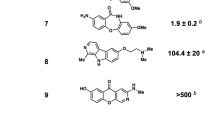
Abstract
Potent peptide ligands for therapeutically relevant targets are regularly returned from screening trillion-member libraries of ribosomally synthesized peptides containing non-canonical amino acids and macrocyclic architectures. Yet the chemical space explored by these peptides is a fraction of that embodied by natural products and pharmaceuticals, and most peptide leads require exhaustive medicinal chemistry optimization to improve potency and physicochemistry. To address the need for strategies to introduce chemical complexity and conformational control into peptide macrocycles, we report here that linear peptides with a reactive N-terminal β-keto or γ-keto amide can be synthesized ribosomally. Subsequent Friedländer reactions generate quinoline–peptide hybrids, some of which contain stable biaryl atropisomeric axes. We also demonstrate intramolecular Friedländer macrocyclization reactions—sufficiently mild to be employed on unprotected and in vitro-translated peptides—that embed a quinoline pharmacophore directly within the peptide backbone. The introduction of N-terminal ketone motifs into genetically encoded materials and their post-translational derivatization provides a paradigm for the programmed synthesis of peptide-derived materials that more closely resemble complex natural products.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
FAIR data for this paper, including .fid files and other primary characterization data for all figures, are available via Figshare at https://doi.org/10.6084/m9.figshare.25968364 (ref. 112). Crystallographic data for the structures reported in this Article have been deposited at the Cambridge Crystallographic Data Centre (CCDC) under deposition numbers CCDC 2341552 ((M,S)-29), 2341553 ((P,S)-29) and 2395208 ((P)-CPa). Copies of these data can be obtained free of charge via www.ccdc.cam.ac.uk/structures. Source data are provided with this paper.
References
Sims, E. K., Carr, A. L. J., Oram, R. A., DiMeglio, L. A. & Evans-Molina, C. 100 years of insulin: celebrating the past, present and future of diabetes therapy. Nat. Med. 27, 1154–1164 (2021).
Banting, F. G., Best, C. H., Collip, J. B., Campbell, W. R. & Fletcher, A. A. Pancreatic extracts in the treatment of diabetes mellitus. Can. Med. Assoc. J. 12, 141–146 (1922).
Wilding, J. P. H. et al. Once-weekly semaglutide in adults with overweight or obesity. N. Engl. J. Med. 384, 989–1002 (2021).
Müller, T. D., Blüher, M., Tschöp, M. H. & DiMarchi, R. D. Anti-obesity drug discovery: advances and challenges. Nat. Rev. Drug Discov. 21, 201–223 (2022).
Schepartz, A. & Kim, P. S. Interaction, assembly and processing at the chemistry—biology interface. Curr. Opin. Chem. Biol. 2, 9–10 (1998).
Salveson, P. J. et al. Expansive discovery of chemically diverse structured macrocyclic oligoamides. Science 384, 420–428 (2024).
Muttenthaler, M., King, G. F., Adams, D. J. & Alewood, P. F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 20, 309–325 (2021).
Costa, L., Sousa, E. & Fernandes, C. Cyclic peptides in pipeline: what future for these great molecules? Pharmaceuticals 16, 996 (2023).
Zorzi, A., Deyle, K. & Heinis, C. Cyclic peptide therapeutics: past, present and future. Curr. Opin. Chem. Biol. 38, 24–29 (2017).
Yang, J. et al. Utilization of macrocyclic peptides to target protein-protein interactions in cancer. Front. Oncol. 12, 992171 (2022).
Ohta, A. et al. Validation of a new methodology to create oral drugs beyond the rule of 5 for intracellular tough targets. J. Am. Chem. Soc. 145, 24035–24051 (2023).
Tucker, T. J. et al. A series of novel, highly potent, and orally bioavailable next-generation tricyclic peptide PCSK9 inhibitors. J. Med. Chem. 64, 16770–16800 (2021).
Li, X., Craven, T. W. & Levine, P. M. Cyclic peptide screening methods for preclinical drug discovery. J. Med. Chem. 65, 11913–11926 (2022).
Sawyer, T. in Contemporary Accounts in Drug Discovery and Development (eds Huang, X. et al.) 273–292 (Wiley, 2022).
Howard, J. F. et al. Safety and efficacy of zilucoplan in patients with generalised myasthenia gravis (RAISE): a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Neurol. 22, 395–406 (2023).
Rezai, T., Yu, B., Millhauser, G. L., Jacobson, M. P. & Lokey, R. S. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J. Am. Chem. Soc. 128, 2510–2511 (2006).
Rossi Sebastiano, M. et al. Impact of dynamically exposed polarity on permeability and solubility of chameleonic drugs beyond the rule of 5. J. Med. Chem. 61, 4189–4202 (2018).
Vinogradov, A. A., Yin, Y. & Suga, H. Macrocyclic peptides as drug candidates: recent progress and remaining challenges. J. Am. Chem. Soc. 141, 4167–4181 (2019).
Kawakami, T., Murakami, H. & Suga, H. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem. Biol. 15, 32–42 (2008).
Katoh, T., Tajima, K. & Suga, H. Consecutive elongation of D-amino acids in translation. Cell Chem. Biol. 24, 46–54 (2017).
Katoh, T. & Suga, H. Ribosomal incorporation of consecutive β-amino acids. J. Am. Chem. Soc. 140, 12159–12167 (2018).
Iskandar, S. E. et al. Enabling genetic code expansion and peptide macrocyclization in mRNA display via a promiscuous orthogonal aminoacyl-tRNA synthetase. J. Am. Chem. Soc. 145, 1512–1517 (2023).
Ekanayake, A. I. et al. Genetically encoded fragment-based discovery from phage-displayed macrocyclic libraries with genetically encoded unnatural pharmacophores. J. Am. Chem. Soc. 143, 5497–5507 (2021).
Schissel, C. K. et al. Peptide backbone editing via post-translational O to C acyl shift. J. Am. Chem. Soc. 147, 6503–6513 (2025).
Roe, L. T. et al. Site-selective protein editing by backbone extension acyl rearrangements. Nat. Chem. Bio. https://doi.org/10.1038/s41589-025-01999-w (2025).
He, J., Ghosh, P. & Nitsche, C. Biocompatible strategies for peptide macrocyclisation. Chem. Sci. 15, 2300–2322 (2024).
White, C. J. & Yudin, A. K. Contemporary strategies for peptide macrocyclization. Nat. Chem. 3, 509–524 (2011).
Bechtler, C. & Lamers, C. Macrocyclization strategies for cyclic peptides and peptidomimetics. RSC Med. Chem. 12, 1325–1351 (2021).
Miller, S. J., Blackwell, H. E. & Grubbs, R. H. Application of ring-closing metathesis to the synthesis of rigidified amino acids and peptides. J. Am. Chem. Soc. 118, 9606–9614 (1996).
Walensky, L. D. & Bird, G. H. Hydrocarbon-stapled peptides: principles, practice, and progress. J. Med. Chem. 57, 6275–6288 (2014).
Rivera, D. G., Ojeda-Carralero, G. M., Reguera, L. & Eycken, E. V. Vder Peptide macrocyclization by transition metal catalysis. Chem. Soc. Rev. 49, 2039–2059 (2020).
Liu, M. et al. Selective thiazoline peptide cyclisation compatible with mRNA display and efficient synthesis. Chem. Sci. 14, 10561–10569 (2023).
Liu, M. et al. An efficient, site-selective and spontaneous peptide macrocyclisation during in vitro translation. Chem. Eur. J. 29, e202203923 (2023).
Abdelkader, E. H. et al. Genetic encoding of cyanopyridylalanine for in-cell protein macrocyclization by the nitrile–aminothiol click reaction. Angew. Chem. Int. Ed. 61, e202114154 (2022).
Patil, N. A. et al. 2-Cyanoisonicotinamide conjugation: a facile approach to generate potent peptide inhibitors of the Zika virus protease. ACS Med. Chem. Lett. 12, 732–737 (2021).
Thombare, V. J. et al. Advancing nitrile-aminothiol strategy for dual and sequential bioconjugation. Chem. Eur. J. 30, e202401674 (2024).
Fleming, M. C., Bowler, M. M., Park, R., Popov, K. I. & Bowers, A. A. Tyrosinase-catalyzed peptide macrocyclization for mRNA display. J. Am. Chem. Soc. 145, 10445–10450 (2023).
Bowler, M. M., Glavatskikh, M., Pecot, C. V., Kireev, D. & Bowers, A. A. Enzymatic macrolactamization of mRNA display libraries for inhibitor selection. ACS Chem. Biol. 18, 166–175 (2023).
Vinogradov, A. A. et al. De novo discovery of thiopeptide pseudo-natural products acting as potent and selective TNIK kinase inhibitors. J. Am. Chem. Soc. 144, 20332–20341 (2022).
Passioura, T. The road ahead for the development of macrocyclic peptide ligands. Biochemistry 59, 139–145 (2020).
Melsen, P. R. A., Yoshisada, R. & Jongkees, S. A. K. Opportunities for expanding encoded chemical diversification and improving hit enrichment in mRNA-displayed peptide libraries. ChemBioChem 23, e202100685 (2022).
Smolyar, I. V., Yudin, A. K. & Nenajdenko, V. G. Heteroaryl rings in peptide macrocycles. Chem. Rev. 119, 10032–10240 (2019).
Dengler, S. et al. Display selection of a hybrid foldamer–peptide macrocycle. Angew. Chem. Int. Ed. 62, e202308408 (2023).
Diaz, D. B. et al. Illuminating the dark conformational space of macrocycles using dominant rotors. Nat. Chem. 13, 218–225 (2021).
Saunders, G. J. & Yudin, A. K. Property-driven development of passively permeable macrocyclic scaffolds using heterocycles. Angew. Chem. Int. Ed. 61, e202206866 (2022).
Yadav, P. & Shah, K. Quinolines, a perpetual, multipurpose scaffold in medicinal chemistry. Bioorg. Chem. 109, 104639 (2021).
Musiol, R. An overview of quinoline as a privileged scaffold in cancer drug discovery. Expert Opin. Drug Discov. 12, 583–597 (2017).
Childs-Disney, J. L. et al. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 21, 736–762 (2022).
Costales, M. G., Childs-Disney, J. L., Haniff, H. S. & Disney, M. D. How we think about targeting RNA with small molecules. J. Med. Chem. 63, 8880–8900 (2020).
Haniff, H. S., Knerr, L., Chen, J. L., Disney, M. D. & Lightfoot, H. L. Target-directed approaches for screening small molecules against RNA targets. SLAS Discov. 25, 869–894 (2020).
Donlic, A. et al. R-BIND 2.0: an updated database of bioactive RNA-targeting small molecules and associated RNA secondary structures. ACS Chem. Biol. 17, 1556–1566 (2022).
Krishnamurthy, M., Gooch, B. D. & Beal, P. A. Peptide quinoline conjugates: a new class of RNA-binding molecules. Org. Lett. 6, 63–66 (2004).
Krishnamurthy, M., Gooch, B. D. & Beal, P. A. RNA binding and thiolytic stability of a quinoline-containing helix-threading peptide. Org. Biomol. Chem. 4, 639–645 (2006).
Chen, J. L. et al. Design, optimization, and study of small molecules that target tau pre-mRNA and affect splicing. J. Am. Chem. Soc. 142, 8706–8727 (2020).
Li, G., Zhu, D., Xue, L. & Jiang, H. Quinoline-based fluorescent probe for ratiometric detection of lysosomal pH. Org. Lett. 15, 5020–5023 (2013).
Fu, C. et al. Genetically encoding quinoline reverses chromophore charge and enables fluorescent protein brightening in acidic vesicles. J. Am. Chem. Soc. 140, 11058–11066 (2018).
Shao, Y.-D. et al. Organocatalytic atroposelective Friedländer quinoline heteroannulation. Org. Lett. 21, 4831–4836 (2019).
Ad, O. et al. Translation of diverse aramid- and 1,3-dicarbonyl-peptides by wild type ribosomes in vitro. ACS Cent. Sci. 5, 1289–1294 (2019).
Fricke, R. et al. Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-α-amino acids in vitro and in vivo. Nat. Chem. 15, 960–971 (2023).
Murakami, H., Ohta, A., Ashigai, H. & Suga, H. A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods 3, 357–359 (2006).
Fricke, R., Knudson, I., Swenson, C. V., Smaga, S. & Schepartz, A. Direct and quantitative analysis of tRNA acylation using intact tRNA liquid chromatography–mass spectrometry. Nat. Protoc. 20, 1246–1274 (2025).
Lee, J. et al. Ribosome-mediated biosynthesis of pyridazinone oligomers in vitro. Nat. Commun. 13, 6322 (2022).
Li, W., Zheng, Y., Qu, E., Bai, J. & Deng, Q. β-Keto amides: a Jack-of-all-trades building block in organic chemistry. Eur. J. Org. Chem. 2021, 5151–5192 (2021).
Wu, X. & Li, W. The applications of β-keto amides for heterocycle synthesis. J. Heterocycl. Chem. 59, 1445–1490 (2022).
Amarnath, V. & Amarnath, K. Intermediates in the Paal-Knorr synthesis of furans. J. Org. Chem. 60, 301–307 (1995).
Friedlaender, P. Ueber o-amidobenzaldehyd. Ber. Dtsch. Chem. Ges. 15, 2572–2575 (1882).
Cheng, C.-C. & Yan, S.-J. in Organic Reactions (ed. Dauben, W. G.) Vol. 28, 37–201 (John Wiley & Sons, 2005).
Marco-Contelles, J. et al. Recent advances in the Friedländer reaction. Chem. Rev. 109, 2652–2671 (2009).
Meléndez, A. et al. Straightforward synthesis of novel 4-styrylquinolines/4-styrylquinolin-2-ones and 9-styryldihydroacridin-1(2H)-ones from substituted 2′-aminochalcones. Synthesis 52, 1804–1822 (2020).
Shen, Q. et al. Synthesis of quinolines via Friedländer reaction in water and under catalyst-free conditions. Synthesis 44, 389–392 (2012).
Ţînţaş, M.-L. et al. Rational design of carbamate-based dual binding site and central AChE inhibitors by a “biooxidisable” prodrug approach: synthesis, in vitro evaluation and docking studies. Eur. J. Med. Chem. 155, 171–182 (2018).
Atechian, S. et al. New vistas in quinoline synthesis. Tetrahedron 63, 2811–2823 (2007).
Gopi, P. & Sarveswari, S. Effective water mediated green synthesis of polysubstituted quinolines without energy expenditure. Monatsh. Chem. 148, 1043–1049 (2017).
Bose, D. S., Idrees, M., Jakka, N. M. & Rao, J. V. Diversity-oriented synthesis of quinolines via Friedländer annulation reaction under mild catalytic conditions. J. Comb. Chem. 12, 100–110 (2010).
Albert, A. & Yamamoto, H. The structures of the anhydro-polymers of 2-aminobenzaldehyde. J. Chem. Soc. B 1966, 956–963 (1966).
Christie, G. H. & Kenner, J. LXXI.—The molecular configurations of polynuclear aromatic compounds. Part I. The resolution of γ-6:6′-dinitro- and 4:6:4′:6′-tetranitro-diphenic acids into optically active components. J. Chem. Soc. Trans. 121, 614–620 (1922).
Perreault, S., Chandrasekhar, J. & Patel, L. Atropisomerism in drug discovery: a medicinal chemistry perspective inspired by atropisomeric class I PI3K inhibitors. Acc. Chem. Res. 55, 2581–2593 (2022).
Basilaia, M., Chen, M. H., Secka, J. & Gustafson, J. L. Atropisomerism in the pharmaceutically relevant realm. Acc. Chem. Res. 55, 2904–2919 (2022).
LaPlante, S. R. et al. Assessing atropisomer axial chirality in drug discovery and development. J. Med. Chem. 54, 7005–7022 (2011).
Toenjes, S. T. & Gustafson, J. L. Atropisomerism in medicinal chemistry: challenges and opportunities. Future Med. Chem. 10, 409–422 (2018).
Janes, M. R. et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018).
Lanman, B. A., Parsons, A. T. & Zech, S. G. Addressing atropisomerism in the development of sotorasib, a covalent inhibitor of KRAS G12C: structural, analytical, and synthetic considerations. Acc. Chem. Res. 55, 2892–2903 (2022).
Fader, L. D. et al. Discovery of BI 224436, a noncatalytic site integrase inhibitor (NCINI) of HIV-1. ACS Med. Chem. Lett. 5, 422–427 (2014).
Nanudorn, P. et al. Atropopeptides are a novel family of ribosomally synthesized and posttranslationally modified peptides with a complex molecular shape. Angew. Chem. Int. Ed. 61, e202208361 (2022).
Reisberg, S. H. et al. Total synthesis reveals atypical atropisomerism in a small-molecule natural product, tryptorubin A. Science 367, 458–463 (2020).
Yao, G. et al. The occurrence of ansamers in the synthesis of cyclic peptides. Nat. Commun. 13, 6488 (2022).
Hegemann, J. D., Zimmermann, M., Xie, X. & Marahiel, M. A. Lasso peptides: an intriguing class of bacterial natural products. Acc. Chem. Res. 48, 1909–1919 (2015).
de Veer, S. J., Kan, M.-W. & Craik, D. J. Cyclotides: from structure to function. Chem. Rev. 119, 12375–12421 (2019).
Moore, M. J. et al. Next-generation total synthesis of vancomycin. J. Am. Chem. Soc. 142, 16039–16050 (2020).
Bose, D. S. & Kumar, R. K. An efficient, high yielding protocol for the synthesis of functionalized quinolines via the tandem addition/annulation reaction of o-aminoaryl ketones with α-methylene ketones. Tetrahedron Lett. 47, 813–816 (2006).
Wu, J., Zhang, L. & Diao, T.-N. An expeditious approach to quinolines via Friedländer synthesis catalyzed by FeCl3 or Mg(ClO4)2. Synlett 2005, 2653–2657 (2005).
Muchowski, J. M. & Maddox, M. L. Concerning the mechanism of the Friedländer quinoline synthesis. Can. J. Chem. 82, 461–478 (2004).
De, S. K. & Gibbs, R. A. A mild and efficient one-step synthesis of quinolines. Tetrahedron Lett. 46, 1647–1649 (2005).
Lekhok, K. C., Bhuyan, D., Prajapati, D. & Boruah, R. C. Zinc triflate: a highly efficient reusable catalyst in the synthesis of functionalized quinolines via Friedlander annulation. Mol. Diversity 14, 841–846 (2010).
Pohl, E. et al. Structure of octreotide, a somatostatin analogue. Acta Crystallogr. D Biol. Crystallogr. 51, 48–59 (1995).
Weckbecker, G. et al. Opportunities in somatostatin research: biological, chemical and therapeutic aspects. Nat. Rev. Drug Discov. 2, 999–1017 (2003).
Blankenstein, J. & Zhu, J. Conformation-directed macrocyclization reactions. Eur. J. Org. Chem. 2005, 1949–1964 (2005).
Wang, J. et al. Kinetically guided radical-based synthesis of C(sp3)−C(sp3) linkages on DNA. Proc. Natl Acad. Sci. USA 115, E6404–E6410 (2018).
Kondo, T. et al. cDNA TRAP display for rapid and stable in vitro selection of antibody-like proteins. Chem. Commun. 57, 2416–2419 (2021).
Yoshisada, R., Weller, S., Çobanoğlu, T. S., de Kock, H. N. & Jongkees, S. A. K. Chemical stability of mRNA/cDNA complexes: defining the limits of mRNA display. Chem. Asian J. 19, e202400336 (2024).
Katoh, T., Iwane, Y. & Suga, H. tRNA engineering for manipulating genetic code. RNA Biol. 15, 453–460 (2017).
Cui, Z., Wu, Y., Mureev, S. & Alexandrov, K. Oligonucleotide-mediated tRNA sequestration enables one-pot sense codon reassignment in vitro. Nucleic Acids Res. 46, 6387–6400 (2018).
Barrett, K. T., Metrano, A. J., Rablen, P. R. & Miller, S. J. Spontaneous transfer of chirality in an atropisomerically enriched two-axis system. Nature 509, 71–75 (2014).
Širvinskas, M. J., Saunders, G. J., Mitrache, M. & Yudin, A. K. Stabilization of 310-helices in macrocycles using dominant rotor methodology. J. Am. Chem. Soc. 146, 24085–24093 (2024).
Danelius, E., Bu, G., Wieske, L. H. E. & Gonen, T. MicroED as a powerful tool for structure determination of macrocyclic drug compounds directly from their powder formulations. ACS Chem. Biol. 18, 2582–2589 (2023).
Shapovalov, M., Vucetic, S. & Dunbrack, R. L. Jr. A new clustering and nomenclature for beta turns derived from high-resolution protein structures. PLoS Comput. Biol. 15, e1006844 (2019).
Hantzsch, A. Condensationsprodukte aus aldehydammoniak und ketonartigen verbindungen. Ber. Dtsch. Chem. Ges. 14, 1637–1638 (1881).
Knorr, L. Einwirkung von acetessigester auf phenylhydrazin. Ber. Dtsch. Chem. Ges. 16, 2597–2599 (1883).
Knorr, L. Synthese von pyrrolderivaten. Ber. Dtsch. Chem. Ges. 17, 1635–1642 (1884).
Bose, D. S., Fatima, L. & Mereyala, H. B. Green chemistry approaches to the synthesis of 5-alkoxycarbonyl-4-aryl-3,4-dihydropyrimidin-2(1H)-ones by a three-component coupling of one-pot condensation reaction: comparison of ethanol, water, and solvent-free conditions. J. Org. Chem. 68, 587–590 (2003).
Kennedy-Smith, J. J., Staben, S. T. & Toste, F. D. Gold(I)-catalyzed Conia-ene reaction of β-ketoesters with alkynes. J. Am. Chem. Soc. 126, 4526–4527 (2004).
Knudson, I. J. et al. Chemical and ribosomal synthesis of atropisomeric and macrocyclic peptides with embedded quinolines. figshare https://doi.org/10.6084/m9.figshare.25968364 (2025).
Tajima, K., Katoh, T. & Suga, H. Drop-off-reinitiation triggered by EF-G-driven mistranslocation and its alleviation by EF-P. Nucleic Acids Res. 50, 2736–2753 (2022).
Katoh, T. & Suga, H. Drop-off-reinitiation at the N-termini of nascent peptides and its regulation by IF3, EF-G, and RRF. RNA 29, 663–674 (2023).
Katoh, T. & Suga, H. Translation initiation with exotic amino acids using EF-P-responsive artificial initiator tRNA. Nucleic Acids Res. 51, 8169–8180 (2023).
Lee, J. et al. Expanding the limits of the second genetic code with ribozymes. Nat. Commun. 10, 5097 (2019).
Acknowledgements
This work was supported by the National Science Foundation (NSF) Center for Genetically Encoded Materials (C-GEM) grant CHE-2002182 (I.J.K., T.L.D., D.A.D., C.P., H.C., A.S. and S.J.M.) and the NSF Graduate Research Fellowship Program grants DGE-2139841 (T.L.D.) and DGE-1122492 (D.A.D.). C-GEM supported acylation and IVT experiments, as well as the synthesis, development and characterization of linear and macrocyclic peptides. MicroED experiments were supported by the National Institutes of Health (NIH) grant P41GM136508 (O.P., J.L. and T.G.) and funds from the Howard Hughes Medical Institute (O.P. and T.G.). This research made use of the Pines Magnetic Resonance Center’s Core NMR Facility (PMRC Core), as well as the Chemical and Biophysical Instrumentation Center (CBIC) at Yale University (RRID:SCR_021738), with equipment purchased with funds from NIH grant S10OD024998 (PMRC) or Yale University. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
We acknowledge H. Celik (PMRC Core), R. Giovine (PMRC Core) and R. E. Berry (CBIC) for assistance with NMR data acquisition; B. Q. Mercado (CBIC) and M. Houck (CBIC) for assistance with X-ray structural determination; and F. S. Menges (CBIC) for assistance with HRMS, MALDI-TOF and infrared data acquisition. We also acknowledge M. C. Guo (Yale) for additional assistance with infrared data acquisition, A. T. Champlin (Yale) for assistance with chiral semi-preparative HPLC, S. J. Hasnain (Yale) for preliminary contributions and members of the Schepartz and Miller groups for fruitful discussion.
Author information
Authors and Affiliations
Contributions
I.J.K., D.A.D., A.S. and S.J.M. designed the project. I.J.K. led acylation experiments, Friedländer reaction optimization, IVT, SPPS and post-translational reactions. I.J.K., T.L.D. and D.A.D. synthesized materials and analysed/interpreted results. H.C. and C.P. assisted with acylation experiments. O.P., J.L. and T.G. collected and processed microED data. I.J.K., T.L.D., A.S. and S.J.M. prepared the paper.
Corresponding authors
Ethics declarations
Competing interests
I.J.K., D.A.D., A.S. and S.J.M. have submitted a patent application, ‘Peptide Macrocycles with Embedded Heterocycles’ (2024, US patent application no. 63/559,854), related to the work disclosed here. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks Julien Tailhades and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Flexizyme-promoted acylation of MH with DNB esters of fMet (fM) and monomers 1-4.
a-e, Abs260 chromatograms, extracted ion chromatograms, and deconvoluted mass spectra of the products resulting from flexizyme-promoted acylation of MH by the DNB esters of fMet (fM) and 1-4. In EIC, red traces correspond to the acylated MH, while black traces represent the unacylated MH. The red stars on the mass spectra indicate the acyl-MH peak in the deconvolution.
Extended Data Fig. 2 Peptides initiated with 2-tRNAfMet or 3-tRNAfMet were translated at levels comparable to those initiated with fMet-tRNAfMet.
Total ion chromatogram of IVT reactions programmed with a duplex DNA template encoding the FLAG-containing polypeptide MGVDYKDDDDK (MGV-flag) in the presence or absence of tRNAfMet acylated with the DNB esters of fMet (fM) or monomers 2 or 3 following anti-FLAG purification as described in the Supplementary Information, Sections III–IV. As previously described by others, we also observe side-products resulting from “drop-off-reinitiation”32,59,113,114,115,116.
Extended Data Fig. 3 Extracted ion chromatograms of Friedländer reactions of peptides synthesized using in vitro translation (IVT) methods.
a-d, Extracted mass chromatograms for 2-P1 (highlighted in grey) and each quinoline peptide (pink) after reaction in AcOH at 40 °C for 24 h. e, Extracted mass chromatograms for 2-P1 (grey) and presumed atropisomer quinoline peptide Q26-P1 (teal) after reaction in AcOH or in the presence of Yb(OTf)3 in EtOH or in MeCN. All reactions were run for 24 h at 40 °C.
Extended Data Fig. 4 Peptide stability and conversion in various Friedländer conditions.
a, Scheme of IVT reaction to produce peptides H2N-P1 and 2-P1, which were then exposed to either no reaction, AcOH at 40 °C for 24 h with no 2-aminoarylcarbonyl added, or AcOH at 40 °C for 24 h with 6 added. Extracted ion chromatogram (EIC) abundance for major ions of the starting material 2-P1 and product Q6-P1 (b), as well as the bystander peptide H2N-P1 (c). Bystander peptide H2N-P1 functions as an internal standard to demonstrate the stability of a nonreactive peptide under reaction conditions. AcOH reactions may result in mild degradation of 2-P1 but have little impact on H2N-P1. All reactions performed in triplicate, and all LC-HRMS measurements taken on the same day with the assumption that minimal changes in ionization efficiency occur during this timeframe. Error bars represent standard deviation (SD) d, Scheme showing reaction of IVT peptides with various 2-aminoarylcarbonyl substrates under three different Friedländer conditions. e, EIC abundance measurements show that Yb(OTf)3 reactions in organic solvents result in a more limited recovery of bystander peptide H2N-P1. EtOH conditions result in the complete disappearance of 2-P1, with a higher abundance of Q26-P1, while MeCN conditions show sizeable amounts of residual 2-P1 and a lower abundance of Q26-P1. All LC-HRMS measurements taken on the same day with the assumption that minimal changes in ionization efficiency occur across this timeframe. Note: experiments from a-c and d-e were performed separately with different batches of tRNA, IVT kit, monomer, and anti-FLAG magnetic beads and should not be directly compared.
Extended Data Fig. 5 The diastereomeric atropisomers of biaryl 29 show distinct chemical shifts and are chromatographically separable.
a, The crude 1H NMR spectrum in CDCl3 of biaryl 29 from Preparation II. The signals arising from the red protons in the accompanying structures were used to determine the diastereomeric ratio. b, The 1H NMR spectrum of purified biaryl 29 as a mixture of diastereomeric atropisomers in CDCl3. c, The 1H NMR spectrum of the minor diastereomeric atropisomer (M, S)-29 in CDCl3. d, The 1H NMR spectrum of the major diastereomeric atropisomer (P, S)-29 in CDCl3. The spectra shown are truncated for clarity. Additionally, the spectra have been manually aligned to the peaks at δ 2.70 and 2.68 ppm in panel (b) to account for minor deviations in chemical shift, presumably arising from differences in concentration and intermolecular interactions. Processed full spectra are provided within the Supplementary Information: “Processed NMR Spectra,” and raw .fid files are provided in the FAIR data.
Extended Data Fig. 6 Diagnostic shifts in the NMR spectra upon Friedländer macrocyclization.
a, Proton NMR of LP4 (red) and CP4 (teal) show the emergence of a distinct set of downfield aromatic protons (blue box) corresponding to the quinoline ring protons. b, gCOSY NMR spectra of LP4 (red, left) and CP4 (teal, right) display the correlated spin-couplings of the quinoline system. c, Carbon NMR of LP4 (red) and CP4 (teal) show the distinct loss of two ketone carbons around 200-210 ppm. Note: the tops of the solvent residuals have been truncated to save space; the truncation point is indicated with a dashed line.
Extended Data Fig. 7 Friedländer macrocyclization and DNA stability with high equivalents of Yb(OTf)3 in aqueous acetonitrile.
Scheme (a) and Total Ion Chromatograms (b-e) of Friedländer macrocyclization of LP2 (1 mM or 10 µM) with Yb(OTf)3 (1 mM, 10 mM, 100 mM or 200 mM) in H2O/MeCN (1:1 v/v). TIC conversions are averages of three independent replicates. Data is representative. Conversion of Friedländer macrocyclization is dramatically reduced in highly aqueous environments, but reactivity can be rescued with high equivalents of Yb(OTf)3. f, Stability of an oligonucleotide DNA:RNA duplex was assessed under aqueous acetonitrile Friedländer macrocyclization conditions in the presence of 200 mM Yb(OTf)3. Absorbance chromatogram (g) and extracted ion chromatogram (h) showing that RNA is degraded while DNA remains stable under reaction conditions. dFx was used as an internal standard to control for ionization efficiency across samples. Samples were run in triplicate, and representative traces are shown. i, Stability of RNA and DNA when exposed to reaction conditions. We suspect that the higher recovery of the DNA post-reaction is a result of improved DNA precipitation and column retention because of the Yb(OTf)3 salt compared to the unreacted sample. Error bars represent standard deviation (SD). j, Mass spectra of DNA before and after reaction are identical.
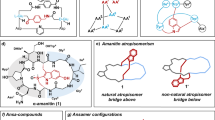
Extended Data Fig. 8 Structure and conversion between conformational isomers CPa1 and (P)-CPa.
a, Ramachandran plot of solid-state (P)-CPa. b, MicroED structure showing a double beta-turn motif (type II’ followed by type I). Cyclic backbone carbons highlighted in green. N-Methyl highlighted in orange. Intramolecular H-bonds indicated by orange dashed lines. H-bond distance is 2.30 Å between i + 3 Phe N-H and i carbonyl and 2.17 Å between i + 4 Kyn and i + 1 N-Me-Ala carbonyl. c, Scheme showing the thermal conversion of CPa1 to (P)-CPa. d, 1H NMR spectra show complete conversion of CPa1 to (P)-CPa after heating. The signal at δ 6.53 ppm in the bottom spectrum arises from the putative quinolinium proton.
Supplementary information
Supplementary Information
Supplementary Sections VI–VIII, Figs. 1–29, Tables 1–22, Methods, Discussion and processed NMR spectra.
Source data
Source Data Fig. 1
Tabulated A260 chromatograms, extracted ion chromatograms and mass spectra for Fig. 1c–h.
Source Data Fig. 2
Tabulated replicates of normalized A214 conversions.
Source Data Fig. 3
Tabulated extracted ion chromatograms and mass spectra for Fig. 3b–e.
Source Data Fig. 4
Tabulated extracted ion chromatograms and mass spectrum for Fig. 4c.
Source Data Fig. 5
Tabulated HPLC chromatograms for Fig. 5b–e and extracted ion chromatograms for Fig. 5g.
Source Data Fig. 6
Tabulated HPLC chromatogram for Fig. 6b and total ion chromatogram time points for Fig. 6c.
Source Data Extended Data Fig. 1
Tabulated A260 chromatograms, extracted ion chromatograms and mass spectra for Extended Data Fig. 1a–e.
Source Data Extended Data Fig. 2
Tabulated total ion chromatograms for Extended Data Fig. 2.
Source Data Extended Data Fig. 3
Tabulated extracted ion chromatograms for Extended Data Fig. 3a–e.
Source Data Extended Data Fig. 4
Tabulated extracted ion abundances for Extended Data Fig. 4b,c,e.
Source Data Extended Data Fig. 7
Tabulated total ion chromatograms for Extended Data Fig. 7b–e; and tabulated absorbance chromatograms, extracted ion chromatograms and mass spectra for Extended Data Fig. 7g–j.
Source Data Extended Data Fig. 8
Tabulated phi and psi angles for the Ramachandran plot in Extended Data Fig. 8a.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Knudson, I.J., Dover, T.L., Dilworth, D.A. et al. Chemical and ribosomal synthesis of atropisomeric and macrocyclic peptides with embedded quinolines. Nat. Chem. 18, 61–72 (2026). https://doi.org/10.1038/s41557-025-01935-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41557-025-01935-4
This article is cited by
-
Redefining peptide chemistry beyond accumulating analogues
Nature Reviews Chemistry (2025)