin protein family. Genome Biol. 12, 222 (2011).
Dechat, T., Gesson, K. & Foisner, R. Lamina-independent lamins in the nuclear interior serve important functions. Cold Spring Harb. Symp. Quant. Biol. 75, 533–543 (2010).
Kirby, T. J. & Lammerding, J. Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell Biol. 20, 373–381 (2018).
Dahl, K. N., Kahn, S. M., Wilson, K. L. & Discher, D. E. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 117, 4779–4786 (2004).
Klaasen, S. J. et al. Nuclear chromosome locations dictate segregation error frequencies. Nature 607, 604–609 (2022).
Swift, J. et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 (2013).
Vahabikashi, A., Adam, S. A., Medalia, O. & Goldman, R. D. Nuclear lamins: structure and function in mechanobiology. APL Bioeng. 6, 011503 (2022).
Irianto, J., Pfeifer, C. R., Ivanovska, I. L., Swift, J. & Discher, D. E. Nuclear lamins in cancer. Cell. Mol. Bioeng. 9, 258–267 (2016).
Bell, E. S. & Lammerding, J. Causes and consequences of nuclear envelope alterations in tumour progression. Eur. J. Cell Biol. 95, 449–464 (2016).
Singh, I. & Lele, T. P. Nuclear morphological abnormalities in cancer: a search for unifying mechanisms. Results Probl. Cell Differ. 70, 443–467 (2022).
Fischer, E. G. Nuclear morphology and the biology of cancer cells. Acta Cytol. 64, 511–519 (2020).
Janssen, A. F. J., Breusegem, S. Y. & Larrieu, D. Current methods and pipelines for image-based quantitation of nuclear shape and nuclear envelope abnormalities. Cells 11, 347 (2022).
Mason, F. M. et al. SETD2 safeguards the genome against isochromosome formation. Proc. Natl Acad. Sci. USA 120, e2303752120 (2023).
Nicetto, D. & Zaret, K. S. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 55, 1–10 (2019).
Schibler, A. C., Jevtic, P., Pegoraro, G., Levy, D. L. & Misteli, T. Identification of epigenetic modulators as determinants of nuclear size and shape. eLife 12, e80653 (2023).
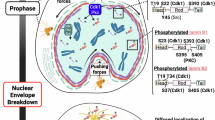
Liu, S. Y. & Ikegami, K. Nuclear lamin phosphorylation: an emerging role in gene regulation and pathogenesis of laminopathies. Nucleus 11, 299–314 (2020).
Heald, R. & McKeon, F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 61, 579–589 (1990).
Mall, M. et al. Mitotic lamin disassembly is triggered by lipid-mediated signaling. J. Cell Biol. 198, 981–990 (2012).
Dronamraju, R. et al. Set2 methyltransferase facilitates cell cycle progression by maintaining transcriptional fidelity. Nucleic Acids Res. 46, 1331–1344 (2018).
Ikegami, K., Secchia, S., Almakki, O., Lieb, J. D. & Moskowitz, I. P. Phosphorylated lamin A/C in the nuclear interior binds active enhancers associated with abnormal transcription in progeria. Dev. Cell 52, 699–713 (2020).
Park, I. Y. et al. Dual chromatin and cytoskeletal remodeling by SETD2. Cell 166, 950–962 (2016).
Lampe, J. W. et al. Discovery of a first-in-class inhibitor of the histone methyltransferase SETD2 suitable for preclinical studies. ACS Med. Chem. Lett. 12, 1539–1545 (2021).
Hacker, K. E. et al. Structure/function analysis of recurrent mutations in SETD2 protein reveals a critical and conserved role for a SET domain residue in maintaining protein stability and histone H3 Lys-36 trimethylation. J. Biol. Chem. 291, 21283–21295 (2016).
Wong, X. et al. Lamin C is required to establish genome organization after mitosis. Genome Biol. 22, 305 (2021).
Smits, V. A. & Medema, R. H. Checking out the G(2)/M transition. Biochim. Biophys. Acta 1519, 1–12 (2001).
Mehta, S. & Zhang, J. Liquid–liquid phase separation drives cellular function and dysfunction in cancer. Nat. Rev. Cancer 22, 239–252 (2022).
Zhou, P., Lugovskoy, A. A. & Wagner, G. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J. Biomol. NMR 20, 11–14 (2001).
Schulze-Gahmen, U. et al. The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat. eLife 2, e00327 (2013).
Michowski, W. et al. Cdk1 controls global epigenetic landscape in embryonic stem cells. Mol. Cell 78, 459–476 (2020).
Xie, Y. et al. SETD2 loss perturbs the kidney cancer epigenetic landscape to promote metastasis and engenders actionable dependencies on histone chaperone complexes. Nat. Cancer 3, 188–202 (2022).
Reilly, A. et al. Lamin B1 deletion in myeloid neoplasms causes nuclear anomaly and altered hematopoietic stem cell function. Cell Stem Cell 29, 577–592 (2022).
Baluapuri, A. et al. MYC recruits SPT5 to RNA polymerase II to promote processive transcription elongation. Mol. Cell 74, 674–687 (2019).
Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinform. 22, 433 (2021).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Wickham, H. et al. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 (2019).
Zhang, C. et al. Von Hippel Lindau tumor suppressor controls m6A-dependent gene expression in renal tumorigenesis. J. Clin. Invest. 134, e175703 (2024).
Acknowledgements
We thank all members of the Strahl and Davis laboratories for their valuable input. We thank S. Prasanth and M. Emanuele for critical comments on the manuscript, R. Berlow and M. Begley for advice related to protein expression, W. Salmon for advice and helpful suggestions related to microscopy, K. Ikegami for sharing LMNA mutant cell lines, W. Legant and F. Rahman for help with microscopy, J. Cook for sharing cell lines and reagents and Z. Mayo for help with the initial experiments. We thank the UNC Proteomics Core Facility and the UNC Hooker Imaging Core Facility, which are supported in part by a NCI Cancer Center Core Support Grant (grant number 2P30CA016086-45) to the UNC Lineberger Comprehensive Cancer Center. This work was supported by an NIH grant to B.D.S. (grant number GM126900), a DOD grant (grant number W81XWH2110786) to F.M.M., an NIH grant (grant number R01CA275082) to F.M.M. and W.K.R., and a CPRIT award (grant number RR190058) to Q.Z. L.V. is supported by NIH 5T32CA009592.
Author information
Authors and Affiliations
Contributions
A.K. and B.D.S. conceived the study. A.K., P.G.N. and J.M.M. performed all the experiments with help from L.C.C. and A.L.B. C.Z. performed the 3D colony growth assays and mice experiments. K.J. purified proteins. W.K.R., F.M.M. and L.V. shared dTAG-related reagents. B.M.B. and M.B.M. helped with initial mass spectrometry experiments. C.A.M. and L.E.H. performed mass spectrometry analyses. K.L. and J.A. synthesized EPZ-719. I.J.D. and Q.Z. provided technical resources and conceptual ideas. A.K. and B.D.S. analysed data and wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
B.D.S. is a co-founder and board member of EpiCypher, Inc. and holds equity in the company. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 SETD2 interacts with nuclear lamina proteins.
a, Schematic of SETD2 with its indicated domains. b, Fluorescence intensity profiles of SETD2, lamin A/C and DAPI in interphase or mitotic cells. Scale bar, 10 µm. c, Proximity ligation assay (PLA) on SETD2-KD cells expressing Halo-Flag-tagged SETD2 using anti-SETD2 and anti-lamin A/C antibodies. PLA pairs are in green; SETD2 (red) visualized using fluorescent Halo ligand JFX-549. DNA was counterstained with DAPI. Scale bar, 10 µm. d, Quantitative analysis of PLAs between SETD2 and lamin A/C shown in c. Two-tailed unpaired Student’s t-test was performed from two independent biological replicates, with n (left to right) = 55, 41, 7 cells; mean ± s.e.m.
Extended Data Fig. 2 SETD2 loss during G2/M leads to nuclear lamina defects.
a, Representative confocal microscopy images of control and SETD2-KD HKC cells immunostained with anti-lamin A/C and lamin B1 antibodies. DNA counterstained with DAPI. b, Representative confocal microscopy images of control and Setd2-KO MEF cells immunostained with anti-lamin B1 antibody. DNA counterstained with DAPI. c, Schematic of cell synchronization by double thymidine block and release method used in d–f. d, Immunoblot analysis of indicated proteins in untreated and dTAG-47 treated (at 6 h post release) cells. e, Representative images of either untreated or dTAG-47 treated SETD2–FKBP cells, fixed at the indicated timepoints post release from thymidine block and immunostained with anti-lamin A/C antibody. DNA counterstained with DAPI. f, Quantification of nuclear defects observed in e. Ordinary one-way ANOVA with Sidak’s multiple comparison test was performed on data from two independent biological replicates, with n (left to right) = 614, 558, 735, 654, 754, 693 cells; mean ± s.e.m. g, Schematic of cell synchronization by serum starvation of Setd2 (w/flox) MEFs. h, Immunoblot analysis of indicated proteins in untreated and tamoxifen treated (Setd2-KO) MEF cells upon release from G0/G1 block. i, Representative images of either control or tamoxifen-treated MEF cells at the indicated timepoints post release from serum starvation, immunostained with anti-lamin A/C antibody. j, Quantification of nuclear defects from data in i. Two-way ANOVA with Sidak’s multiple comparison test was performed on data from three independent replicates with n (left to right) = 184, 166, 247, 223, 256, 247, 387, 348 cells; mean ± s.d. All scale bars are 10 µm.
Extended Data Fig. 3 Nuclear lamina defects caused by SETD2 loss arise from events dysregulated during G2/M.
a, Western blot analysis of indicated proteins from control and SETD2-KD RPE cells that are either arrested in G0/G1 or growing asynchronously (Async). b, Representative images of control and SETD2-KD RPE cells that are either proliferating (Async) or arrested in G0/G1, immunostained with anti-lamin A/C antibody. DNA counterstained with DAPI. Scale bars, 10 µm. c, Quantification of nuclear defects observed in b. Two-tailed unpaired Student’s t-test was performed from two independent biological replicates with n (left to right) = 357, 318, 395, 424 cells; mean ± s.d. d, Representative flow cytometry analysis of control and SETD2-KD RPE cells growing asynchronously. e, Representative flow cytometry analysis of control and SETD2-KD RPE cells arrested in G0/G1. Y-axis is EdU intensity and x-axis DNA content. f, Immunoblot analysis of indicated proteins in two independent replicates of control and SETD2-KD HKC cells. g, Quantitative analysis of γH2AX intensity in untreated (control) and dTAG-47 treated SETD2–FKBP HKC cells that were synchronized by the double thymidine block method and released for indicated timepoints. Integrated intensity per nucleus was measured from n (left to right) = 2,349, 2,180, 2,044, 2,061, 3,188, 3,191 cells. Kruskal–Wallis with Dunn’s multiple comparisons test was performed on data from two independent biological replicates; medians in red.
Extended Data Fig. 4 SETD2 loss impairs lamin phosphorylation during G2/M.
a, Representative time-lapse images of control and SETD2-depleted HKC expressing Emerald-lamin A (green). DNA was labelled with SiR-Hoechst (red). Arrowheads point to partially depolymerized lamin filaments observed throughout mitosis. b, Representative flow cytometry analysis of control and SETD2-KD cells to determine cell-cycle distribution. Y-axis is EdU intensity and x-axis DNA content. c, Quantitation of flow cytometry analysis data showing cell-cycle distribution of control and SETD2-KD cells. Two-tailed unpaired Student’s t-test was performed from three independent biological replicates; mean ± s.d. d, Quantitative analysis of immunofluorescence data from control and SETD2-KD RPE cells. Integrated fluorescence intensity of either S22ph- or S392ph-lamin A signal from mitotic cells normalized to integrated intensity of pan lamin A/C signal in control (n = 505, S22ph; n = 390, S392ph) and shSETD2 (n = 490, S22ph; n = 382, S392ph) cells from two independent biological replicates. Two-tailed non-parametric Mann–Whitney U-test was performed; medians in red. e, Quantitative analysis of immunofluorescence data from control and Setd2-KO MEF cells. Integrated fluorescence intensity of either S22ph- or S392ph-lamin A signal from mitotic cells normalized to integrated intensity of pan lamin A/C signal in control (n = 515, S22ph; n = 407, S392ph) and Setd2-KO (n = 457, S22ph; n = 327, S392ph) cells from two independent biological replicates. Two-tailed non-parametric Mann–Whitney U-test was performed; medians in red. f, Representative images of LMNA KO human fibroblasts cells expressing either WT, S- > A (S22A; S392A) or S- > D (S22D; S392D) mutant lamin A. Cells were immunostained with lamin A/C antibody (red) and DNA counterstained with DAPI. All scale bars are 10 µm. g, qRT-PCR analysis of SETD2 and Cyclin B1 mRNA levels during G2/M, normalized to Actin mRNA. X-axis is time (h) post release from a double thymidine block, similar to Fig. 3e. Y-axis is fold change relative to 0 hr timepoint. Paired Student’s t-test comparing different timepoints for each gene was performed from two independent biological replicates.
Extended Data Fig. 5 Non-enzymatic function of SETD2 in maintenance of nuclear morphology.
a, Immunoblot analyses of indicated proteins from control, Tamoxifen (Setd2-KO) and SETD2i (EPZ-719)-treated MEF cells for 48 h. b, Representative images of control, Setd2-KO and SETD2i-treated MEF cells immunostained with lamin B1 antibody. c, Quantitative analysis of nuclear defects from b. Two-tailed unpaired Student’s t-test was performed from two independent biological replicates; mean ± s.d.; DNA counterstained with DAPI. d, Representative images of control, SETD2-KD or SETD2i (EPZ-719) treated HKC cells for 2 d or 6 d, immunostained with lamin B1 antibody. DNA counterstained with DAPI. Scale bars, 10 µm.
Extended Data Fig. 6 The N terminus of SETD2 regulates nuclear lamina stability.
a, Immunoblot analysis for the indicated proteins following immunoprecipitation of endogenous SETD2 from asynchronous or nocodazole-arrested mitotic cells, showing SETD2 associates with CDK1 more robustly during mitosis. b, Representative images of Proximity Ligation Assay (PLA) analysis using anti-lamin B1 and anti-CDK1 antibodies in SETD2-KD cells expressing either WT-SETD2 or tSETD2. PLA pairs are green and DNA was counterstained with DAPI. c, Representative images of SETD2-depleted cells expressing either WT SETD2 or the indicated N-terminal mutants, immunostained with lamin B1 (green). SETD2 (red) visualized using Halo ligand JFX-549. DNA was counterstained with DAPI. Scale bars, 10 µm.
Extended Data Fig. 7 SETD2 N terminus enhances CDK1-mediated lamin phosphorylation and is required to suppress ccRCC tumour growth.
a, Long exposure of western blot analyses (from Fig. 6a) with anti-SUMO antibody following in vitro GST pulldown assays using either GST-tagged lamin A N-terminus (NT) or C-terminus (CT) or free GST (control). b, Immunoblot analyses using indicated antibodies following GST pulldown assays using either GST-tagged CDK1/Cyclin A2 complex or free GST (control). Anti-SUMO antibody was used to detect SUMO-tagged SETD2 (B/C) proteins. Asterisk indicates cross-reacting band of His–SUMO-B–GB1 in the anti-GST immunoblot. This is because the GB1 (B1 domain of Protein G) exhibits significant affinity towards most immunoglobulin IgG. c, Immunoblot analysis of indicated proteins following an in vitro kinase assay using lamin A-NT as substrate and CDK1/Cyclin B1 as kinase in the presence of either SETD2-B or SETD2-C or a control His–SUMO-GB1 protein. Red asterisk highlights cross-reacting band of His–SUMO-B–GB1 due to aforementioned reasons. d, Western blot with indicated antibodies from UMRC2 cells expressing either Halo (control) or Halo-3xFlag tagged WT-SETD2 or tSTED2 or BC-tSETD2. e, Representative images of UMRC2 cells expressing either Halo (control) or Halo-tagged WT SETD2 or tSETD2 or BC-tSETD2, immunostained with pan-lamin A/C antibody. DNA counterstained with DAPI; scale bar, 10 µm. f, Representative images of the full 35 mm cell culture well with 3D colony growth assay of UMRC2 cells expressing the indicated transgenes; scale bar is 10 mm. g, Xenograft tumour growth measurements (volume) of UMRC2 cells expressing the indicated transgene (n = 9). Related to Fig. 6j; mean ± s.e.m.
Extended Data Fig. 8
Flow analysis gating strategy.
Supplementary information
Source data
Source Data
Unprocessed western blots for all figures.
Source Data
Numerical source data for all figures.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khan, A., Zhang, C., Nguyen, P.G. et al. A SETD2–CDK1–lamin axis maintains nuclear morphology and genome stability. Nat Cell Biol 27, 1327–1341 (2025). https://doi.org/10.1038/s41556-025-01723-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01723-9
This article is cited by
-
Molecular mechanism of co-transcriptional H3K36 methylation by SETD2
Nature Communications (2025)