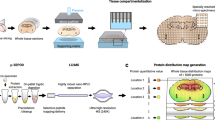
Abstract
The number of spatial omics technologies being developed is increasing1. However, a missing tool is one that can locate proteins in tissues in an untargeted manner at high spatial resolution and coverage. Here we present in situ imaging proteomics via expansion (iPEX), which integrates isotropic tissue magnification2 with matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging. iPEX provides scalable spatial resolution down to the micrometre scale and substantially increases the sensitivity of protein identification by 10–100-fold. Using the retina as a model, iPEX enabled the construction of spatial proteomic maps with high precision, the visualization of single-cell layers and extrasomatic structures and the identification of colocalized proteins. iPEX was readily applied to diverse tissues, including brain, intestine, liver and organoids, detecting 600–1,500 proteins at 1–5-µm effective pixel size. The application of iPEX to depict spatial proteomic maps in brains of mice with 5xFAD Alzheimer’s disease revealed an early-onset mitochondrial aberrancy. Notably, in young mice, the peroxisomal acetyl-CoA acyltransferase ACAA1A—of which the N392S mutant is a monogenic risk factor in Alzheimer’s disease3—was downregulated. ACAA1 depletion blocked the biosynthesis of long-chain polyunsaturated fatty acids, including docosahexaenoic acid, in multiple cellular contexts. These lipidome alterations were restored in cells overexpressing wild-type ACAA1 but not ACAA1(N392S), which suggests that the dysregulation of long-chain polyunsaturated fatty acids has an early role in neurodegeneration. Together, these results demonstrate that iPEX facilitates untargeted spatial proteomics at micrometre resolution for diverse applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Reference libraries, ion lists and annotations from iPEX data analyses and LC–MS/MS proteomic, lipidomic and metabolomic datasets included in this study are presented as Supplementary Datasets. Raw lipidomic datasets have been deposited into Zenodo (https://doi.org/10.5281/zenodo.17011514)87. Anatomical annotations of brain images were acquired from the Allen Mouse Brain Reference Atlas (https://mouse.brain-map.org) and the Allen Brain Reference Atlases (https://atlas.brain-map.org). The external dataset of proteomic analysis in 5-month-old and 10-month-old wild-type mice and 5xFAD mice used in Extended Data Fig. 6j can be found in ref. 46. Source data are provided with this paper.
Code availability
Original code for data analysis is presented as a Supplementary Code and has been deposited into GitHub (https://github.com/zouyilonglab/ipex), which is also accessible at Zenodo (https://doi.org/10.5281/zenodo.17061817)88.
References
Bressan, D., Battistoni, G. & Hannon, G. J. The dawn of spatial omics. Science 381, eabq4964 (2023).
Li, L. et al. Spatially resolved proteomics via tissue expansion. Nat. Commun. 13, 7242 (2022).
Luo, R. et al. A novel missense variant in ACAA1 contributes to early-onset Alzheimer’s disease, impairs lysosomal function, and facilitates amyloid-β pathology and cognitive decline. Signal. Transduct. Target. Ther. 6, 325 (2021).
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 (2018).
Hickey, J. W. et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat. Methods 19, 284–295 (2022).
Mund, A. et al. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 40, 1231–1240 (2022).
Bhatia, H. S. et al. Spatial proteomics in three-dimensional intact specimens. Cell 185, 5040–5058 (2022).
Ma, M. et al. In-depth mapping of protein localizations in whole tissue by micro-scaffold assisted spatial proteomics (MASP). Nat. Commun. 13, 7736 (2022).
Zubair, F. MALDI mass spectrometry based proteomics for drug discovery & development. Drug Discov. Today Technol. 40, 29–35 (2021).
Gorman, B. L. et al. Imaging and spatially resolved mass spectrometry applications in nephrology. Nat. Rev. Nephrol. 21, 399–416 (2025).
McMillen, J. C., Gutierrez, D. B., Judd, A. M., Spraggins, J. M. & Caprioli, R. M. Enhancement of tryptic peptide signals from tissue sections using MALDI IMS postionization (MALDI-2). J. Am. Soc. Mass. Spectrom. 32, 2583–2591 (2021).
Spraggins, J. M. et al. High-performance molecular imaging with MALDI trapped ion-mobility time-of-flight (timsTOF) mass spectrometry. Anal. Chem. 91, 14552–14560 (2019).
Soltwisch, J. et al. Mass spectrometry imaging with laser-induced postionization. Science 348, 211–215 (2015).
Heiles, S., Kompauer, M., Müller, M. A. & Spengler, B. Atmospheric-pressure MALDI mass spectrometry imaging at 213 nm laser wavelength. J. Am. Soc. Mass Spectrom. 31, 326–335 (2020).
Drelich, L. et al. Toward high spatially resolved proteomics using expansion microscopy. Anal. Chem. 93, 12195–12203 (2021).
Ryan, D. J., Spraggins, J. M. & Caprioli, R. M. Protein identification strategies in MALDI imaging mass spectrometry: a brief review. Curr. Opin. Chem. Biol. 48, 64–72 (2019).
Chen, F., Tillberg, P. W. & Boyden, E. S. Optical imaging. Expansion microscopy. Science 347, 543–548 (2015).
Wen, G., Leen, V., Rohand, T., Sauer, M. & Hofkens, J. Current progress in expansion microscopy: chemical strategies and applications. Chem. Rev. 123, 3299–3323 (2023).
Masland, R. H. The neuronal organization of the retina. Neuron 76, 266–280 (2012).
Uhlén, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Dahl, T. M., Reed, M., Gerstner, C. D., Ying, G. & Baehr, W. Effect of conditional deletion of cytoplasmic dynein heavy chain DYNC1H1 on postnatal photoreceptors. PLoS ONE 16, e0248354 (2021).
Rose, K., Walston, S. T. & Chen, J. Separation of photoreceptor cell compartments in mouse retina for protein analysis. Mol. Neurodegener. 12, 28 (2017).
Kizhatil, K., Sandhu, N. K., Peachey, N. S. & Bennett, V. Ankyrin-B is required for coordinated expression of beta-2-spectrin, the Na/K-ATPase and the Na/Ca exchanger in the inner segment of rod photoreceptors. Exp. Eye Res. 88, 57–64 (2009).
Ferguson, L. R., Dominguez, J. M. 2nd, Balaiya, S., Grover, S. & Chalam, K. V. Retinal thickness normative data in wild-type mice using customized miniature SD-OCT. PLoS ONE 8, e67265 (2013).
Sherry, D. M. & Heidelberger, R. Distribution of proteins associated with synaptic vesicle endocytosis in the mouse and goldfish retina. J. Comp. Neurol. 484, 440–457 (2005).
Chidlow, G., Wood, J. P. M., Knoops, B. & Casson, R. J. Expression and distribution of peroxiredoxins in the retina and optic nerve. Brain Struct. Funct. 221, 3903–3925 (2016).
Campbell, W. A. et al. Chromatin access regulates the formation of Müller glia-derived progenitor cells in the retina. Glia 71, 1729–1754 (2023).
Voogd, J. & Glickstein, M. The anatomy of the cerebellum. Trends Neurosci. 21, 370–375 (1998).
Aldinger, K. A. et al. Spatial and cell type transcriptional landscape of human cerebellar development. Nat. Neurosci. 24, 1163–1175 (2021).
Laughton, J. D. et al. Differential messenger RNA distribution of lactate dehydrogenase LDH-1 and LDH-5 isoforms in the rat brain. Neuroscience 96, 619–625 (2000).
Sjöstedt, E. et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367, eaay5947 (2020).
Allen Institute for Brain Science. Allen Reference Atlas—Mouse Brain https://mouse.brain-map.org (2011).
Hol, E. M. & Pekny, M. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr. Opin. Cell Biol. 32, 121–130 (2015).
Cesca, F., Baldelli, P., Valtorta, F. & Benfenati, F. The synapsins: key actors of synapse function and plasticity. Prog. Neurobiol. 91, 313–348 (2010).
McCarthy, N., Kraiczy, J. & Shivdasani, R. A. Cellular and molecular architecture of the intestinal stem cell niche. Nat. Cell Biol. 22, 1033–1041 (2020).
Sumigray, K. D. & Lechler, T. Desmoplakin controls microvilli length but not cell adhesion or keratin organization in the intestinal epithelium. Mol. Biol. Cell 23, 792–799 (2012).
Chen, T. et al. Myosin VI mediates the movement of NHE3 down the microvillus in intestinal epithelial cells. J. Cell Sci. 127, 3535–3545 (2014).
Lancaster, M. A & Knoblich, J. A Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340 (2014).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Tischfield, M. A. et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell 140, 74–87 (2010).
Oakley, H. et al. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006).
Depp, C. et al. Myelin dysfunction drives amyloid-β deposition in models of Alzheimer’s disease. Nature 618, 349–357 (2023).
Jafarian, Z. et al. Association of glutathione S-transferases M1, P1 and T1 variations and risk of late-onset Alzheimer’s disease. Neurol. Res. 40, 41–44 (2018).
Mao, H. et al. The expression patterns of septin-9 after traumatic brain injury in rat brain. J. Mol. Neurosci. 51, 558–566 (2013).
Kim, D. K. et al. Deep proteome profiling of the hippocampus in the 5XFAD mouse model reveals biological process alterations and a novel biomarker of Alzheimer’s disease. Exp. Mol. Med. 51, 1–17 (2019).
Yang, Z. et al. Cyclooctatetraene-conjugated cyanine mitochondrial probes minimize phototoxicity in fluorescence and nanoscopic imaging. Chem. Sci. 11, 8506–8516 (2020).
Zhang, H. et al. Bright and stable monomeric green fluorescent protein derived from StayGold. Nat. Methods 21, 657–665 (2024).
Schram, A. W. et al. Human peroxisomal 3-oxoacyl-coenzyme A thiolase deficiency. Proc. Natl Acad. Sci. USA 84, 2494–2496 (1987).
Ferdinandusse, S. et al. Identification of the peroxisomal β-oxidation enzymes involved in the biosynthesis of docosahexaenoic acid. J. Lipid Res. 42, 1987–1995 (2001).
Moore, S. A., Hurt, E., Yoder, E., Sprecher, H. & Spector, A. A. Docosahexaenoic acid synthesis in human skin fibroblasts involves peroxisomal retroconversion of tetracosahexaenoic acid. J. Lipid Res. 36, 2433–2443 (1995).
Salem, N. Jr, Litman, B., Kim, H. Y. & Gawrisch, K. Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids 36, 945–959 (2001).
Bazinet, R. P. & Layé, S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 15, 771–785 (2014).
Hein, M. Y. et al. Global organelle profiling reveals subcellular localization and remodeling at proteome scale. Cell 188, 1137–1155 (2025).
Lapillonne, A. & Moltu, S. J. Long-chain polyunsaturated fatty acids and clinical outcomes of preterm infants. Ann. Nutr. Metab. 69, 35–44 (2016).
Neumann, E. K., Djambazova, K. V., Caprioli, R. M. & Spraggins, J. M. Multimodal imaging mass spectrometry: next generation molecular mapping in biology and medicine. J. Am. Soc. Mass. Spectrom. 31, 2401–2415 (2020).
Feodorova, Y., Koch, M., Bultman, S., Michalakis, S. & Solovei, I. Quick and reliable method for retina dissociation and separation of rod photoreceptor perikarya from adult mice. MethodsX 2, 39–46 (2015).
Yang, J. et al. A quick protocol for the preparation of mouse retinal cryosections for immunohistochemistry. Open Biol. 11, 210076 (2021).
Hankin, J. A., Barkley, R. M. & Murphy, R. C. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass. Spectrom. 18, 1646–1652 (2007).
Yang, J. & Caprioli, R. M. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 83, 5728–5734 (2011).
Nakamura, R. et al. A simple method for in-depth proteome analysis of mammalian cell culture conditioned media containing fetal bovine serum. Int. J. Mol. Sci. 55, 2565 (2021).
Wang, F., Naowarojna, N. & Zou, Y. Stratifying ferroptosis sensitivity in cells and mouse tissues by photochemical activation of lipid peroxidation and fluorescent imaging. STAR Protoc. 3, 101189 (2022).
Luu, G. T. et al. TIMSCONVERT: a workflow to convert trapped ion mobility data to open data formats. Bioinformatics 38, 4046–4047 (2022).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Kreutzer, L. et al. Simultaneous metabolite MALDI-MSI, whole exome and transcriptome analysis from formalin-fixed paraffin-embedded tissue sections. Lab. Invest. 102, 1400–1405 (2022).
Heuckeroth, S. et al. On-tissue dataset-dependent MALDI-TIMS-MS2 bioimaging. Nat. Commun. 14, 7495 (2023).
Zhang, H. et al. Single-cell lipidomics enabled by dual-polarity ionization and ion mobility-mass spectrometry imaging. Nat. Commun. 14, 5185 (2023).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Thomas, P. D. et al. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22 (2022).
Tchieu, J. et al. A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell 21, 399–410 (2017).
Loh, K. M. et al. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell 166, 451–467 (2016).
Chen, S. et al. Hepatic spheroids derived from human induced pluripotent stem cells in bio-artificial liver rescue porcine acute liver failure. Cell Res. 30, 95–97 (2020).
Zhang, S. C., Wernig, M., Duncan, I. D., Brüstle, O. & Thomson, J. A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 19, 1129–1133 (2001).
Busskamp, V. et al. Rapid neurogenesis through transcriptional activation in human stem cells. Mol. Syst. Biol. 10, 760 (2014).
Rosa, F. et al. In vitro differentiated human stem cell-derived neurons reproduce synaptic synchronicity arising during neurodevelopment. Stem Cell Rep. 15, 22–37 (2020).
Schafer, S. T. et al. An in vivo neuroimmune organoid model to study human microglia phenotypes. Cell 186, 2111–2126 (2023).
Mancuso, R. et al. Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat. Neurosci. 22, 2111–2116 (2019).
Kadoshima, T. et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl Acad. Sci. USA 110, 20284–20289 (2013).
Shi, Y. et al. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 18, e3000705 (2020).
Alcamo, E. A. et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377 (2008).
Britanova, O. et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378–392 (2008).
Wang, Y. et al. ACSL4 and polyunsaturated lipids support metastatic extravasation and colonization. Cell 188, 412–429 (2025).
Yang, A. et al. Regulation of RAS palmitoyltransferases by accessory proteins and palmitoylation. Nat. Struct. Mol. Biol. 31, 436–446 (2024).
Rakotomamonjy, J. & Guemez-Gamboa, A. Purkinje cell survival in organotypic cerebellar slice cultures. J. Vis. Exp. 154, e60353 (2019).
Paxinos, G. & Franklin, K. B. J. Paxinos and Franklin’s The Mouse Brain in Stereotaxic Coordinates (Elsevier Science, 2019).
Westlake Universit & Zou, Y. iPEX enables micron-resolution spatial proteomics with hydrogel expansion [Data set]. Zenodo https://doi.org/10.5281/zenodo.17011514 (2025).
yilongzou-lab. zouyilonglab/ipex: iPEX code v1.0 (v1.0.0). Zenodo https://doi.org/10.5281/zenodo.17061817 (2025).
Acknowledgements
We thank staff at the Westlake University Mass Spectrometry and Metabolomics Core Facility, especially M. Fan, X. Bai, W. Zhang and J. Pan, for providing assistance in operations of MS instruments; staff at the High-Performance Computing Center, especially N. Li and K. Hu, for setting up the computer hardware for data analyses; P. Ji at the University of Chinese Academy of Sciences for assisting MSI experimentation and data analyses; L.Wu at Create (Beijing) Technology for support in AP-SMALDI10 MSI experimentation and data analyses; the engineers at Bruker (Beijing) Scientific Technology, including M. Wang, L. Han, X. Liu and D. Ye, for providing MALDI hardware support; C. Wang and J. Yu at Shanghai Applied Protein Technology for assisting the use of the timsTOF fleX MALDI instrument; Y.-G. Yao at the Kunming Institute of Zoology, Chinese Academy of Sciences, and R. Luo at Lanzhou University for the wild-type and ACAA–/– U251MG cells; W. Zhang for assistance with the mBaoJin fluorescent labelling plasmid; X. Li, Y. Wang, X. Wang, H. Yu, S. Cai, B. Dang, T. Guo and R. Sun at Westlake University for discussions; and members of the Zou Laboratory, especially Y. Wang, T. Shi, N. Naowarojna and Q. Peng, as well as all members from the Piatkevich Laboratory at Westlake University, for technical assistance, daily support and discussions. This work is supported in part by the National Key Research and Development Program of China (awarded to Y.Z., project number 2023YFA0914900; awarded to X.L., project numbers 2022YFA1105700 and 2022YFC3400400), in part by the ‘Pioneer’ and ‘Leading Goose’ R&D Program of Zhejiang (2024SSYS0036), in part by the Research Center for Industries of the Future of Westlake University (awarded to Y.Z., K.D.P. and X.L.), in part by the National Natural Science Foundation of China (awarded to Y.Z., project number 82273257; awarded to K.D.P., project number 32171093; awarded to X.L., project number 32370784; and awarded to F.W., project number 323B2023), in part by startup funds from Westlake Education Foundation (awarded to Y.Z., K.D.P. and X.L.), and in part by Westlake Laboratory of Life Sciences and Biomedicine (awarded to Y.Z., K.D.P. and X.L.).
Author information
Authors and Affiliations
Contributions
Y.Z. and K.D.P. conceived and supervised the project. F.W. and C.S. designed and performed the experiments with assistance from Y. Fan, S. Zhao, K.H., Z.P., J.R.H., L.Z., T.C., S. Zhang, C.Y., S.S.M., X.G., W.L., X.S., J.H., S.F., P.L. and L.D. T.W.W., H.L., F.W., S.A. and Y.Z. performed data analyses and visualization. Y. Fu, S.J. and X.L. assisted with the iPS cell culture and differentiation experiments. Y.L. assisted with the molecular biology and cell characterizations. Y.Z., K.D.P., F.W. and T.W.W. wrote the manuscript with input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
K.D.P. is the co-founder of a company that pursues commercial applications of expansion microscopy and is listed as an inventor on several patent applications concerning the development of new expansion microscopy methods. X.L. is a co-founder of iCamuno Biotherapeutics. All other authors declare that they have no competing interests related to this study. Y.Z., K.D.P., F.W., C.S., T.W.W. and H.L. are named as inventors on a pending patent application for the iPEX technology (PCT/CN2025/119421).
Peer review
Peer review information
Nature thanks Li Gan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Development of the iPEX spatial proteomic analysis method.
a-b, Expected and observed m/z values of a standard peptide mix (a) or digested bovine serum albumin (b). c, Left, key steps of iPEX sample preparation process. Right, bright-field images of mouse retina at the states of pre-expansion, post-expansion with Coomassie blue staining, and post-expansion with hydrogel dehydrated on MALDI slide. ∅, abbreviated symbol for linear expansion factor (LEF); V, abbreviated symbol for area expansion factor (AEF). d, Workflow for processing MALDI-MSI data acquired via iPEX. e, The MS1 spectra of expanded mouse retina iPEX sample. Right top, average ion intensity of iPEX detected peptides in mouse retina. f, Intensity distributions of top 30 most abundant peptide ions in mouse retina sections analysed by iPEX. g, Number of shared peptides and proteins in iPEX analysis of mouse retina sections. h, Venn diagram and scatter plot showing the overlap and correlation of proteins identified in three replicate mouse retina samples analyzed by iPEX at different EPS. i, Number of detected ions, peptides and proteins of MALDI analysis in mouse retina sections analysed via iPEX. j, Folds of relative enrichment and cross-pixel detection frequency of representative peptides from each layer of mouse retina. Scale bars indicated in images. INL: inner nuclear layer, OPL: outer plexiform layer, ONL: outer nuclear layer, RGL: retinal ganglion layer, IPL: inner plexiform layer. The schematics in a, c and d were created using BioRender (https://www.biorender.com).
Extended Data Fig. 2 iPEX enables identification of colocalized protein groups and extra-somatic structures in the mouse retina and tandem MS validations of selected peptide ions.
a-b, Ion information (a) and the MS2 spectra of the matched fragment ions acquired on the timsTOF fleX MALDI-2 platform (b) for 10 selected proteolysis-dependent precursor ions of which the MS2 spectra are matching with the MS1 annotated peptide sequences. c, Immunofluorescence (IF) staining of Dync1h1 and Sag expression in mouse retina tissues. d, Intensity distributions of indicated peptide ions in conventional MALDI and iPEX analysed mouse retina segments. Lines inside each violin represents quantile intensity value cross pixels. e-f, Correlation coefficient analysis of all annotated peptide ions (e) and top colocalized peptides (f) with distributions of the target ion (m/z 1,607.959) in the mouse retina iPEX sample. g, Ion intensity maps of the target ion and some top colocalized peptides. h-i, Correlation coefficient analysis of all annotated peptide ions (h) and top colocalized peptides (i) with distributions of the target ion (m/z 1,578.791) in the mouse retina iPEX sample. j, Ion intensity maps of some top colocalized peptides. k, IF image of Cltc expression in mouse retina tissues. l, Ion intensity maps of indicated peptide ions enriched in the neural synapse layers (OPL and IPL). The black arrow points to the outer plexiform layer. m, Experimental procedure evaluating the dynamic range of iPEX protein detection using the protein extracts from fetal bovine serum (FBS). n, Number of detected peptides and proteins in precipitated and serial-diluted FBS proteins analysed via iPEX or conventional MALDI-MSI. o, Average ion intensity (solid line) and standard deviation (in the background) of peptides detected in precipitated FBS protein analyzed via iPEX. Scale bars indicated in images. The schematic in m was created using BioRender (https://www.biorender.com).
Extended Data Fig. 3 iPEX analysis of mouse cerebellum tissues and tandem MS validations of selected peptide ions.
a, H&E staining of mouse cerebellum. b, Coomassie blue staining and bright-field image of mouse cerebellum sections. c, The MS1 spectra of expanded mouse cerebellum iPEX sample. Scatter plots showing the average ion intensity of iPEX detected peptides in mouse cerebellum. d, Peptide delocalization assessment using Cy5-NHS ester-mediated protein labeling. e, Ion intensity heatmap of representative ions with enhanced expression in the Purkinje neuron soma alignment in two iPEX sections of mouse cerebellum. The white arrow points to Purkinje neuron soma. f, Ion information for 8 selected proteolysis-dependent precursor ions of which the MS2 spectra are matching with the MS1 annotated peptide sequences. GL: granule layer; FT: fibre tract; CBN: cerebellar nuclei; ML: molecular layer. Scale bars indicated in images. The schematic in d was created using BioRender (https://www.biorender.com).
Extended Data Fig. 4 Assessments for the annotation accuracy, subcellular compartments, dynamic ranges and pixel distributions of iPEX detected proteins.
a, Left, Coomassie blue staining of post-expansion mouse cerebellum before and after microdissection on the LCM sample slide. Right, sample information for LCM-LC-MS/MS spatial proteomics. b, Distribution of ion-peptide matching count in indicated mouse cerebellum layers when using LCM- and bulk tissue-derived MS spectral libraries. c, Abundance of LCM–LC-MS/MS detected peptides in indicated regions of mouse cerebellum samples. Gray dots represent all proteins detected in LCM-LC-MS/MS and red dots represent protein detected in both LCM-LC-MS/MS and iPEX analysis. Box-and-whisker plot inside each plot indicates median value and range of corresponding scatter plot. d, Cumulative distribution function of overlapped peptides in LCM–LC-MS/MS and iPEX analysis across the abundance rank in LCM–LC-MS/MS. e, Top 10 abundant high-confidence (≥2 UPs) proteins in LCM library and iPEX analysis in mouse cerebellum. f, Percentage of iPEX or LC-MS/MS detected proteins belonging to the indicated gene ontology (GO) cellular compartment terms. g, Distribution of detected peptides and proteins (≥2 UPs) across pixels in the mouse retina iPEX sample, mouse brain iPEX sample, and human cerebral organoid iPEX sample, respectively. Box-and-whisker plot inside each violin plot indicates median and quantile value of corresponding scatter plot. h, Coomassie blue staining and bright-field images of mouse cerebral coronal sections. i, Average ion intensity of iPEX detected peptides in mouse brain samples. j, Estimated anatomical divisions suggested by the Allen Mouse Brain Atlas database33 for the iPEX brain slice. Scale bars indicated in images. Brain images in j were reproduced from the Allen Mouse Brain Reference Atlas (https://mouse.brain-map.org)33.
Extended Data Fig. 5 iPEX analysis of various mouse tissues and human cerebral organoids.
a, Number of detected ions, peptides and proteins from iPEX analysis of mouse intestine, mouse liver, and human cerebral organoids. b, The MS1 spectra and average ion intensity of iPEX detected peptides in the expanded mouse intestine iPEX sample. c, Relative enrichment fold and cross-pixel detection frequency of representative peptides from each intestinal region. d, Ion intensity heatmap of representative ions with enhanced expression in the villus, muscularis, and crypt in two mouse intestine segments via iPEX. e, Coomassie blue staining and bright-field images of mouse liver slice. f, Average ion intensity of iPEX detected peptides in mouse liver samples. g, Correlation scores of intensities of commonly detected peptides across mouse liver iPEX replicates. h, Numbers of commonly detected peptides across mouse liver iPEX replicates. i, IF images of cerebral organoids (d-91) stained for intermediate progenitor markers. j, Ion intensity heatmaps of selected ions matched to the cell type markers. k, Average ion intensity of iPEX detected peptides in the cerebral organoid. l, Top, pixel clustering map and number of detected ions, peptides and proteins of human brain organoid analysed via iPEX. Bottom, pseudo-colored ion intensity maps of ions 1st matched to validated cell type marker proteins. m, Ion intensity heatmap of selected ions matched to the cell type markers. Scale bars indicated in images.
Extended Data Fig. 6 iPEX uncovers spatial proteomic profiles of brain tissues in 5xFAD mouse models of Alzheimer’s Disease.
a-b, Fluorescent images of plaques and tangles indicated by Thioflavin-S staining (in green) of WT and 5xFAD mouse brain at indicated ages. Scale bars are indicated in the images. c, Quantification of plaques and tangles in 5xFAD mouse brain at indicated ages. For quantification, each data point represents an ROI (see Methods section) n = 14, 33, and 24 ROIs for 2-, 6-, and 8-mo, respectively. Data are presented as mean± s.d. d, Scatter plot showing the overlap and correlation of proteins identified in WT and AD mouse brain. e, The brain anatomical structures of mouse brain coronal sections shown in Fig. 3 annotated from the Allen Mouse Brain Atlas database33. f, Ion intensity heatmaps of representative ions with enhanced expression in specific brain anatomical regions of 11-mo-old WT and 5xFAD mouse revealed by iPEX. g, Folds of relative enrichment and cross-pixel detection frequency of representative peptides from each cerebral region. h, Intensity distribution of ion markers in each cerebral region. i, Differential expression analysis in corresponding brain region clusters between 11-mo-old WT and 5xFAD mouse brain section. j, Protein intensity levels of the top, indicated upregulated proteins identified in 11-mo-old 5xFAD mouse brain via iPEX analysis in an external dataset from 5-mo and 10-mo wild-type and 5xFAD mouse cohorts46. n = 6 mice for each condition. Data are presented as mean± s.d. k, STRING protein-protein interaction analysis of top 114 downregulated proteins in the fibre tract of 5xFAD mouse brain compared with that in the WT mouse. Scale bars indicated in images. Brain images in e were reproduced from the Allen Mouse Brain Reference Atlas (https://mouse.brain-map.org)33.
Extended Data Fig. 7 Mitochondrial alterations in brain sections of 5xFAD mouse model at different stages of AD pathogenesis.
a, IF images showing expression and localization of the Tom20 protein in mouse brain sections of WT and 5x FAD at 2-, 6-, and 8-mo of ages. b, Fluorescence images of 10-mo wild-type and 5x FAD mouse brains following live-tissue PK-mito-red staining. c, Fluorescence images of 2-mo wild-type and 5x FAD mouse brains following live-tissue PK-mito-red staining. d, Fluorescence images of 12-mo wild-type and 5x FAD mouse brains following live-tissue PK-mito-red staining. e, Fluorescence images of 5-mo wild-type and 5x FAD mouse brains following intracerebral injection of pAAV-Ascl1-CAGdMito2-COX8-mBaoJin. f, Transmission electron microscope (TEM) images showing the ultrastructures of brain tissues in WT and 5xFAD brain tissues at different ages including mitochondrial morphologies in the fibre tracts (top) and hippocampus (bottom) regions. Scale bars indicated in images.
Extended Data Fig. 8 iPEX uncovers early stage proteomic alterations during AD progression in 5xFAD mice.
a, The brain anatomical structures of mouse brain coronal sections shown in Fig. 4a–d annotated from the Mouse Brain in Stereotaxic Coordinates 3rd Edition86. b, Ion intensity maps of selected ions that are dysregulated in the 5xFAD mouse brain. c, Information for Acaa1 precursor ions detected from the mouse cerebrum iPEX analysis. d, The MS2 spectra of the matched fragment ions for Acaa1 precursor ions identified in the mouse cerebrum. e, Normal MALDI validation for ion quantification in one 2-mo-old WT and one 5xFAD mouse brain regions including hippocampus (HPF), fibre tracts (FT), and cortex (CTX). f-g, IF image and quantification of Acaa1 (Acaa1a) protein expression and localization in brain sections of WT and 5xFAD mice at indicated ages. For quantification, each data point represents an ROI (see Methods section). P-values are calculated using unpaired, two-tailed Student’s t-test. n = 1399, 1717, 1686, 1447, 1270, 1138, 470, 548, 485, 472, 469, 743, 369, 426, 267, 346, 272, 412, 445, 560, 448, 344, 478, 634, 205, 193, 166, 246, 493, 253, 366, 442, 324, 297, 473, 456, 165, 156, 73, 119, 177, 163, 79, 27, 34, 47, 64, 69 ROIs for each bar. Scale bars indicated in images. Lines inside each violin represents quantile intensity values among pixels.
Extended Data Fig. 9 ACAA1 contributes to homeostasis of fatty acids and membrane lipids in U251MG cells and iPS cells.
a, Biochemical reactions involved in ACAA1-mediated β-oxidation of tetracosahexaenoic acid (24:6, THA) to docosahexaenoic acid (22:6, DHA). b, Co-localizing protein network of human ACAA1 protein. The dataset was adapted from a recent report54. c, Alterations in free fatty acid (FFA) levels in U251MG cells across indicated experimental groups. d, Representative lipid species from the lipidomics analysis of phospholipid levels in U251MG cells across indicated experimental groups. e, IF images of pluripotency markers OCT3/4 and Stage-Specific Embryonic Antigen 4 (SSEA4) in single cell progenies (SCP) of WT and ACAA1 KO human iPS cells. f, Immunoblot analysis of ACAA1 expression in WT and ACAA1 KO iPS cells. g-h, Alterations in free fatty acid levels (g) and phospholipids (h) between WT and ACAA1 KO iPS cells. i, Indicated lipid species in the lipidomics analysis of phospholipid levels in iPS cells between ACAA1 WT and KO conditions in three single cell progenies (SCP). j, Alterations in FFA levels in iPS cells across indicated experimental groups. k, Percentage distribution of lipid classes identified via LC-MS/MS based lipidomics analysis in U251MG cells and iPS cells in negative (NEG) ion mode. l, Relative intensities of indicated lipid species in the lipidomics analysis of phospholipid levels in iPS cells across indicated experimental groups. All lipidomics data are presented as mean± s.d. (d,i,l). For the volcano plot (h), FDR was calculated using Benjamini-Hochberg procedure. For iPSC ACAA1 KO lipidomic analysis (i), unpaired, two-tailed Student’s t-test was used. n = 3 biologically independent cell replicates for each condition specified (d,h,i,l). Scale bars indicated in images.
Extended Data Fig. 10 Involvement of ACAA1 in fatty acid and membrane lipid metabolism in iPSCs-differentiated brain cell types.
a, Induced differentiation strategies for generating cortical neurons, glutamatergic neurons and microglia from iPS cells. b, IF images of TUBB3 and VGLUT2 in cells differentiated from indicated WT and ACAA1 KO iPS cell SCPs. c, IF images of IBA1 and P2RY12 in cells differentiated from indicated WT and ACAA1 KO iPS cell SCPs. d, IF images of neural SOX2, TBR2 and CTIP2 in cells differentiated from indicated WT and ACAA1 KO iPS cell SCPs. e, Alterations in free fatty acid species between WT and ACAA1 KO iPSC-derived cortical neurons. f, Percentage distribution of lipid classes identified in the LC-MS/MS based lipidomics analysis in the indicated three lineages of iPSC-derived neural cells in negative ion mode (NEG). g-i, Indicated lipid species in the lipidomics analysis of phospholipid levels in iPSC-derived glutamatergic neuron (g), microglia (h) and cortical neuron (i) between WT and ACAA1 KO conditions in SCP#1 (left) and SCP#2 (right). Lipidomics data are presented as mean± s.d. (g,h,i). n = 3 biologically independent cell replicates for all conditions of glutamatergic neuron, ACAA1 WT microglia SCP#1 and all conditions of cortical neurons (g,h,i). n = 4 biologically independent cell replicates for ACAA1 KO microglia SCP#1 and all conditions of microglia SCP#2 (h). Scale bars indicated in images. The schematic in a was created using BioRender (https://www.biorender.com).
Supplementary information
Supplementary Fig. 1
Raw immunoblot images presented in this study.
Supplementary Datasets
Supplementary Dataset 1–30.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, F., Sun, C., Wu, T.W. et al. iPEX enables micrometre-resolution deep spatial proteomics via tissue expansion. Nature 649, 505–514 (2026). https://doi.org/10.1038/s41586-025-09734-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09734-0