Abstract
Chemoresistance is the leading cause of cancer-related death. How chemotherapy subjugates the cellular crosstalk in the tumour microenvironment to cause chemoresistance remains to be defined. Here we find chemotherapy enables immunosuppressive SDF1+ endothelial niche to evade immunosurveillance in ovarian and breast cancers. We integrated human patient data and mouse models to show that chemotherapy selectively activates PARP1–SDF1 axis in tumour endothelial cells (ECs). This angiocrine SDF1 interferes with antitumour interplay between CXCL10+ macrophages and CXCR3+CD8+ T cells and promotes tumour progression in ovarian and breast cancers. Proteome-based screening revealed that endothelial PARP1 PARylates MLKL, a key necroptosis effector to upregulate angiocrine SDF1 in ECs. In sum, we identify PARylation-dependent necroptosis in tumour ECs as an important step in subverting the tumour microenvironment to evade immunosurveillance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
scRNA-seq and bulk RNA sequencing that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE298945. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD064443 and PXD064394. All other data supporting the findings of this study are available from the corresponding authors on reasonable request. Source data are provided with this paper.
References
Nakasone, E. S. et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell 21, 488–503 (2012).
Muranen, T. et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell 21, 227–239 (2012).
Keklikoglou, I. et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat. Cell Biol. 21, 190–202 (2019).
Patch, A. M. et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521, 489–494 (2015).
Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308 (2018).
Hayakawa, Y. et al. Mist1 expressing gastric stem cells maintain the normal and neoplastic gastric epithelium and are supported by a perivascular stem cell niche. Cancer Cell 28, 800–814 (2015).
Pitt, L. A. et al. CXCL12-producing vascular endothelial niches control acute T cell leukemia maintenance. Cancer Cell 27, 755–768 (2015).
Ghajar, C. M. et al. The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15, 807–817 (2013).
Wynn, T. A., Chawla, A. & Pollard, J. W. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455 (2013).
Ruffell, B. & Coussens, L. M. Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472 (2015).
Locati, M., Curtale, G. & Mantovani, A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev. Pathol. 15, 123–147 (2020).
Tavazoie, M. F. et al. LXR/ApoE activation restricts innate immune suppression in cancer. Cell 172, 825–840 (2018).
Nalio Ramos, R. et al. Tissue-resident FOLR2+ macrophages associate with CD8+ T cell infiltration in human breast cancer. Cell 185, 1189–1207 (2022).
Sanmamed, M. F. & Chen, L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell 175, 313–326 (2018).
Littman, D. R. Releasing the brakes on cancer immunotherapy. Cell 162, 1186–1190 (2015).
Chow, M. T. et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity 50, 1498–1512 (2019).
Lord, C. J., Garrett, M. D. & Ashworth, A. Targeting the double-strand DNA break repair pathway as a therapeutic strategy. Clin. Cancer Res. 12, 4463–4468 (2006).
Thomas, A., Murai, J. & Pommier, Y. The evolving landscape of predictive biomarkers of response to PARP inhibitors. J. Clin. Investig. 128, 1727–1730 (2018).
Murai, J. et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 72, 5588–5599 (2012).
Pettitt, S. J. et al. Genome-wide and high-density CRISPR–Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018).
Suskiewicz, M. J. et al. HPF1 completes the PARP active site for DNA damage-induced ADP-ribosylation. Nature 579, 598–602 (2020).
Li, Z. et al. Shigella evades pyroptosis by arginine ADP-riboxanation of caspase-11. Nature 599, 290–295 (2021).
Fu, H. et al. Poly(ADP-ribosylation) of P-TEFb by PARP1 disrupts phase separation to inhibit global transcription after DNA damage. Nat. Cell Biol. 24, 513–525 (2022).
Cao, Z. et al. Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemoresistance. Cancer Cell 25, 350–365 (2014).
Pantelidou, C. et al. PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Newton, K. et al. Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 574, 428–431 (2019).
Sun, L. & Wang, X. A new kind of cell suicide: mechanisms and functions of programmed necrosis. Trends Biochem. Sci. 39, 587–593 (2014).
Strilic, B. et al. Tumour-cell-induced endothelial cell necroptosis via death receptor 6 promotes metastasis. Nature 536, 215–218 (2016).
Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012).
Ying, Z. et al. Mixed lineage kinase domain-like protein MLKL breaks down myelin following nerve injury. Mol. Cell 72, 457–468 (2018).
Jiao, S. et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin. Cancer Res. 23, 3711–3720 (2017).
Muvarak, N. E. et al. Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents-a potential therapy for cancer. Cancer Cell 30, 637–650 (2016).
Ding, X. et al. Synthetic viability by BRCA2 and PARP1/ARTD1 deficiencies. Nat. Commun. 7, 12425 (2016).
Jungmichel, S. et al. Proteome-wide identification of poly(ADP-ribosyl)ation targets in different genotoxic stress responses. Mol. Cell 52, 272–285 (2013).
Grimaldi, G., Catara, G., Valente, C. & Corda, D. In vitro techniques for ADP-ribosylated substrate identification. Methods Mol. Biol. 1813, 25–40 (2018).
Pasparakis, M. & Vandenabeele, P. Necroptosis and its role in inflammation. Nature 517, 311–320 (2015).
Ozga, A. J. et al. CXCL10 chemokine regulates heterogeneity of the CD8+ T cell response and viral set point during chronic infection. Immunity 55, 82–97 (2022).
House, I. G. et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res. 26, 487–504 (2020).
Ma, X. et al. CD36-mediated ferroptosis dampens intratumoral CD8+ T cell effector function and impairs their antitumor ability. Cell Metab. 33, 1001–1012 e1005 (2021).
Klug, F. et al. Low-dose irradiation programs macrophage differentiation to an iNOS+/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell 24, 589–602 (2013).
Barker, K. T. & Yogev, O. Establishment of in vivo acquired resistance to chemotherapy via individual dose escalation treatment regime. Methods Mol. Biol. https://doi.org/10.1007/978-1-0716-2513-2_6 (2022).
Clark, M., Kollara, A., Brown, T. J. & May, T. A mouse model of neoadjuvant chemotherapy followed by interval cytoreductive surgery indicates impaired efficacy of perioperative cisplatin. J. Ovarian Res. 14, 157 (2021).
Mañas, A. et al. Clinically relevant treatment of PDX models reveals patterns of neuroblastoma chemoresistance. Sci. Adv. 8, eabq4617 (2022).
Alexander, E. T., Minton, A. R., Peters, M. C., van Ryn, J. & Gilmour, S. K. Thrombin inhibition and cisplatin block tumor progression in ovarian cancer by alleviating the immunosuppressive microenvironment. Oncotarget 7, 85291–85305 (2016).
El-Khamisy, S. F., Masutani, M., Suzuki, H. & Caldecott, K. W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31, 5526–5533 (2003).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019).
Jin, S. et al. Inference and analysis of cell–cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Gong, Y. et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer 18, 100 (2019).
Kusnadi, A. et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8+ T cells. Sci. Immunol. 6, eabe4782 (2021).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Büttner, M., Ostner, J., Müller, C. L., Theis, F. J. & Schubert, B. scCODA is a Bayesian model for compositional single-cell data analysis. Nat. Commun. 12, 6876 (2021).
Rafii, S. et al. Platelet-derived SDF-1 primes the pulmonary capillary vascular niche to drive lung alveolar regeneration. Nat. Cell Biol. 17, 123–136 (2015).
Cao, Z. et al. Molecular checkpoint decisions made by subverted vascular niche transform indolent tumor cells into chemoresistant cancer stem cells. Cancer Cell 31, 110–126 (2017).
Zhang, H. et al. Targeting epigenetically maladapted vascular niche alleviates liver fibrosis in nonalcoholic steatohepatitis. Sci. Transl. Med. 13, eabd1206 (2021).
Acknowledgements
This work was supported by grants from Noncommunicable Chronic Diseases-National Science and Technology Major Project (grant no. 2023ZD0506004 to B.-S.D.), the National Natural Science Foundation of China (grant nos. 82125002 and 92268201 to Z.C., 82525101 to B.-S.D. and 82203652 to G.J.), the Natural Science Foundation of Sichuan Province (grant nos. 2023NSFSC0003 to Z.C. and 2024NSFTD0021 to B.-S.D.), the Key Research and Development Program Project of Ningxia Hui Autonomous Region (grant no. 2024BEG01001 to B.-S.D.), launching project from Ningxia Basic Medical Research Center, Ningxia Medical University (to B.-S.D.), and Open Competition Mechanism to Select the Best Candidates for Key Research Projects of Ningxia Medical University (to X.L.). We acknowledge X. Wang for generously providing the MlklloxP/loxP mouse strain.
Author information
Authors and Affiliations
Contributions
These authors contributed equally: N.Y., X.L., W.H., G.J., W.L. and F.J. Z.C., B.-S.D., N.Y., X.L., W.H., G.J., W.L. and F.J. designed experiments and wrote and edited the paper. Z.C., B.-S.D., N.Y., X.L., W.H., G.J., W.L. and H.Z. performed experiments and analysed the data for all figures. Yali Chen, Yao Chen, L.Q., L.C., S.R., W.W. and A.Z. provided technical support, comments and suggestions.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
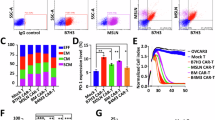
Extended Data Fig. 1 Proteome based identification of PARP1 target protein.
a, Flowchart of ADP-ribose modified protein target screening in human primary ECs. HUVECs were pretreated with 5 μg/mL CBP for 48 hours to induce PARP1 activation and lysed. Cell lysates were incubated with agarose conjugated with MAR/PAR-binding module GST-Af1521-WT or GST-Af1521-Mutant without binding activity. ADP-ribose modified proteins were then identified by mass spectrometry. b, GST-tagged Af1521, RIPK1, RIPK3, and MLKL was incubated with 20 pmol PAR in reaction buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1% NP-40) respectively. Individual protein was pull down with anti-GST antibody and analyzed by immunoblotting with anti-PAR antibody. Representative blot is shown in figures. c, Identification of MLKL as a PARP1 substrate protein by mass spectrometry (MS) analysis. HUVECs lysates retrieved in step (a) were analyzed with SDS-PAGE. Gel slices were digested and fractionated by high PH reverse-phase HPLC equipped with Agilent 300Extend C18 column. LC-MS/MS was performed with Q Exactive Plus mass spectrometer (Thermo). Raw data were processed with Maxquant search engine.d, Co-immunoprecipitation shows interaction between endogenous MLKL and PARP1 in HUVECs. HUVECs were treated with 5 μg/mL CBP for 48 hours. Endogenous PARP1 was immunoprecipitated by anti-PARP1 (Abcam), and PARP1 associated endogenous MLKL was detected by MLKL antibody (Huabio). Endogenous MLKL was immunoprecipitated by MLKL antibody (Huabio) and MLKL associated endogenous PARP1 was detected by PARP1 antibody (Abcam). Experiments were repeated three times, and representative western blot image is shown. Each lane represents one biological sample, and three biological replicates were used.
Extended Data Fig. 2 Identification of MLKL PARylation sites by PARP1.
a, Schema illustrating mono(ADP-ribosyl)ated (MARylated) MLKL with mass spectrometry. Recombinant MLKL and recombinant PARP1 were incubated together in cell free reaction buffer and treated with poly(ADP-ribose) glycohydrolase (PARG) to acquire MARylated MLKL. MARylated MLKL was analyzed with mass spectrometry to identify amino acid residue that is modified by ADP-ribose. b, Identification of MLKL PARylation sites. MARylated MLKL was isolated with SDS-PAGE. The gel slices containing desired proteins were analyzed with mass spectrometry to identify amino acid residue that is modified by ADP-ribose. c, Schema illustrating the construction of MLKLWT ECs, MLKLE351A ECs, MLKLT253A ECs or MLKLE351A/T253A ECs (HUVECs). shRNA targeting the intron of MLKL mRNA was introduced into HUVECs to construct MLKL-null ECs. MLKLWT, MLKLE351A or MLKLE351A/T253A double mutants were introduced into MLKL-null ECs via lentivirus to construct MLKLWT ECs, MLKLE351A ECs, MLKLT253A ECs or MLKLE351A/T253A ECs. d, MLKL protein structure was analyzed with AlphaFold2. It is predicted that modification on MLKL E351 and T253 alter MLKL protein conformation to facilitate MLKL phosphorylation. e, f, MLKLWT ECs or MLKLE351A/T253A ECs (HUVECs) were treated with or without 5 μg/mL CBP for 48 hours. The level of cell death was determined by PI staining and analyzed with flow cytometry (e). Quantification of percentage of PI+ cells was shown (f) (n = 5 biologically replicates). ****P < 0.0001 was calculated using two-way ANOVA followed by Tukey’s test as post-hoc analysis for f.
Extended Data Fig. 3 Chemotherapy enhances SDF1 expression via upregulating c-JUN.
a, SCENIC package was used to analyze ECs from breast cancer patients and ovarian cancer patients with or without chemotherapy. The activity score of each transcription factor (TF) regulon was calculated and ranked. Top 10 TF regulons are marked. b, The gene sets representing upregulated TF activity after chemotherapy in breast and ovarian cancer patients were identified and then overlapped with necroptosis pathway (GSE121149) to perform Venn diagram analysis. Top 10 genes were used in each gene set. c, The scatter plot showed the activity of c-JUN transcription factor in the ECs of breast and ovarian cancer patients from scRNA-seq data. The black horizontal line in the plot represents the average value of the two groups. d, Cut&Tag assay of SDF1 promoter was performed with anti-c-JUN antibody. Promoter binding ability was determined with qPCR (n = 5 biological replicates). e, c-JUN was overexpressed in HUVECs. SDF1 promoter activity was determined by luciferase reporter assay (n = 4 biological replicates). f, HUVECs stimulated by CBP were treated with increasing doses of T5224. SDF1 promoter activity was determined by luciferase reporter assay (n = 4 biological replicates). g-j, PARP1 or MLKL was silenced with shRNA in HUVECs and treated with CBP. The expression levels of SDF1 and c-JUN were measured with western blot (g, i) and quantified by ImageJ (h, j) (n = 6 biological replicates). k, Schema illustrating the mechanism by which c-JUN binds to the SDF1 promoter, leading to increased SDF1 expression in ECs post-chemotherapy. ****P < 0.0001 was calculated using two-sided unpaired Wilcoxon rank sum test for c or two-way ANOVA followed by Tukey’s test as post-hoc analysis for d, e, f, h and j.
Extended Data Fig. 4 Chemotherapy decrease CXCL10+ macrophage population in breast and ovarian tumors through CXCR4 pathway.
a, Cell-type compositional analysis of denoted macrophage subclusters. Statistical difference is revealed by scCODA and indicated by red bars. Each dot represents a sample (n = 5 patients for BC Chemo-, BC chemo + , OC chemo- groups; n = 4 patients for OC chemo+ group). The box represents the interquartile range with median, minimum and maximum represented by the box centre line and whiskers, respectively. b, scRNA-seq analysis showing chemotherapy decreased CXCL10 expression in macrophages derived from breast cancer and ovarian cancer patient tumors. c, Schema illustrating generation of myeloid cell specific deletion of Cxcr4. Cxcr4loxP/loxP mice were crossed with mice expressing myeloid cell-specific LysM-Cre to generate Cxcr4ΔM/ΔM mice, enabling specific deletion of Cxcr4 in macrophages. d, e, Characterization of myeloid cell specific deletion of CXCR4 (Cxcr4ΔM/ΔM) mice. Breast cancer cells (E0771) were transplanted into the mouse mammary fat pad. Tumor associated macrophages were isolated. CXCR4 expression was analyzed by western blot (d) and optical density was quantified by ImageJ (e) (n = 8 biological replicates). f, g, Reduction of CXCL10+ macrophage population in mouse breast tumor after CBP was recovered in Cxcr4ΔM/ΔM mice. Breast cancer cells (E0771) were transplanted into the mouse mammary fat pad. Percentage of CXCL10+ macrophage population from indicated mice was analyzed by flow cytometry (f) and compared (g) (n = 8 biological replicates). ****P < 0.0001 were calculated using two-way ANOVA followed by Tukey’s test as post-hoc analysis for e and g.
Extended Data Fig. 5 Depletion of CD8 T cells enhanced the growth of transplanted tumor cells.
a, EC-specific deletion of Parp1 increased CD8+ T cell number in the presence of CBP. CD8 T cell number (per mg of tumor) was quantified by flow cytometry. Each dot represents one individual animal (n = 8 biological replicates). b, Flowchart of CD8+ T cell depletion by anti-CD8a antibody in E0771 tumor-bearing mice. Depletion of CD8+ T cells was achieved by intraperitoneal injections of 100 μg of anti-mouse CD8a antibody suspended in 200 μL of PBS every 4 days, starting one day before tumor cell injection. Depletion of CD8 T cells was confirmed by flow cytometry analyses of peripheral blood at day 5 and day 13. c, Flow-cytometry plots of CD8+ populations in peripheral blood at day 5 and 13 demonstrate that CD8+ T cell population was depleted by αCD8 antibody (n = 8 biological replicates). d, e, Breast tumor tissues were excised from Sdf1iΔEC/iΔEC or control Sdf1loxP/loxP mice and analyzed with flow cytometry (d). Percentage of CXCR3+CD8+ T cell population in the retrieved CD8+ T cells from indicated mice was compared (e). Each dot represents sample from individual animals (n = 8 biological replicates). **P < 0.01, ****P < 0.0001 and N.S. were calculated using two-sided unpaired t-test for a or two-way ANOVA followed by Tukey’s test as post-hoc analysis for c and e.
Extended Data Fig. 6 Genetic deletion of Mlkl in ECs effectively prevented the development of chemoresistance in ovarian tumor.
a, b, Genetic deletion of Mlkl in ECs (MlkliΔEC/iΔEC) attenuated the development of chemoresistance in orthotopic ovarian tumor model after repeated CBP treatments. Tumor growth curves were generated by averaging whole-body bioluminescence imaging in MlkliΔEC/iΔEC or control (MlklloxP/loxP) mice (a) (n = 13 biological replicates). The duration of each chemotherapy cycle was determined and compared between MlkliΔEC/iΔEC and control mice (b). c, d, Genetic deletion of Mlkl in ECs inhibited increased Sdf1 expression in ECs after CBP treatment. Tumor associated ECs were isolated at each chemotherapy cycle and the transcriptional levels of Sdf1 were measured by qPCR (n = 10 biological replicates). e-g, Genetic deletion of Mlkl in ECs recovered CXCL10+ macrophages that were decreased by CBP treatment. Tumor associated macrophages were isolated at the end of each chemotherapy cycle and CXCL10+ macrophage percentage was determined by flow cytometry (n = 10 biological replicates). h-j, Genetic deletion of Mlkl in ECs recovered the number of CXCR3+CD8+ T cells that were decreased by CBP. Tumor associated T cells were isolated at the end of each chemotherapy cycle and CXCR3+CD8+ T cell percentage was determined by flow cytometry (n = 10 biological replicates). The data are presented as the mean ± S.E.M. for a, d, g and j. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 and N.S. were calculated using one-way ANOVA followed by Tukey’s test as post-hoc analysis for f and i or two-way ANOVA followed by Tukey’s test as post-hoc analysis for a, c, d, g and j.
Supplementary information
Supplementary Information
Supplementary Figs. 1–22, methods, methods-only references and unprocessed blots.
Supplementary Tables 1–7
Supplementary Table 1: heat map genes related to Fig. 3f. Supplementary Table 2: gene list related to Figs. 3i, 7f, 7i, 7j, 7k and 7l. Supplementary Table 3: heat map genes related to Fig. 4l. Supplementary Table 4: heat map genes related to Fig. 5i. Supplementary Table 5: heat map genes related to Fig. 6d. Supplementary Table 6: representative T cell subpopulation marker genes scale expression, related to Fig. 7c. Supplementary Table 7: list of shRNA targets, related to Methods. Supplementary Table 8: summary statistics for all quantified data presented in the figures.
Supplementary Data 1
Statistical source data for Supplementary Figs. 1–22.
Supplementary Data 2
Summary statistics for all quantified data presented in Supplementary Figs. 1–22.
Supplementary Data 3
List of qPCR primers.
Source data
Source Data Fig. 1
Unprocessed western blots for Fig. 1.
Source Data Fig. 1
Statistical source data for Fig. 1.
Source Data Fig. 2
Unprocessed western blots for Fig. 2.
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 3
Statistical source data for Fig. 3.
Source Data Fig. 4
Unprocessed western blots for Fig. 4.
Source Data Fig. 4
Statistical source data for Fig. 4.
Source Data Fig. 5
Statistical source data for Fig. 5.
Source Data Fig. 6
Statistical source data for Fig. 6.
Source Data Fig. 7
Statistical source data for Fig. 7.
Source Data Fig. 8
Statistical source data for Fig. 8.
Source Data Extended Data Fig. 1
Unprocessed western blots for Extended Data Fig. 1.
Source Data Extended Data Fig. 2
Statistical source data for Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Unprocessed western blots for Extended Data Fig. 3.
Source Data Extended Data Fig. 3
Statistical source data for Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Unprocessed western blots for Extended Data Fig. 4.
Source Data Extended Data Fig. 4
Statistical source data for Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Statistical source data for Extended Data Fig. 5.
Source Data Extended Data Fig. 6
Statistical source data for Extended Data Fig. 6.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, N., Li, X., Huang, W. et al. MLKL PARylation in the endothelial niche triggers angiocrine necroptosis to evade cancer immunosurveillance and chemotherapy. Nat Cell Biol 27, 1526–1542 (2025). https://doi.org/10.1038/s41556-025-01740-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-025-01740-8