Abstract
Pseudomonas aeruginosa frequently causes antibiotic-recalcitrant pneumonia, but the mechanisms driving its adaptation during human infections remain unclear. To reveal the selective pressures and adaptation strategies at the mucosal surface, here we investigated P. aeruginosa growth and antibiotic tolerance in tissue-engineered airways by transposon insertion sequencing (Tn-seq). Metabolic modelling based on Tn-seq data revealed the nutritional requirements for P. aeruginosa growth, highlighting reliance on glucose and lactate and varying requirements for amino acid biosynthesis. Tn-seq also revealed selection against biofilm formation during mucosal growth in the absence of antibiotics. Live imaging in engineered organoids showed that biofilm-dwelling cells remained sessile while colonizing the mucosal surface, limiting nutrient foraging and reduced growth. Conversely, biofilm formation increased antibiotic tolerance at the mucosal surface. Moreover, mutants with exacerbated biofilm phenotypes protected less tolerant but more cytotoxic strains, contributing to phenotypic heterogeneity. P. aeruginosa must therefore navigate conflicting physical and biological selective pressures to establish chronic infections.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Spreadsheets containing the source data used to generate each plot (main and extended data figures), the code used during metabolic modelling, the microscopy data displayed in all the figures and all files related to Tn-seq (that is, WIG and annotation files used during analyses with TRANSIT) are openly available via Zenodo at https://doi.org/10.5281/zenodo.13629466 (ref. 129). The Tn-seq sequencing data have been submitted to the NCBI Sequence Read Archive under accession number PRJNA1156351. The custom image analysis software used for quantification of fitness ratios and cluster sizes distribution is available via GitHub under the following repository: https://github.com/PersatLab/CompetitionAssay. Additional files, such as the raw files of the dozens of videos recorded during AirGel experiments as well as all strains and plasmids used in this study, are available from the corresponding author upon request.
References
Breidenstein, E. B. M., de la Fuente-Núñez, C. & Hancock, R. E. W. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19, 419–426 (2011).
Horcajada, J. P. et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32, e00031–19 (2019).
Valderrey, A. D. et al. Chronic colonization by Pseudomonas aeruginosa of patients with obstructive lung diseases: cystic fibrosis, bronchiectasis, and chronic obstructive pulmonary disease. Diagn. Microbiol. Infect. Dis. 68, 20–27 (2010).
Fahy, J. V. & Dickey, B. F. Airway mucus function and dysfunction. N. Engl. J. Med. 363, 2233–2247 (2010).
Dolan, S. K. Current knowledge and future directions in developing strategies to combat Pseudomonas aeruginosa infection. J. Mol. Biol. 432, 5509–5528 (2020).
Hibbert, T. M., Whiteley, M., Renshaw, S. A., Neill, D. R. & Fothergill, J. L. Emerging strategies to target virulence in Pseudomonas aeruginosa respiratory infections. Crit. Rev. Microbiol. https://doi.org/10.1080/1040841X.2023.2285995 (2023).
Rossi, E. et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-020-00477-5 (2020).
Martínez-Solano, L., Macia, M. D., Fajardo, A., Oliver, A. & Martinez, J. L. Chronic Pseudomonas aeruginosa infection in chronic obstructive pulmonary disease. Clin. Infect. Dis. 47, 1526–1533 (2008).
Cao, P. et al. A Pseudomonas aeruginosa small RNA regulates chronic and acute infection. Nature 618, 358–364 (2023).
Jorth, P. et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18, 307–319 (2015).
Friedman, L. & Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186, 4457–4465 (2004).
Jenal, U. & Malone, J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40, 385–407 (2006).
Hickman, J. W., Tifrea, D. F. & Harwood, C. S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl Acad. Sci. USA 102, 14422–14427 (2005).
Ceri, H. et al. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37, 1771–1776 (1999).
Ciofu, O. & Tolker-Nielsen, T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents—how P. aeruginosa can escape antibiotics. Front. Microbiol. 10, 913 (2019).
de la Fuente-Nunez, C., Cesaro, A. & Hancock, R. E. W. Antibiotic failure: beyond antimicrobial resistance. Drug Resist. Updat. 71, 101012 (2023).
Walters, M. C., Roe, F., Bugnicourt, A., Franklin, M. J. & Stewart, P. S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47, 317–323 (2003).
Levin-Reisman, I. et al. Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017).
Santi, I., Manfredi, P., Maffei, E., Egli, A. & Jenal, U. Evolution of antibiotic tolerance shapes resistance development in chronic Pseudomonas aeruginosa infections. MBio 12, e03482–20 (2021).
Harrison, J. J. et al. Elevated exopolysaccharide levels in Pseudomonas aeruginosa flagellar mutants have implications for biofilm growth and chronic infections. PLoS Genet. 16, e1008848 (2020).
Jennings, L. K. et al. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep. 34, 108782 (2021).
Marvig, R. L., Sommer, L. M., Molin, S. & Johansen, H. K. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47, 57–64 (2015).
López-Jiménez, A. T. & Mostowy, S. Emerging technologies and infection models in cellular microbiology. Nat. Commun. 12, 6764 (2021).
Shi, D., Mi, G., Wang, M. & Webster, T. J. In vitro and ex vivo systems at the forefront of infection modeling and drug discovery. Biomaterials 198, 228–249 (2019).
Wagner, C. E., Wheeler, K. M. & Ribbeck, K. Mucins and their role in shaping the functions of mucus barriers. Annu. Rev. Cell Dev. Biol. 34, 189–215 (2018).
Roy, M. G. et al. Muc5b is required for airway defence. Nature 505, 412–416 (2014).
Rossy, T. et al. Pseudomonas aeruginosa type IV pili actively induce mucus contraction to form biofilms in tissue-engineered human airways. PLoS Biol. 21, e3002209 (2023).
Lewin, G. R. et al. Application of a quantitative framework to improve the accuracy of a bacterial infection model. Proc. Natl Acad. Sci. USA 120, e2221542120 (2023).
Turner, K. H., Wessel, A. K., Palmer, G. C., Murray, J. L. & Whiteley, M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl Acad. Sci. USA 112, 4110–4115 (2015).
Palmer, K. L., Mashburn, L. M., Singh, P. K. & Whiteley, M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187, 5267–5277 (2005).
Palmer, K. L., Aye, L. M. & Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087 (2007).
Leoni Swart, A. et al. Pseudomonas aeruginosa breaches respiratory epithelia through goblet cell invasion in a microtissue model. Nat. Microbiol. https://doi.org/10.1038/s41564-024-01718-6 (2024).
Hasan, S., Sebo, P. & Osicka, R. A guide to polarized airway epithelial models for studies of host–pathogen interactions. FEBS J. 285, 4343–4358 (2018).
Cain, A. K. et al. A decade of advances in transposon-insertion sequencing. Nat. Rev. Genet. 21, 526–540 (2020).
Winstanley, C., O’Brien, S. & Brockhurst, M. A. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 24, 327–337 (2016).
Gao, C. et al. Lactate utilization is regulated by the FadR-type regulator LldR in Pseudomonas aeruginosa. J. Bacteriol. 194, 2687–2692 (2012).
El Husseini, N. et al. Characterization of the Entner–Doudoroff pathway in Pseudomonas aeruginosa catheter-associated urinary tract infections. J. Bacteriol. https://doi.org/10.1128/jb.00361-23 (2023).
McMorran, B. J., Merriman, M. E., Rombel, I. T. & Lamont, I. L. Characterisation of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene 176, 55–59 (1996).
Watson, A. R. et al. Metabolic independence drives gut microbial colonization and resilience in health and disease. Genome Biol. 24, 78 (2023).
Bartell, J. A. et al. Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nat. Commun. 8, 14631 (2017).
McShane, A. et al. Mucus. Curr. Biol. 31, R938–R945 (2021).
Maurice, N. M., Bedi, B. & Sadikot, R. T. Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am. J. Respir. Cell Mol. Biol. 58, 428–439 (2018).
Kong, W. et al. ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa. Nucleic Acids Res. 43, 8268–8282 (2015).
Petrova, O. E., Cherny, K. E. & Sauer, K. The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J. Bacteriol. 196, 2827–2841 (2014).
Kuchma, S. L. et al. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 8165–8178 (2007).
Manner, C. et al. A genetic switch controls Pseudomonas aeruginosa surface colonization. Nat. Microbiol. 8, 1520–1533 (2023).
Roy, A. B., Petrova, O. E. & Sauer, K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194, 2904–2915 (2012).
Muggeo, A., Coraux, C. & Guillard, T. Current concepts on Pseudomonas aeruginosa interaction with human airway epithelium. PLoS Pathog. 19, e1011221 (2023).
Cont, A., Rossy, T., Al-Mayyah, Z. & Persat, A. Biofilms deform soft surfaces and disrupt epithelia. Elife 9, e56533 (2020).
Meirelles, L. A., Perry, E. K., Bergkessel, M. & Newman, D. K. Bacterial defenses against a natural antibiotic promote collateral resilience to clinical antibiotics. PLoS Biol. 19, e3001093 (2021).
Purssell, A. & Poole, K. Functional characterization of the NfxB repressor of the mexCD-oprJ multidrug efflux operon of Pseudomonas aeruginosa. Microbiology 159, 2058–2073 (2013).
Matsuo, Y., Eda, S., Gotoh, N., Yoshihara, E. & Nakae, T. MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol. Lett. 238, 23–28 (2004).
Laborda, P. et al. Mutations in the efflux pump regulator MexZ shift tissue colonization by Pseudomonas aeruginosa to a state of antibiotic tolerance. Nat. Commun. 15, 2584 (2024).
Lister, P. D., Wolter, D. J. & Hanson, N. D. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610 (2009).
Fair, R. J. & Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 6, 25–64 (2014).
Bulitta, J. B. et al. Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob. Agents Chemother. 59, 2315–2327 (2015).
Clark, S. T. et al. Phenotypic diversity within a Pseudomonas aeruginosa population infecting an adult with cystic fibrosis. Sci. Rep. 5, 10932 (2015).
Mowat, E. et al. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am. J. Respir. Crit. Care Med. 183, 1674–1679 (2011).
O’Brien, S. et al. High virulence sub-populations in Pseudomonas aeruginosa long-term cystic fibrosis airway infections. BMC Microbiol. 17, 30 (2017).
Bartell, J. A. et al. Evolutionary highways to persistent bacterial infection. Nat. Commun. 10, 629 (2019).
Armbruster, C. R. et al. Adaptation and genomic erosion in fragmented Pseudomonas aeruginosa populations in the sinuses of people with cystic fibrosis. Cell Rep. 37, 109829 (2021).
Bensel, T. et al. Lactate in cystic fibrosis sputum. J. Cyst. Fibros. 10, 37–44 (2011).
Davey, L. E. et al. A genetic system for Akkermansia muciniphila reveals a role for mucin foraging in gut colonization and host sterol biosynthesis gene expression. Nat. Microbiol. 8, 1450–1467 (2023).
Flynn, J. M., Phan, C. & Hunter, R. C. Genome-wide survey of Pseudomonas aeruginosa PA14 reveals a role for the glyoxylate pathway and extracellular proteases in the utilization of mucin. Infect. Immun. 85, e00182–17 (2017).
Hoet, P. H. & Nemery, B. Polyamines in the lung: polyamine uptake and polyamine-linked pathological or toxicological conditions. Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L417–L433 (2000).
Liu, Z., Hossain, S. S., Morales Moreira, Z. & Haney, C. H. Putrescine and its metabolic precursor arginine promote biofilm and c-di-GMP synthesis in Pseudomonas aeruginosa. J. Bacteriol. 204, e0029721 (2022).
Barth, A. L. & Pitt, T. L. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45, 110–119 (1996).
Thomas, S. R., Ray, A., Hodson, M. E. & Pitt, T. L. Increased sputum amino acid concentrations and auxotrophy of Pseudomonas aeruginosa in severe cystic fibrosis lung disease. Thorax 55, 795–797 (2000).
DePas, W. H. et al. Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. MBio 7, e00796–16 (2016).
Racanelli, A. C., Kikkers, S. A., Choi, A. M. K. & Cloonan, S. M. Autophagy and inflammation in chronic respiratory disease. Autophagy 14, 221–232 (2018).
Cantin, A. M., Hartl, D., Konstan, M. W. & Chmiel, J. F. Inflammation in cystic fibrosis lung disease: pathogenesis and therapy. J. Cyst. Fibros. 14, 419–430 (2015).
Marvig, R. L. et al. Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients. BMC Microbiol. 15, 218 (2015).
Stanford, G. E., Dave, K. & Simmonds, N. J. Pulmonary exacerbations in adults with cystic fibrosis: a grown-up issue in a changing cystic fibrosis landscape. Chest 159, 93–102 (2021).
Woo, J. K. K., Webb, J. S., Kirov, S. M., Kjelleberg, S. & Rice, S. A. Biofilm dispersal cells of a cystic fibrosis Pseudomonas aeruginosa isolate exhibit variability in functional traits likely to contribute to persistent infection. FEMS Immunol. Med. Microbiol. 66, 251–264 (2012).
Sousa, A. M. & Pereira, M. O. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens 3, 680–703 (2014).
Stewart, P. S. et al. Conceptual model of biofilm antibiotic tolerance that integrates phenomena of diffusion, metabolism, gene expression, and physiology. J. Bacteriol. 201, e00307–e00319 (2019).
Tseng, B. S. et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 15, 2865–2878 (2013).
Winans, J. B., Wucher, B. R. & Nadell, C. D. Multispecies biofilm architecture determines bacterial exposure to phages. PLoS Biol. 20, e3001913 (2022).
Thurlow, L. R. et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 186, 6585–6596 (2011).
Rowe, W. J., Lebman, D. A. & Ohman, D. E. Mechanism of resistance to phagocytosis and pulmonary persistence in mucoid Pseudomonas aeruginosa. Front. Cell Infect. Microbiol. 13, 1125901 (2023).
Mishra, M. et al. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 14, 95–106 (2012).
Malhotra, S., Limoli, D. H., English, A. E., Parsek, M. R. & Wozniak, D. J. Mixed communities of mucoid and nonmucoid Pseudomonas aeruginosa exhibit enhanced resistance to host antimicrobials. MBio 9, e00275–18 (2018).
Köhler, T. et al. Personalized aerosolised bacteriophage treatment of a chronic lung infection due to multidrug-resistant Pseudomonas aeruginosa. Nat. Commun. 14, 3629 (2023).
Strathdee, S. A., Hatfull, G. F., Mutalik, V. K. & Schooley, R. T. Phage therapy: from biological mechanisms to future directions. Cell 186, 17–31 (2023).
Mishra, R. et al. Mechanopathology of biofilm-like Mycobacterium tuberculosis cords. Cell 186, 5135–5150.e28 (2023).
Luckett, K. A. & Ganesh, K. Engineering the immune microenvironment into organoid models. Annu. Rev. Cancer Biol. 7, 1972–1988.e16 (2023).
Holloway, B. W. & Morgan, A. F. Genome organization in Pseudomonas. Annu. Rev. Microbiol. 40, 79–105 (1986).
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J. & Schweizer, H. P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 (1998).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Simon, R., Priefer, U. & Pühler, A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791 (1983).
Choi, K.-H. & Schweizer, H. P. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat. Protoc. 1, 153–161 (2006).
Rybtke, M. T. et al. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78, 5060–5069 (2012).
Jacobs, M. A. et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 100, 14339–14344 (2003).
Basta, D. W., Bergkessel, M. & Newman, D. K. Identification of fitness determinants during energy-limited growth arrest in Pseudomonas aeruginosa. MBio 8, e01170–17 (2017).
Fulcher, M. L. & Randell, S. H. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol. 945, 109–121 (2013).
Jiménez-Torres, J. A., Peery, S. L., Sung, K. E. & Beebe, D. J. Lumenext: a practical method to pattern luminal structures in ECM gels. Adv. Health. Mater. 5, 198–204 (2016).
Chen, Y. et al. Validation of human small airway measurements using endobronchial optical coherence tomography. Respir. Med. 109, 1446–1453 (2015).
Horsfield, K. & Cumming, G. Morphology of the bronchial tree in man. J. Appl. Physiol. 24, 373–383 (1968).
DeJesus, M. A., Ambadipudi, C., Baker, R., Sassetti, C. & Ioerger, T. R. TRANSIT—a software tool for Himar1 TnSeq analysis. PLoS Comput. Biol. 11, e1004401 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DeJesus, M. A. & Ioerger, T. R. Normalization of transposon-mutant library sequencing datasets to improve identification of conditionally essential genes. J. Bioinform. Comput. Biol. 14, 1642004 (2016).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
The pandas development team. pandas-dev/pandas: Pandas 1.0.3. Zenodo https://doi.org/10.5281/zenodo.3509134 (2020).
McKinney, W. Data structures for statistical computing in python. in Proceedings of the 9th Python in Science Conference 56–61. https://doi.org/10.25080/Majora-92bf1922-00a (SciPy, 2010).
Pedregosa, F. et al. AnchorScikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
EUCAST Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 9, ix–xv (2003).
Meirelles, L. A. & Newman, D. K. Phenazines and toxoflavin act as interspecies modulators of resilience to diverse antibiotics. Mol. Microbiol. 117, 1384–1404 (2022).
Masid, M., Ataman, M. & Hatzimanikatis, V. Analysis of human metabolism by reducing the complexity of the genome-scale models using redHUMAN. Nat. Commun. 11, 2821 (2020).
Jankowski, M. D., Henry, C. S., Broadbelt, L. J. & Hatzimanikatis, V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys. J. 95, 1487–1499 (2008).
Salvy, P. et al. pyTFA and matTFA: a Python package and a Matlab toolbox for thermodynamics-based flux analysis. Bioinformatics 35, 167–169 (2019).
Soh, K. C. & Hatzimanikatis, V. Network thermodynamics in the post-genomic era. Curr. Opin. Microbiol. 13, 350–357 (2010).
Soh, K. C. & Hatzimanikatis, V. Constraining the flux space using thermodynamics and integration of metabolomics data. Methods Mol. Biol. 1191, 49–63 (2014).
Thiele, I. et al. A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2. BMC Syst. Biol. 5, 8 (2011).
Dulbecco, R. & Freeman, G. Plaque production by the polyoma virus. Virology 8, 396–397 (1959).
Wheeler, K. M. et al. Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nat. Microbiol. 4, 2146–2154 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Krull, A., Buchholz, T.-O. & Jug, F. Noise2Void - Learning denoising from single noisy images. in 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2124–2132 (IEEE, 2019).
Ho, T. K. Random decision forests. in Proceedings of 3rd International Conference on Document Analysis and Recognition 278–282 (IEEE Comput. Soc. Press, 1995).
van der Walt, S. et al. scikit-image: image processing in Python. PeerJ 2, e453 (2014).
Fiorio, C. & Gustedt, J. Two linear time Union-Find strategies for image processing. Theor. Comput. Sci. 154, 165–181 (1996).
Wu, K., Otoo, E. & Shoshani, A. Optimizing connected component labeling algorithms. in Medical Imaging 2005: Image Processing (eds Fitzpatrick, J. M. & Reinhardt, J. M.) 5747, 1965 (SPIE, 2005).
Burger, W. & Burge, M. J. Principles of Digital Image Processing (Springer, 2009); https://doi.org/10.1007/978-1-84800-195-4
Reiss, T. H. Recognizing Planar Objects Using Invariant Image Features (Springer, 1993); https://doi.org/10.1007/BFb0017553
Harris et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Waskom, M. seaborn: statistical data visualization. JOSS 6, 3021 (2021).
Meirelles, L. A. et al. Pseudomonas aeruginosa faces a fitness trade-off between mucosal colonization and antibiotic tolerance during airway infection. Zenodo https://doi.org/10.5281/zenodo.13629466 (2024).
Acknowledgements
We thank Z. Al-Mayyah for laboratory technical assistance, the Lausanne Genomic Technologies Facility for sequencing, A. Martins Bravo for initial feedback on Tn-seq analysis and members of the Persat lab for constructive feedback throughout the development of the project. We also thank D. K. Newman, S. Saunders, E. Perry, M. Bergkessel, C. Nadell and N. Wespe (National Centres of Competence in Research (NCCR) AntiResist Research Data Officer) for their comments on the manuscript. Funding supporting this work was from Swiss National Science Foundation: 310030_189084 (A.P.), NCCR AntiResist (A.P.), European Molecular Biology Organization Postdoctoral Fellowship ALTF 12-2022 (L.A.M.), Swiss National Science Foundation: 200021_188623 (V.H.) and NCCR Microbiomes (V.H.).
Author information
Authors and Affiliations
Contributions
Conceptualization: L.A.M. and A.P. Data curation: L.A.M., E.V. and E.S. Formal analysis: L.A.M., E.V. and E.S. Funding acquisition: V.H. and A.P. Investigation: L.A.M. and E.V. Methodology: L.A.M., E.V., A.D., E.S., T.R. and T.D. Project administration: L.A.M. and A.P. Supervision: V.H. and A.P. Visualization: L.A.M., E.V. and E.S. Writing—original draft: L.A.M. and A.P. Writing—review and editing: L.A.M., E.V., V.H. and A.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Liang Li, Janne Thöming, Ben Vezina and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Detailed procedure for the Tn-seq during mucosal colonization.
A. Representative confocal image of (z-slice) showing intact epithe lium after 11 h of infection with the Tn-library. Experiment repeated twice with similar results. B. Exhaustive illustration of experiments and control conditions for the Tn-seq during mucosal colonization. Briefly, an aliquot of Tn-library was grown overnight in LB (starting OD = 0.25) and used as inoculum for infection or a new LB control culture. Infections or growth in the LB control proceeded for ~11 h, followed by sample collection and sequencing. For all details, including ODs used, inoculum sizes, and number of generations measured, see the Methods section. All five samples were sequenced (Tn-library, population used for inoculum, healthy HBE, CF HBE, and LB control). Blue arrows represent the comparisons (#1-3) made using the TRANSIT software to assess the conditional essentiality of genes. In comparison #1, the inoculum was used as the control; in comparison #2 and #3, the “LB control” condition was used as the control. Red arrows represent the two “quality control – QC” comparisons made using the TRANSIT software to check for bias in the inoculum used for infection. Abbreviations: CF, cystic fibrosis; HBE cells, human bronchial epithelial cells; LB, Luria-Bertani broth. The data underlying this figure can be found in the source data available with this manuscript (see Data Availability session).
Extended Data Fig. 2 Functional annotation of Tn-seq data using Database for Annotation, Visualization and Integrated Discovery (DAVID).
We used the list of significant genes found in comparison #1 of our Tn-seq analysis (see Supplementary Table 1) as input for DAVID and used the web tool to calculate the fold enrichment of Gene Ontology (GOTERM_BP_DIRECT) terms. Disruption of processes found on top leads to decreased fitness (magenta), while disruptions of processes found on the bottom lead to increased fitness (green). The results of the same analysis but using KEGG Pathways can be found in Fig. 1B. For full parameters calculated in the analyses (for example, gene counts, % ID, p-values) displayed here and in Fig. 1B, see Supplementary Table 2. The data underlying this figure can be found in the source data available with this manuscript (see Data Availability session).
Extended Data Fig. 3 Schematic of the metabolic modeling approach used in our study.
A computational approach based on genome-scale metabolic modeling and constraint-based optimization was used to assist in the interpretation of Tn-seq data. To this end, a genome-scale metabolic model was used to simulate metabolism and perform in silico single gene deletion experiments on 66 different media compositions of varying complexity (that is, single carbon media, double carbon source media, and rich media). Media selection was guided by the assumed mucus composition and Tn-seq data. Dimensionality reduction techniques and clustering were then used to compare the in silico data with the Tn-seq data to infer carbon utilization on the mucosal surface. Abbreviations: LB, Luria-Bertani broth; CF, cystic fibrosis; SCFM, synthetic cystic fibrosis sputum medium.
Extended Data Fig. 4 Relative cdGMP levels of strains used in the study.
Relative values of cdGMP were measured using a previously published fluorescence-based reporter (GFP under the control of the cdrA promoter by Rybtke et. al.92). See Methods section for details. Each data point represents an independent biological replicate (n = 3); horizontal black lines mark their mean. Statistics: 1-way ANOVA with Tukey HSD multiple comparison test, with asterisks showing significant differences relative to WT (* p < 0.05, *** p < 0.001). Due to the large difference in magnitude of the cdGMP levels for strains with the ∆wspF deletion, the comparisons were made in two parts (part 1: WT vs ∆algR or ∆bifA; part 2: WT vs. ∆wspF or ∆wspF∆pel∆psl). The data underlying this figure and information on statistical analyses can be found in the source data available with this manuscript (see Data Availability session).
Extended Data Fig. 5 Competition between WT and an alginate overproducing strain (mucoid).
A. Pictures of the mucoid PAO1 strain, a mucA merodiploid. B. CFU-based validation of the fitness changes mediated by alginate overproduction. Competitions of WT and mucoid strain at the mucosa (left) and in LB (right). Each data point represents the CFUs ratio (mucoid divided by WT) of a single replicate (n = 4 for each condition). These ratios were calculated for the inoculum (0 h) and at the end of competition (8 h), for both conditions (mucosa vs LB). The horizontal black lines mark their mean. C. Fitness ratio of ∆wspF competed against WT, displayed for comparison. These ratios were calculated the same way using the CFU values shown in Fig. 3b. All experiments were performed with CF HBE cells. Statistics: panels B and C, Welch unpaired t-test (two-sided) (** p < 0.01, * p < 0.05, n.s. p > 0.05). Abbreviations: CFU, colony-forming units; LB, Luria-Bertani broth; WT, wild-type. The data underlying this figure and information on statistical analyses can be found in the source data available with this manuscript (see Data Availability session).
Extended Data Fig. 6 Biofilm fitness quantification during mucosal colonization.
A. Fitness ratio based on the quantification of the total area measured from surface coverage by ∆bifA-mNeonGreen relative to WT-Scarlet during competition assays at the mucosal surface (HBE cultures). Each data point represents one imaged field of view (n = 15 for WT vs. WT; n = 12 for WT vs. ∆bifA) distributed within three biological replicates. The data for the WT-mNeonGreen vs. WT-mScarlet competition is the same from Fig. 3E but is shown here for comparison. Horizontal black lines mark the mean fitness ratio for each condition. B. Representative images of WT and ∆bifA competition assays in HBE cultures (maximum intensity projection; same field of view). For additional representative images, see Supplementary Fig. 1C. C. Cumulative distributions for cluster size formed by WT and ∆bifA. Distributions include three biological replicates. Vertical lines represent the median cluster-size for each strain. D. Percentage of small clusters (clusters smaller than 20 µm2) that comes from WT or ∆bifA populations. Each data point represents an independent biological replicate (n = 3); horizontal black lines mark their mean. Statistics: panel A and D, Welch unpaired t-test (two-sided) (*** p < 0.001, ** p < 0.01). All experiments were performed with CF HBE cells. Abbreviations: HBE cells, human bronchial epithelial cells; WT, wild-type. The data underlying this figure and information on statistical analyses can be found in the source data available with this manuscript (see Data Availability session).
Extended Data Fig. 7 Biofilms mechanically damage epithelia while constraining pathogenicity.
A. Experimental design for imaging epithelial damage upon WT and ∆wspF infections in AirGels. AirGels were infected with WT or ∆wspF expressing mNeonGreen, and infections were monitored for 11 hours for the observation of lysis and cell viability (propidium iodide staining). B. Representative maximum intensity projection images showing differences in growth and subsequent epithelial cell lysis by WT (top) and ∆wspF (bottom). Note that after 10 h of infection, WT had completely taken over the surface and had lysed the epithelial cells. By contrast, ∆wspF formed large expanding biofilms that opened up nodules that stretched out within the tissue (white circles). See Supplementary Movies 2 and 3 for full time-lapse visualization of these images. C. Representative maximum intensity projection images showing differences in growth and the killing of epithelial cells by WT (top) and ∆wspF (bottom). Note the uniform cytotoxic effect to the WT due to its faster spreading, while ∆wspF biofilms showed only cytotoxic activity towards epithelial cells in the immediate vicinity of the nodules (white circles). See Supplementary Movie 4 for a full time-lapse visualization of these images. All experiments were performed with CF HBE cells. In B-C, for each strain, infections of two distinct AirGels were visualized, with similar patterns. Abbreviations: PI, propidium iodide; WT, wild-type. The data underlying this figure can be found in the source data available with this manuscript (see Data Availability session).
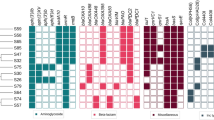
Extended Data Fig. 8 Tn-seq analysis identifies genetic determinants of antibiotic adaptation at the mucosal surface.
A. Full experimental design of the Tn-seq during antibiotic tolerance at the mucosal surface. The library was grown on CF HBE cells, treated with CIP or TOB (or no treatment for the control), and then an outgrowth step (7 to 12 hours) on LB was performed for all samples. The three outgrowth samples were sequenced. Blue lines represent the comparisons made using the TRANSIT software to assess the conditional essentiality of genes (comparisons #4-5), and the “no antibiotic” condition was used as the control (see Methods for full experimental details) B. Fitness effects of transposon insertions in representative genes and their categories discovered in our antibiotic tolerance Tn-seq. Genes are separated by CIP and TOB-specific hits. These do not represent all the genes that made the significance cutoff. See Supplementary Table 6 for the complete dataset. All experiments were performed with CF HBE cells. Abbreviations: CIP, ciprofloxacin; CF, cystic fibrosis; HBE cells, human bronchial epithelial cells; TOB, tobramycin. The data underlying this figure can be found in the source data available with this manuscript (see Data Availability session).
Extended Data Fig. 9 Matrix production mediates tolerance to CIP in high cdGMP strains.
A. Validation of the growth defects caused by cdGMP-mediated matrix overproduction at the mucosal surface using CFUs. Each data point represents an independent biological replicate (that is, one transwell, n = 3); the horizontal black lines mark their mean. B. Tolerance to CIP at the mucosal surface measured by CFUs for different strains used in our study. For each strain, tolerance is calculated based on total CFUs recovered before and after CIP treatment. Each data point represents an independent biological replicate (that is, one transwell, n = 3); the horizontal black lines mark their mean. Statistics: panel A, 1-way ANOVA with Tukey HSD multiple comparison test, with asterisks showing significant differences relative to WT (** p < 0.01, *** p < 0.001, n.s. p > 0.05); panel B, 1-way ANOVA with Tukey HSD multiple comparison test, with asterisks displaying significant differences for highlighted comparisons (* p < 0.05, n.s. p > 0.05). In panel B, statistical analyses were run in two parts (part 1: WT vs ∆bifA or ∆wspF∆pel∆psl; part 2: ∆wspF vs ∆wspF∆pel∆psl or WT). All experiments were performed with CF HBE cells. Abbreviations: CFUs, colony-forming units; CIP, ciprofloxacin. The data underlying this figure and information on statistical analyses can be found in the source data available with this manuscript (see Data Availability session).
Extended Data Fig. 10 Model of the colonization vs tolerance trade-offs for biofilms and its consequences for phenotypic diversification during chronic infections.
Left. During colonization, planktonic behavior is beneficial as it allows better spread on the mucosa, eventually leading to the killing of epithelial cells. However, upon antibiotic treatment selection, the biofilm lifestyle, which minimizes tissue damage, is selected. Right. Long-term infection leads to genetic diversification, where strains displaying different genotypes co-exist. Such genotypes display phenotypic lifestyles (acute or chronic behavior). During antibiotic treatment selection, biofilm-forming strains may protect genotypes associated with acute behavior. We hypothesize that upon removal of the antibiotic selection (either by treatment break and/or mutation leading to resistance), acute behavior is again beneficial, potentially leading to lung exacerbations.
Supplementary information
Supplementary Information
Supplementary Fig. 1, Full legends for Tables 1–8 and Full legends for Videos 1–7.
Supplementary Tables
Supplementary Tables 1–8.
Supplementary Video 1
Growth of WT (orange) and ∆wspF (green) during AirGel co-infections. Representative case for data used in the quantification shown in Fig. 3f.
Supplementary Video 2
Growth of WT (top) and ∆wspF (bottom), both in green, during separate AirGel infections. Note the different growth patterns of each strain, with WT quickly spreading over the mucosal surface while ∆wspF expands as compact biofilms. The full time lapse for images shown in Extended Data Fig. 7b.
Supplementary Video 3
Visualization of tissue lysis by WT (top) and ∆wspF (bottom). Note quick tissue lysis caused by WT (that is, the tissue completely disintegrates), while ∆wspF formed large expanding biofilms that opened up nodules that stretched out the tissue. The full time lapse for images shown in Extended Data Fig. 7b.
Supplementary Video 4
Epithelium cell death measured with propidium iodide over the course of AirGel infections by WT (top) or ∆wspF (bottom). Note the faster and uniform cytotoxic effect caused by WT in comparison to ∆wspF (that is, complete killing of HBE cells in the displayed area by 7 h of infection). The full time lapse for images shown in Extended Data Fig. 7c.
Supplementary Video 5
Representative videos displaying the tolerance levels WT (top) and ∆bifA (bottom) upon ciprofloxacin treatment in AirGels. Both strains are shown in green. The full time lapse for images shown in Fig. 5e. Note the heterogeneous survival of ∆bifA, where rapidly expanding regions (top left corner) collapses faster than compact slower-growing biofilms (bottom right corner).
Supplementary Video 6
Representative 3D rendering videos showing differences in tolerance to ciprofloxacin of mixed infections forming large (left) or small (right) biofilms. The two distinct regions shown were collected in the same AirGel, located millimeters apart. WT is shown in orange, ∆wspF is shown in green, and the epithelium is shown in magenta. The full time-lapse rendering of the imaging data shown in Fig. 6c.
Supplementary Video 7
Representative maximum intensity projection videos showing differences in tolerance to ciprofloxacin of mixed infections forming large (left) or small (right) biofilms. The two distinct regions shown were collected in the same AirGel, located millimeters apart. WT is shown in orange, and ∆wspF is shown in green. Note the increased survival of large biofilms. The full time lapse for images shown in Fig. 6c.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Meirelles, L.A., Vayena, E., Debache, A. et al. Pseudomonas aeruginosa faces a fitness trade-off between mucosal colonization and antibiotic tolerance during airway infection. Nat Microbiol 9, 3284–3303 (2024). https://doi.org/10.1038/s41564-024-01842-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-024-01842-3
This article is cited by
-
Pseudomonas aeruginosa: ecology, evolution, pathogenesis and antimicrobial susceptibility
Nature Reviews Microbiology (2025)
-
Effect of host microenvironment and bacterial lifestyles on antimicrobial sensitivity and implications for susceptibility testing
npj Antimicrobials and Resistance (2025)
-
Decoding host-microbe interactions with engineered human organoids
The EMBO Journal (2025)