Abstract
Spatial transcriptomic techniques offer unprecedented insights into the molecular organization of complex tissues. However, integrating cost-effectiveness, high throughput, a wide field of view and compatibility with three-dimensional (3D) volumes has been challenging. Here we introduce microfluidics-assisted grid chips for spatial transcriptome sequencing (MAGIC-seq), a new method that combines carbodiimide chemistry, spatial combinatorial indexing and innovative microfluidics design. This technique allows sensitive and reproducible profiling of diverse tissue types, achieving an eightfold increase in throughput, minimal cost and reduced batch effects. MAGIC-seq breaks conventional microfluidics limits by enhancing barcoding efficiency and enables analysis of whole postnatal mouse sections, providing comprehensive cellular structure elucidation at near single-cell resolution, uncovering transcriptional variations and dynamic trajectories of mouse organogenesis. Our 3D transcriptomic atlas of the developing mouse brain, consisting of 93 sections, reveals the molecular and cellular landscape, serving as a valuable resource for neuroscience and developmental biology. Overall, MAGIC-seq is a high-throughput, cost-effective, large field of view and versatile method for spatial transcriptomic studies.
Similar content being viewed by others
Main
Spatial transcriptomics (ST) empowers researchers to uncover both spatial and molecular characteristics of tissues, offering the potential to create comprehensive three-dimensional (3D) transcriptomic maps, which is vital for understanding many fundamental biological processes including embryogenesis, organogenesis and tumor microenvironments. The construction of 3D spatial atlases typically involves stacking dozens of consecutive ST sections1,2,3,4,5,6,7. Achieving high-quality 3D mapping with this approach requires the ST method to possess the following key attributes: (1) the capacity to efficiently process a large number of ST samples with high throughput and robustness, (2) affordability to make it accessible to a broader spectrum of researchers and (3) a substantial capture area capable of encompassing entire tissue sections. However, it remains challenging to simultaneously satisfy these requirements with current ST methods8,9,10,11,12,13,14,15.
Microfluidics emerges as a promising avenue for 3D spatial atlases. Notably, microfluidics-based DBiT-seq16 has demonstrated remarkable potential in ST with distinguished cost-effectiveness and adaptability for multi-omics applications17,18. Moreover, recent studies have used and further advanced this method19,20, underscoring its widespread accessibility within the research community. Nevertheless, two primary obstacles still impede the broader adoption of microfluidics for 3D omics. First, handling hundreds of samples can be challenging, as it requires delicate manipulation to prevent tissue deformation and cross-contamination between microchannels. Second, the capture area remains limited, especially in high spatial resolution chips (only 1 mm2 for 10 μm microfluidic chip), making it unsuitable for larger tissue sections. Therefore, a microfluidics-based ST method tailored for 3D omics research, with a focus on addressing these challenges and enhancing the applicability of microfluidics to complex tissues is required.
In this study, we introduce microfluidics-assisted grid chips for spatial transcriptome sequencing (MAGIC-seq) to resolve these constraints of current ST methods. By incorporating serpentine intersecting channels, we substantially enhance the number of functional grids generated per run while reducing the average cost compared to previous techniques. Application to various tissue types demonstrated its exceptional higher sensitivity, stability and reduced batch effects. We adapted the concept of splicing screens, referred to as splicing-grid chips, substantially boosted the efficiency of barcode encoding by two orders of magnitude and broadened the capture area. We further evaluated MAGIC-seq’s capacity to resolve single cells and faithfully dissected various cell types and fine structural details from the adult mouse brain. Finally, we generated a spatiotemporal atlas of mouse organogenesis from E17.5 to P4 at the whole mouse level, and reconstructed a high-quality 3D model of the embryonic mouse brain at E18.5 using approximately one hundred interspersed tissue sections.
Results
The MAGIC-seq workflow
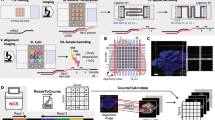
A distinctive and pivotal feature of MAGIC-seq lies in the fusion of multiple-grid microfluidics designs and prefabricated DNA arrays (Fig. 1a). First, MAGIC-seq incorporates serpentine channels to generate multiple duplicated capture areas, resulting in enhanced throughput and reduced average expenses. For instance, we can easily achieve three or nine capture areas on a slide with the multiple-grid chip (Fig. 1a and Extended Data Fig. 1a, b), each area measuring 7 mm × 7 mm and containing 4,900 spots of 50 μm × 50 μm. Second, to further expand the capture area, we reduced the intervals between adjacent grids to combine nine capture grids into a larger entirety, aligning with the concept of splicing LCD screens. Through this unique design, the capture area can be readily expanded to 21.6 mm × 21.6 mm (Extended Data Fig. 1c). Third, the strategy of prefabricating a barcoded DNA array is essential for improving the robustness. On one hand, directly barcoding the slide rather than the tissue section16 prevents potential clogging or deformation of the microfluidic channels caused by the tissue which can substantially affect consistency across channels and reproducibility among different slides. On the other hand, prefabricated slides undergo rigorous inspection before conducting ST experiments, further elevating the reliability and reproducibility. Taken together, MAGIC-seq is the only method to seamlessly combine low cost, high throughput and an extensive capture area (Fig. 1b).
a, Overview of the MAGIC-seq workflow. Custom microfluidic chips with serpentine channels were designed in three variations, triple-grid chip, nine-grid chip and splicing-grid chip. To create barcoded DNA arrays, two microfluidic PDMS chips were sequentially placed on a glass slide with carboxyl group on its surface. b, Comparison between MAGIC-seq and other spatial transcriptomic methods. c, Violin plot showing the number of genes per spot captured by MAGIC-seq in comparison with DBiT-seq, Decoder-seq and our sequenced 10x Visium datasets. The results have been normalized to 100k or 60k raw reads per 50 μm or 15 μm spot. d, Saturation curves of MAGIC-seq and other spatial transcriptomic methods for median gene counts per spot under different sequencing depths. e, Comparison of capture area and containing spots between MAGIC-seq and other methods. f, Cost comparison between MAGIC-seq and other methods excluding sequencing and downstream analysis. The panels display the cost normalized to per section, per mm2 and per spot (∼50 μm). g, Microfluidic device used for the fabrication of the nine-grid chips. Two PDMS chips (70 channels with a channel width of 50 μm) with serpentine channels perpendicular to each other were sequentially attached to the glass slide, resulting in nine barcoded arrays at their intersections. The blue and red dyes were used for illustrating the channels and resulting arrays. h, Schematic representation of the experimental evaluation of batch effect among different samples (left) and correlation of gene expression between sections (right). i, CV of top 100 gene expressions between different sections. Intraregion shows CV of genes within the same section. Intraslide shows CV of genes between two sections on the same slide. Interslide shows CV of genes between two sections from two slides. j, Spatial profiling and distribution of clusters for nine tissue types by the nine-grid chip. Tissues are colored by inferred cluster domains. For c and i, boxes, interquartile range. Centerlines and points, median. Whiskers, values within 1.5× interquartile range of the top and bottom quartiles. HIP, hippocampus; R, Spearman correlation coefficient; CV, coefficient of variation.
The workflow of MAGIC-seq includes three major steps discussed further (Fig. 1a). (1) Customized microfluidic chip design—to accommodate the diverse application requirements, we offer multiple chip architectures, including triple and nine-grid chips for different sample sizes, and splicing-grid chip for large field-of-view samples. (2) Prefabrication of a spatially barcoded DNA array via combinatorial indexing—the first microfluidic chip delivers barcodes X, which are covalently bonded to the glass slide using carbodiimide chemistry; Subsequently, the second microfluidic chip delivers barcodes Y in a perpendicular arrangement and ligated with barcode X. This spatial combinatorial indexing process generates a barcoded DNA array with a unique index for each spot. (3) Sample preparation and sequencing—tissue sections are mounted to the barcoded slide, H&E stained and permeabilized. mRNAs are then released and captured by DNA array. After in situ reverse transcription and cDNA amplification, a library containing spatial barcode, UMI and cDNA insert is constructed and subjected to paired-end sequencing. Spatial information and gene expression data are then extracted from the sequencing reads and mapped to the tissue section based on the alignment markers in the corners of each grid (Extended Data Fig. 2a).
Evaluation and benchmark of MAGIC-seq
To evaluate the performance of MAGIC-seq, we systematically benchmarked it against other ST methods including commercial platforms 10x Visium V2, and microfluidics-based approaches like DBiT-seq16 and Decoder-seq21 in gene counts, sequencing efficiency, capture area/spots, throughput and cost at approximately 50 μm or 15 μm resolution. We first compared the number of UMIs and genes detected per spot for MAGIC-seq with other methods. Notably, the fluorescence intensities across spatially barcoded arrays on the same MAGIC-seq slide were uniform (Extended Data Fig. 2b), indicating consistent modification efficiency. Similarly, the median number of UMIs and genes per spot for mouse brain sections by MAGIC-seq remained consistent across different grids, with a median of 15,563 UMIs and 5,576 genes per 50 μm spot (Fig. 1c and Extended Data Fig. 2c). MAGIC-seq outperforms other methods in gene counts per spot irrespective of 50 μm or higher resolution. After normalizing the gene counts to different sequencing depths, MAGIC-seq still excelled in brain and hippocampus samples, demonstrating its exceptional sequencing efficiency (Fig. 1d and Extended Data Fig. 2d–f). This superiority may be attributed to the high-density carboxylic group and effective modification by EDC/NHS reagent on the slide surface. Unsupervised clustering and mapping of consecutive brain sections revealed similar patterns between MAGIC-seq and 10x Visium, aligning well with distinct brain regions, such as cortex, fiber tract, hippocampus, thalamus and striatum (Extended Data Fig. 3a,b). Specifically, a single pyramidal layer consistent with its actual width suggests the excellent spatial resolution of MAGIC-seq. Moreover, correlation coefficient of three replicates on the same slide (above 0.997) and spatial expression of representative marker genes indicated its extreme reproducibility (Extended Data Figs. 3c,d and 4).
For capture area, MAGIC-seq demonstrated the capacity to cover an extensive capture area of 21.6 mm × 21.6 mm at 50 μm resolution and ~4 cm2 even at near single-cell resolution (20 μm) accommodating 202,500 spots (Fig. 1e). This represents approximately 80-fold, 40-fold and 20-fold more coverage than DBiT-seq, 10x Visium and Decoder-seq, respectively, exceeding the capacity of most existing ST methods. In terms of throughput, MAGIC-seq offers the flexibility to map multiple tissue sections or diverse tissue types simultaneously compared to single section per slide in conventional microfluidics-based methods (Fig. 1b). Meanwhile, the availability of more grids with similar reagent consumption and labor and deterministic spatial barcoding strategy without the facility-demanding and expensive decoding step resulted in an average cost of US$0.11 per mm2 for the chip fabrication, marking an 89% reduction to the single-grid chip. This reduction confers a substantial advantage over competing technologies, positioning MAGIC-seq favorably for wide adoptions (Fig. 1f).
High-throughput profiling of diverse tissues by MAGIC-seq
As the channel length extended by more than four folds in a nine-grid chip (Fig. 1g), we evaluated the impact of channel length on modification efficiency. The fluorescence measurements showed consistent distribution in the nine grids, comparable to the triple-grid chip (Extended Data Fig. 5a), suggesting minimal effects of channel length on modification efficiency. To evaluate the potential of multiple-grid chip to mitigate batch effects, we distributed four consecutive sections from an E18.5 mouse brain to two slides, with two sections each (Fig. 1h). Correlation analysis of gene expression unveiled strong correlations among the four sections, especially between sections on the same slide (>0.99) and a similar trend was observed in the results from olfactory bulbs, cerebellums and colons (Fig. 1h and Extended Data Fig. 5b,c). When we clustered and mapped spots from four brain sections together, distinct cluster segregation and distribution among sections from different slides became evident (Extended Data Fig. 5d,e). Moreover, the coefficient of variation of genes within the same brain region was higher between sections on different slides (Fig. 1i). Collectively, MAGIC-seq could effectively mitigate the batch effects between different slides through simultaneous processing of multiple sections.
To further affirm the versatility of our method, we applied MAGIC-seq to the following nine different mouse tissues: olfactory bulb, cerebellum, colon, placenta, lung, heart, liver, spleen and kidney (Fig. 1j). Notably, an average of more than 5,900 genes was detected per spot, especially for tissues such as olfactory bulb and cerebellum with more than 8,000 genes per 50 μm spot (Extended Data Fig. 5f), highlighting its higher sensitivity compared to previous methods16,20,22. Even when processing various tissue types with differing permeabilization times on the same slide, decent results were obtained (Extended Data Fig. 5b). Dimensionality reduction and clustering for the nine tissues showed clear spatial distributions that aligned with anatomical histology (Fig. 1j and Extended Data Fig. 5g,h). For example, the maternal–fetal interface in placenta, crucial for fetal nutrition and normal development, exhibited a typical layered structure such as decidua, junctional zone, labyrinth and yolk sac23,24,25,26 (Fig. 1j and Extended Data Fig. 5i). Therefore, MAGIC-seq offers the advantages of high throughput, mitigation of batch effects and adaptability for various applications.
MAGIC-seq enables spatial mapping of samples with large area
In certain situations, there arises a need to profile tissues with substantial volumes, necessitating larger capture areas which calls for an increase in the number of channels in microfluidics-based methods. However, the inherent constraints posed by the size of commonly used silica wafers in microfluidics limit the expansion of channel numbers. To address this challenge, we designed a new splicing microfluidic chip to connect individual grids into a larger whole through exceedingly narrow borders.
To demonstrate the feasibility of the splicing-grid chip, we used a third nine-square chip aligned at the intersection of the previous two chips to introduce an additional barcode Z to encode the nine grids, resulting in an area of 467 mm2 at a spatial resolution of 50 μm (Fig. 2a,b and Extended Data Fig. 1c). Due to the extremely thin walls separating grids, we used Cy3- and Cy5-labeled linker tagging the nine regions in an interleaved manner to monitor possible contamination. No occurrence of the same fluorescence in adjacent grids was detected (Fig. 2c,d), suggesting the effective separation and the preservation of barcode specificity. Furthermore, the fluorescence of spots on the splicing-grid chip was slightly lower than previous chips, and the efficiency of the additional ligation step was approximately 87% (Fig. 2e). To this end, we were able to encode an impressive 44,100 (70 × 70 × 9) spots using only 149 (70 + 70 + 9) unique barcodes. This barcoding strategy greatly improved the encoding efficiency of MAGIC-seq, far surpassing the capabilities of 10x Visium, Decoder-seq and DBiT-seq (Fig. 2f).
a, Schematic representation of the fabrication of the splicing-grid chip and downstream sample preparation. After conjugating barcode X and ligating barcode Y, a nine-square chip with narrow borders was aligned to the intersection of the previous two chips and delivered unique barcode Z to each individual subgrid, resulting in a larger splicing-grid. After H&E staining and permeabilization, mRNA was captured and RT. b, Microfluidic devices used for the fabrication of the splicing-grid chip. The first two PDMS chips had 70 channels with a channel width of 50 μm (left), forming nine subgrids at their intersections (top right). The space between adjacent subgrids was 300 μm. A third nine-square PDMS chip (bottom right) with a 200 μm separation was aligned to the intersections, tiling nine grids into one entirety (21.6 mm × 21.6 mm). c, Evaluation of liquid leakage between adjacent subgrids in the splicing-grid chip. Cy3- and Cy5-labeled linkers Y–Z were used to mark subgrids in an interleaved manner and monitor crosstalk between adjacent grids. d, The distribution of Cy3 and Cy5 fluorescence intensity of spots in each region for leakage evaluation. Boxes, interquartile range. Centerlines, median. Whiskers, values within 1.5× interquartile range of the top and bottom quartiles. The dashed box indicated the background intensity of non-labeled interleaved grids. e, The distribution of fluorescence intensity for two-round barcoding and three-round barcoding. X–Y stands for two-round barcoding, while X–Y–Z stands for three-round barcoding. The dashed lines indicate the mean value of fluorescence intensity. f, Comparison of barcoding efficiency among MAGIC-seq, 10x Visium, Decoder-seq and DBiT-seq. g,h, UMAP visualization (g) and spatial mapping (h) of spots from a P3 mouse section using the splicing-grid chip. The white dashed lines showed the gaps between the subgrids of splicing chip that have been interpolated using nearest-neighbor interpolation. i, Mean expression of marker genes for each cluster in the P3 mouse section. j, Spatial distribution of selected marker genes in the P3 mouse section. Exp., expression.
We next evaluated the performance of the splicing chip using a P3 mouse which could not be fully covered by other ST techniques. Unsupervised clustering revealed distinctive clusters, which were associated with mouse brain, lung, digestive tract, fat tissue (Fig. 2g, h), and these clusters and related marker genes displayed good continuity and spatial coherence in spite of the separation between adjacent grids (Fig. 2i,j). The successful mapping of the P3 mouse underscores the potential of MAGIC-seq in scenarios demanding a comprehensive and large field of view for spatial transcriptomic analysis.
High-definition single-cell profiling of the mouse brain
Traditional high-resolution ST methods often possess a limited field of view, capturing information from part of a mouse tissue section like hippocampus14,21 or an embryonic eye16. To obtain a large and high-resolution field of view, we advanced our splicing-grid chip to near single-cell resolution (15 μm) spanning an area of 6.48 mm × 6.48 mm which could cover the entire hemisphere instead of the hippocampus alone, as in Decoder-seq (Fig. 3a and Extended Data Fig. 1d). With higher resolution, we observed a finer structural detail of the brain which recapitulates its anatomical morphology and more precise distributions of marker genes in various brain regions (Fig. 3b and Extended Data Figs. 6a,b and 7a) compared to 50 μm. After deconvolving cell types present in each spot using single-cell data27, 73% of spots contained only one cell type, while 22% of spots contained two cell types (Fig. 3c). Oligodendrocytes, excitatory neurons, endothelial cells and inhibitory neurons accounted for a relatively large proportion and were concordant with the anatomical structures documented in previous literature27 (Fig. 3b,c).
a, Schematic representation of spatial profiling of mouse brain tissue at a 15 μm resolution. Two sections from a mouse brain were mounted to a splicing-grid chip and a single-grid chip with a spatial resolution of 15 μm. The splicing-grid chip covered the entire brain hemisphere, while the single-grid chip could merely cover an area of the hippocampus. b, Unsupervised clustering of spots (left) and spatial visualization of deconvolved cell types (right) from the mouse hemibrain section by MAGIC-seq. The white dashed lines showed the gaps between the subgrids of splicing chip that have been interpolated using nearest-neighbor interpolation. Spots and cells are colored by their annotation. Scale bar, 1 mm. c, Bottom left, bar plot showing the proportion of the number of cell types assigned per spot. Top right, pie plot showing the proportion of spots for each cell type defined by scRNA-seq data. d, Spatial mapping of selected cell types in hippocampus regions for the single-grid or splicing-grid chip. The plot shows estimated cell abundances (color intensity) of hippocampal neuron types (Ext_Hpc subtypes) and a thalamic habenular neuron type (Inh_6). e, Spatial mapping of hippocampus marker genes in the single-grid chip (top) and the splicing-grid chip (bottom). f, Correlation analysis of gene expression between the single- and splicing-grid chips in the hippocampus region. g, Measurement of cell type clustering/dispersion using Ripley’s L statistics with increasing distance. h, Spatial mapping of oligodendrocytes (Oligo_1 and Oligo_2) in the mouse brain hemisphere. HPC region was marked with dashed box. i, Spatial expression of the oligodendrocyte marker (Mbp) in the mouse brain hemisphere. Oligo, oligodendrocyte; Ext, excitatory neuron; Thal, thalamus; Endo, endothelia cell; Inh, inhibitory neuron; HPC, hippocampus; DG, dentate gyrus; Micro, microglia; Astro, astrocyte.
Next, we specifically compared the distributions of cell types and gene expression within hippocampus between the single and splicing-grid chips, and found that their patterns were strikingly similar, effectively replicating the tissue’s anatomical features (Fig. 3d,e). For example, hippocampal regions such as cornu ammonis area 1 (CA1), CA2, CA3 and the dentate gyrus featured region-specific excitatory neurons, while the habenula region was abundant in inhibitory neurons. In addition, there was a strong correlation of gene expression within the hippocampus between the two chips (Fig. 3f). Nevertheless, investigation of spatial organization of cell types using Ripley’s L statistics revealed differential cluster patterns between two chips (Fig. 3g and Extended Data Fig. 6c). For example, the aggregation level of oligodendrocytes, a widely distributed and clustered cell type expressing Mbp, increased with the distance in splicing chip but were constrained in single-grid chip (Fig. 3h,i), probably due to its limited area. We then quantitatively evaluated the spatial enrichment of different cell types using a neighborhood enrichment score and found that the splicing-grid chip excelled in revealing cellular spatial adjacencies in regions, such as the hypothalamus, olfactory areas and those situated far from the hippocampal region (Extended Data Fig. 7b). Taken together, the high-definition splicing-grid chip enabled us to gain a comprehensive understanding of the brain’s cellular structure and the interactions among different cell types.
Spatiotemporal transcriptomic atlas of the developing mouse
Postnatal development in mice represents a pivotal stage characterized by myriad cellular and molecular processes governing organogenesis, including the formation of alveoli in lungs and the regionalization of the cerebellum. We refined our splicing-grid chip to achieve superior performance in analyzing sagittal sections of mice at three critical stages (E17.5, P0, P4), crucial for lung and cerebellar development. The customized chip spans an area of 18.6 mm × 18.6 mm, containing 202,500 spots at a high resolution of 20 μm. We identified 81,723, 102,329 and 144,434 high-quality tissue spots for E17.5, P0 and P4 mice, respectively, capturing the expression of more than 22,000 genes (Fig. 4a and Extended Data Fig. 8a). Unsupervised spatially constrained clustering accurately delineated transcriptomic clusters corresponding to known tissue and organ locations and boundaries (for example, skin, muscle, thymus, heart, lung, liver, kidney). Furthermore, tissue-specific gene expression, such as Stmn2 (nervous system), Mylpf (muscle), Fabp2 (gastrointestinal tract), Sftpc (lung), Myh6 (heart) and Col2a1 (cartilage primordium), validated the precision of our approach (Fig. 4b,c and Extended Data Fig. 8b). Consistent with previous single-cell studies28, we observed elevated expression levels of hepatoblast markers such as Afp and Ahsg in livers at all three stages (Extended Data Fig. 8b), indicating ongoing hepatic cell proliferation before and after birth. In addition, elevated expression of alveolar type II epithelial (AT2) cell markers Sftpb and Sftpc in the lungs of P0 and P4 mice (Fig. 4d) suggests enhanced surfactant synthesis postnatally, critical for lung function at birth29,30.
a, Unsupervised spatially constrained clustering (top left) and tSNE visualization (bottom left) of spots from the developing mouse of E17.5, P0 and P4 based on the gene expression. Spatial distribution of the captured UMI counts (top right) was also plotted. Spots are colored by their annotation or UMI count. Scale bar, 3 mm. b, Heatmap showing the expression patterns of tissue-specific marker genes for the P4 mouse. c, Spatial visualization of tissue-specific marker genes across E17.5, P0 and P4 mouse sections. d, Stacked violin plot showing the expression levels of the differentially expressed genes in the lungs of E17.5, P0 and P4 mice. e, Unsupervised spatially constrained clustering and H&E images of the developing brain from E17.5, P0 and P4 mice. Spots are colored by brain region annotations. The red squares indicate the cerebellum region in the brain. Scale bar, 1 mm. f, Dot plot showing the expression levels of differentially expressed genes in the cerebellum of E17.5, P0 and P4 mice. g, RNA velocity streamline (left) and latent time analysis (right) of the cerebellum from E17.5, P0 and P4 brain. Spots are colored by the cerebellum annotations (left) or latent time (right). h, Heatmap displaying scaled expression pattern of top 300 likelihood genes along the latent time of cerebellum. i, Spatial expression of the dynamically changed genes across E17.5, P0 and P4 cerebellums. j, GO functional enrichment based on the top 300 dynamically changed genes across E17.5, P0 and P4 cerebellums. The significant P values of GO terms were calculated by Fisher’s exact test and adjusted by false discovery rate. CbN, cerebellar nuclei; CbWM, white matter of the cerebellum; CbXigr, internal granular layer of the cerebellum cortex; CbXpu, Purkinje cell layer of cerebellum cortex; CbXegr, external granular layer of cerebellum cortex; Cere, cerebellum; Chp, choroid plexus; EGZ, external germinal zone; PCC, Purkinje cell clusters; VZ, ventricular zone; Die, diencephalon; FMN, facial motor nucleus; Hb, hindbrain; Hy, hypothalamus; Mb, midbrain; IC, inferior colliculus; OB, olfactory bulb; OLF, olfactory areas; ORB, orbital area; PAL, pallidum; PG, pontine gray; STRd, striatum dorsal region; STRv, striatum ventral region.
The cerebellum, housing over half of the brain’s neurons, undergoes profound developmental changes from E17.5 to P4, orchestrated by intricate processes including cell proliferation, migration, differentiation and layer formation31,32,33. Through precise mapping of gene expression, we identified key brain regions consistent with the Allen Brain Atlas and finer subdivision of cortical areas, olfactory bulb and cerebellum (Fig. 4e). Combining of H&E images and transcriptional clusters, we delineated substantial morphological changes in the cerebellum across the three developmental stages. Particularly noteworthy was the pronounced emergence of the layered structure within the cerebellar cortex at P4, characterized by the inner granular layer, the Purkinje cell layer and the outer granular layer, each exhibiting distinct gene expression profiles (Fig. 4e and Extended Data Fig. 8c). Remarkably, we observed a gradual upregulation of astrocyte markers Tril, Aqp4 and Slc1a3 in the cerebellum from E17.5 to P4 mice (Fig. 4f), indicating their involvement in neuronal differentiation, granule cell migration, Bergmann glial development and cerebellar synaptogenesis at these developmental stages34.
To gain deeper insights into cerebellar cell differentiation, we employed RNA velocity and Monocle analysis at E17.5, P0 and P4 (Fig. 4g and Extended Data Fig. 8d) to identify genes and functions critical for cerebellar development. RNA velocity analysis revealed robust directional flows, consistent with the documented migration pattern of granule cells from the outer granule layer towards the cerebellar interior32, and facilitated accurate mapping of the complex developmental trajectories of the cerebellum from E17.5 to P0 and P4 (Fig. 4g and Extended Data Fig. 8d). Furthermore, pseudotime analysis identified genes undergoing dynamic changes during mouse cerebellar development (Fig. 4h). For example, Calb1, Car8 and Pcp2 showed high expression levels in the Purkinje cell layer, Tnc in the inner granular layer, Slc6a11 in the cerebellar nuclei and Banp in the outer granular layer (Fig. 4i and Extended Data Fig. 8e). Gene Ontology (GO) enrichment analysis of the top 300 dynamically changing genes revealed substantial enrichment for functions associated with neuron projection, cell cycle transition and axon development (Fig. 4j), underscoring their critical roles in postnatal cerebellar development33. Overall, these findings offer a comprehensive spatiotemporal atlas of gene expression dynamics during mouse development.
Three-dimensional spatial transcriptomic atlas of the mouse brain at E18.5
Currently, only a limited number of studies delved into 3D spatial transcriptomes due to technical limitations2,4,5,6,7,35,36,37. Motivated by the abovementioned advantages of MAGIC-seq, we created a comprehensive 3D molecular atlas of an entire E18.5 mouse brain. To establish precise 3D coordinates and capture spatial molecular profiles, we subjected 337 sections to H&E staining, of which 94 sections underwent the MAGIC-seq workflow (Fig. 5a). Notably, the fabrication of all slides (11 slides with 99 capture grids) was completed within three days and incurred a cost of only US$515, a remarkable cost advantage compared to other ST methods. After quality control, 93 sections were retained for further gene expression analysis. After mapping the spatial gene expression data onto these 3D coordinates, we successfully constructed a high-fidelity 3D molecular brain atlas with a total of 98,192 spots (50 μm × 50 μm) and median of 4,880 genes per spot (Fig. 5a).
a, Schematic representation of the workflow for reconstructing a 3D spatially resolved mouse brain. The E18.5 brain was dissected and sectioned at a thickness of 10 μm. Sections every 20 μm were used for H&E staining, while sections every 50–70 μm were subjected to ST by MAGIC-seq at a resolution of 50 μm. H&E images were used for image registration to construct 3D points and meshes. The ST data was then mapped onto the 3D coordinates to generate 3D atlases. In total, there were 337 and 93 sections for H&E staining and MAGIC-seq, respectively. b, Distribution of gene counts per spot (left) and the fraction of neuron and non-neuronal cells (right) for each brain region. The boxplot centers show median values while the minima and maxima show the 25th and 75th percentiles, respectively, and whiskers extend to the most extreme data point within 1.5× interquartile range of the outer hinge of the boxplot. c, Sankey diagram showing the distribution of all spots according to the definitions of major mouse brain regions and deconvoluted neuronal types. d, Spatial distributions of the brain regions and representative marker genes in 3D space and different planes. Spots were colored by brain regions or gene expression levels. e, Developmental trajectory of DPall. DPall in the 3D space consisted of three layers (left). UMAP plot showed the RNA velocity analysis of all spots from DPall (middle). UMAP visualization of spots from different DPall clusters of the E18.5 mouse brain in which colors indicate the latent time (right). f, Fraction of different neuronal types across major brain regions. g, Cell abundance of neuronal cells in different brain regions. h, Spatial distribution of glutamatergic, GABAergic and glycinergic neurons in the 3D model. Four slides (slides 113, 276, 444 and 597) were selected for illustration. TelA, alar plate of evaginated telencephalic vesicle; NIP, neuronal intermediate progenitor.
Next, we integrated information from the clustering analysis and the Allen Developing Mouse Brain Atlas, and systematically annotated 11 distinct subregions, spanning the forebrain, midbrain, hindbrain and meninges (Fig. 5b,c and Extended Data Fig. 9a). The median number of genes per spot across different subregions ranged from 4,220 to 5,747 (Fig. 5b), enabling a clear visualization of distinct brain regions and associated marker gene expression (Extended Data Fig. 9a,b). To validate our 3D reconstruction, we examined marker gene distribution across the whole brain from different planes and confirmed by aligning our data and the Allen Brain in situ hybridization patterns (Fig. 5d and Extended Data Fig. 9c,d). Subsequently, a pseudotime analysis to stratify the spots based on their expression in dorsal pallium (DPall) revealed a strong correlation between the pseudotime of the spots and their cortical depth, with the inner layer emerging earlier in development compared to the outer layer (Fig. 5e and Extended Data Fig. 9e), consistent with prior observations38.
To elucidate the cellular composition across diverse brain regions, we deconvoluted our data using a single-cell atlas of the developing mouse brain31, resulting in 10 classes of neuron types and 19 classes of non-neuronal cell types (Fig. 5f,g and Extended Data Fig. 9f,g). Overall, the whole brain consisted of 90.4% neurons and 9.6% non-neuronal cells (Fig. 5b). The higher proportion of neuronal cells compared to the single-cell dataset31 may be due to the relatively elevated expression of neuronal marker genes in spots containing multiple cells. Notably, this proportion varied across different brain regions, with DPall and Meninges exhibiting the lowest and highest ratios of neuronal cells, respectively (Fig. 5b). In addition, the 11 major brain regions contained different types of neurons, with glutamatergic and GABAergic neurons prevailing as the most abundant types. The glutamatergic-to-GABAergic neuron ratio exhibited marked variation across regions, with most glutamatergic neurons present in DPall and medial pallium (MPall), and GABAergic and glycinergic neurons in CSPall (Fig. 5f,g). Among the glutamatergic neurons, Slc17a7 and Slc17a6 were differentially distributed in different brain regions (Extended Data Fig. 9h). Slc17a7 dominated in the olfactory areas, alar plate of evaginated telencephalic vesicle, DPall and MPall, whereas Slc17a6 was more abundant in the midbrain and hindbrain, akin to adult brain distributions7. Serotonergic neurons were mainly observed in the midbrain, central subpallium (CSPall) and hindbrain, whereas dopaminergic neurons exhibited enrichment in the midbrain, olfactory areas and hypothalamus (Extended Data Fig. 9i). Other neurotransmitter neuron types were predominantly distributed across the hindbrain, midbrain, hypothalamus, olfactory areas and CSPall. Regarding non-neuronal cells, substantial variations in cell composition were evident across regions, with ventricular systems housing predominantly glioblasts and meninges comprising mainly fibroblasts (Extended Data Fig. 9f,g). While existing 3D brain atlases primarily focus on adult mice, our study fills a crucial gap by providing a high-resolution embryonic atlas.
Discussion
With the development of ST technologies, we have gained unprecedented insights into the molecular profiles of and interactions between cells in their native tissue context. However, current methods, despite their remarkable cellular and even subcellular resolution11,15,39,40, are hampered by substantial limitations in terms of accessibility, throughput and trade-off between resolution and field of view. To address these constraints, we combine carbodiimide chemistry, spatial combinatorial indexing and serpentine microfluidics design to develop MAGIC-seq, a sensitive and robust ST approach.
The remarkable parallel capacity and cost-effectiveness of MAGIC-seq can be largely attributed to its serpentine microfluidic channels. In traditional sequencing-based ST methods, one barcode typically corresponds to one spot. The workload involved in the decoding or spotting steps would multiply with more regions or larger areas, considerably constraining the throughput and costs. In contrast, MAGIC-seq generates strips that can crisscross each other multiple times, resulting in multiple spots sharing the same barcode combinations. Notably, this process does not need additional reagent consumption or labor. By using just 70 barcodes and incorporating three crossovers, MAGIC-seq dramatically reduces the number of required barcodes by two orders of magnitude and increases the throughput by eightfold. Reasonably, by introducing more crossovers with larger silicon wafers, the throughput and cost efficiency could be further elevated.
Furthermore, MAGIC-seq offered great compatibility in resolutions and capture areas. Most sequencing-based ST methods offer capture areas of less than 1 cm2. Consequently, profiling large tissue sections, such as entire postnatal mouse samples, has remained nearly impossible. MAGIC-seq overcomes this challenge by splicing different grids into an entirety. With more channels, for example 200 channels and four crossovers, MAGIC-seq could achieve an area of over 3.2 cm × 3.2 cm containing 640,000 spots at a resolution of 20 μm. Moreover, the flexibility of microfluidics empowers MAGIC-seq to adjust the number and width of channels, effectively modifying the layout and resolution of grids to accommodate different sample types. This adaptability is invaluable when working with specimens, such as zebrafish which possess elongated bodies and require specific considerations in designing spatial transcriptomic experiments.
Previously, only a limited number of studies have reported the reconstruction of 3D transcriptome volumes, including the adult mouse brain4,41, macaque cortex1 or early embryos2,3,36, due to various limitations. MAGIC-seq distinguishes itself with an exceptionally low cost (US$0.11 per mm2), high throughput and large field of view. Moreover, MAGIC-seq uses readily available instruments and reagents and requires no intricate manipulations or complex fluidic handling, making it more accessible to researchers. More importantly, the high reproducibility and minimal batch effect across sections and slides ensure accurate quantification of gene expression. On these bases, we have successfully built a comprehensive 3D representation of an entire mouse brain at E18.5, offering an unbiased insight into the organization and developmental trajectory of the central nervous system on a whole-brain scale and enhancing our ability to decipher the establishment of brain regionalization and the gradual acquisition of functional identities by brain subregions.
We employed MAGIC-seq to construct a comprehensive high-resolution spatial transcriptome map, illuminating organogenesis in mice across prenatal and postnatal stages. Notably, an increased presence of AT2 cells in postnatal mice enhances surfactant synthesis, crucial for reducing alveolar surface tension and preventing lung atelectasis. The prevalence of hepatoblasts suggests ongoing hepatic cell proliferation throughout prenatal and postnatal phases. Additionally, an intricate examination of the cerebellum prebirth and postbirth unveiled specific genes and functions associated with cerebellar development in mice. These findings underscore the efficacy of our technology in providing spatial and temporal insight into cellular and molecular processes.
In summary, MAGIC-seq represents a robust, user-friendly and customizable ST framework applicable to multi-omics research17,19,42,43,44. Our study presents insights that greatly enhance our understanding of the spatial and temporal patterns of gene expression during mouse development. Our creation of a 3D transcriptomic atlas of the developing brain, alongside a spatiotemporal atlas covering the entire mouse, represents a valuable resource to the fields of neuroscience and developmental biology.
Methods
Ethics statement
All experimental procedures involving animals in this study were carried out in accordance with the guidelines for procurement and use of laboratory animals and have been approved by the Institutional Animal Ethics Committee at the Institute of Zoology, Chinese Academy of Sciences.
Animal experiments
All mice used in this work were C57BL/6 and were purchased from SiPeiFu Biotechnology. Adult male mice (8–12 weeks old) were used for tissue dissection and pregnant mice (8–12 weeks old) were used for embryo collection. E17.5, E18.5, P0, P3 and P4 mice were produced by timed mating mice. The day of vaginal plug observation was considered E0.5. Animals were bred and maintained under conventional specific pathogen-free conditions and a 12-h light/12-h dark cycle at 25 °C and 40–60% humidity.
Design and fabrication of the microfluidic devices
The microfluidic devices were designed and fabricated using photoresist molds and soft lithography methods. Briefly, the device layout was designed using AutoCAD software and the chrome photolithography masks with 15 μm, 20 μm and 50 μm channels. Silicon molds for polydimethylsiloxane (PDMS) fabrication were then generated by photolithography using an SU-8 photoresist following the manufacturer’s recommendations (Microchem, SU-8 2010). The microfluidic chips were prepared by repetitive casting on the molds. The PDMS components A and B (DOW Corning, 4019862) were mixed at a ratio of 10:1. After stirring, degassing and curing at 70 °C for 4 h, the solidified chips were cut and peeled off from the molds gently. Finally, the inlet and outlet holes were formed with a metal punch. Moreover, a set of acrylic clamps corresponding to each microfluidic chip was custom designed and firmly holds the PDMS against the glass slides to prevent liquid leakage.
Generation of MAGIC-seq chips
In our workflow, we employed two different chip designs as follows: the multiple-grid (triple or nine grid) and splicing-grid chip. The former was prepared using a two-step method, while the latter was modified in a three-step manner. For detailed information on experimental protocols, refer to Supplementary Information.
In the two-step method, chip A was first aligned and placed on a carboxyl-modified glass slide and held firmly with customized acrylic clamps. Each inlet of the chip received 3 μl of one of the 70 amino-modified barcode A solutions (A1–A70, 40 μM) in 0.1 M MES buffer (pH 6.5; Coolaber, SL33002X) containing N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC; TCI, D1601) and N-hydroxysuccinimide (NHS; Thermo Fisher Scientific, 24500). A unique barcode A was introduced into each inlet, and negative pressure, generated by a house vacuum pump, was used to draw the barcode A into the channels. The amino group at the 5′ termini of barcode A reacted with the carboxyl group activated by EDC/NHS on the slide surface. After being incubated at room temperature within an enclosed humid chamber for 2 h, the barcode A solution was removed and the channels were rinsed with 0.1 M MES buffer continuously for 5 min. Then the acrylic clamp was removed and the chip A was detached from the glass slide. The slide was immediately immersed in a blocking buffer (0.1 M Tris–HCl, 100 mM EDC, 100 mM NHS, 50 mM ethanolamine and 0.1% SDS, pH 7.0) at 50 °C for 30 min, washed sequentially by 2× SSC buffer containing 0.1% SDS for 10 min, 0.2× SSC buffer for 2 min, rinsed by MilliQ water at room temperature.
After drying the slide under a stream of clean compressed air, chip B with microfluidic channels perpendicular to the direction of chip A was aligned and attached to the dried slides. Ligation mix of 3 μl, which included 1× T4 ligation buffer, 25 μM barcode B (one unique barcode B for each channel) pre-annealed with linker oligo, 5 μM Cy3-labeled linker, 0.2 mg ml−1 BSA, 20 U μl−1 T4 DNA ligase (NEB, M0202L), was added to the inlet ports of chip B. The ligation reaction was performed in a humid chamber at room temperature for 2 h before being aspirated and washed by 2× SSC + 0.1% SDS, 0.2× SSC and 0.1× SSC. After dried by a stream of clean air, spatially barcoded arrays were formed in the center of the slide at the intersections of two chips. Marks were drawn around the corner of each array using a Tungsten Steel scriber to assist with the following image alignment. Images from the bright field and confocal 561 nm or 640 nm laser were acquired using an Olympus IXplore SpinSR confocal microscope (Olympus) with ×10 objective using the cellSens software.
To generate a splicing-grid slide, there were three rounds of barcode modifications (barcodes X, Y and Z). Amino-modified barcode X was immobilized to the slide through an EDC/NHS-assisted carbodiimide chemistry same as the two-step method. Barcodes Y and Z were then sequentially connected to the slide through a linker-based T4 ligation. After imaging, the prepared slides were stored under inert conditions in an enclosed desiccator and protected from light at room temperature.
Tissue collection and cryosection
After killing, mouse tissues, including cerebrum, cerebellum, olfactory bulb, kidney, colon, placenta, liver, lung, spleen and heart, along with E18.5 embryo brains, were immediately isolated, washed in cold PBS, embedded in Tissue-Tek optimal cutting temperature (OCT) compounds (Sakura Finetek, 4583), snap-frozen in liquid nitrogen prechilled isopentane and transferred to a −80 °C freezer before the experiment. P3 mice were directly embedded in OCT after being killed. For adult brains, cerebellum and cerebrum were embedded separately. For liver, lung, spleen and heart, mice were perfused with PBS before dissection.
For sectioning, the tissues embedded in the cryomolds were equilibrated to the chamber temperature (−20 °C) for at least 30 min in a Leica CM1950 cryostat (Leica Biosystems). The tissue blocks were trimmed and sectioned to a thickness of 10 μm. These sections were placed within the capture area by gently touching the active surface of the slide and adhered to the slide by warming at the backside of the capture area with a finger. After sectioning, the slides were directly used or stored in a sealed package at −80 °C within one week.
H&E staining and imaging
Tissue sections were incubated on a thermocycler adaptor at 37 °C for 1 min and fixed in prechilled methanol for 30 min at −20 °C. Next, the sections were covered with 1 ml isopropanol, incubated for 1 min and air-dried for 5 min at room temperature after removal of remaining isopropanol. Then the sections were stained using H&E staining kit (Solarbio, G1120) with hematoxylin for 2 min, bluing buffer for 20 s and eosin for 1 min, followed by washing with MilliQ water after each stain. When the sections become opaque, incubate the slide at 37 °C for 5 min. Bright-field images were acquired using an Olympus IXplore SpinSR confocal microscope (Olympus) with ×10 objective using the cellSens software.
Tissue permeabilization and reverse transcription
H&E-stained tissue slide was assembled with customized metal clamps to form wells on the slide without leakage for the following reactions. In total, 70 μl permeabilization solution (0.1% pepsin in 0.1 N HCl) was added to each well. After incubation at 37 °C for 6–15 min, the permeabilization solution was removed and the well was washed with 0.1× SSC buffer. Then, 70 μl reverse transcribed (RT) mix, containing 1× RT buffer, 1 mM dNTP (NEB, N0447L), 0.2 mg ml−1 BSA, 2.5 μM template switch oligos, 2 U μl−1 RNase inhibitor (Enzymatics, Y9240L), 10 U μl−1 Maxima H minus reverse transcriptase (Thermo Fisher Scientific, EP0751), was added to the well and reverse transcription was performed at 42 °C for 1.5 h.
Tissue removal and secondary strand synthesis
After reverse transcription, the well was washed with 0.1× SSC and the tissue remainder and mRNA were eliminated using 0.08 M KOH for 5 min. A second-strand synthesis solution, containing 1× isothermal amplification buffer, 1 mM dNTPs, 4 mM MgSO4, 2.5 μM second-strand primer, 0.32 U μl−1 Bst2.0 DNA polymerase (NEB, M0537S), was added and incubated at 65 °C for 15 min. Second-strand DNA was eluted from each well with 70 μl 0.08 M KOH at room temperature for 10 min. The eluent was neutralized by 10 μl 1 M Tris solution (pH 7.0; Invitrogen, AM9850G) and purified using Zymo spin IC column (Zymo Research) according to the manufacturer’s instructions. DNA elution buffer of 27 μl was used to elute cDNA from the column.
DNA amplification
The purified cDNA was amplified with a PCR mixture of Kapa HiFi HotStart Ready Mix (KAPA Biosystems, KK2602) and 1 μM cDNA amplification primers (Primers 1 and 2). PCR reactions were performed using the following conditions: initially incubate at 95 °C for 3 min, cycled at 98 °C for 20 s, 65 °C for 30 s and 72 °C for 2 min, and finally incubate at 72 °C for 5 min. The PCR product was cleaned up with Kapa Pure Beads (KAPA Biosystems, KK8001) according to the manufacturer’s protocol. The concentration of cDNA amplification product was quantified by Qubit dsDNA HS Assay Kit (Invitrogen, Q33230).
Library construction and sequencing
To construct libraries for sequencing, a total of 100 ng DNA from each sample was used as input for NEBNext Ultra II FS DNA Library Prep Kit for Illumina (NEB, E7805S). First, DNA was fragmented at 37 °C for 6 min, after which the reactions were terminated by incubation at 65 °C for 30 min. Then a pre-annealed adapter containing a partial sequence of Illumina Truseq Read 2 was ligated to the cDNA fragment at 20 °C for 15 min. After being cleaned up with Kapa Pure Beads, the ligation product was size selected with a beads-to-sample ratio of 0.65× to 0.5×. Finally, the adapter-ligated DNA was amplified using Q5 master mix (NEB) in the kit and 1 μM library amplification primers (P5 and P7 oligos) with sample index. The PCR products were purified by Kapa Pure Beads at a beads-to-sample ratio of 0.8×, quantified using Qubit dsDNA HS Assay Kit and analyzed using Agilent Bioanalyzer High Sensitivity Chip (Agilent). The resulting libraries were sequenced on a NovaSeq 6000 platform (Illumina) or Element AVITI platform (Element Biosciences) in PE150 or PE75 mode.
DNA barcodes and oligos
DNA barcodes and oligos used for grid chip generation and library constructions were synthesized in Sangon Biotech and the detailed sequences are shown in Supplementary Table 1.
Mapping spots to the H&E image
ImageJ was used to obtain alignment markers and spatial coordinates of corner spots from bright field, fluorescence and H&E-stained images acquired after chip preparation. Next, the OTSU algorithm, specifically the maximum inter-class variance method, was employed to segment and binarize the H&E images to obtain the tissue masks. Custom scripts were developed to align the coordinates of the alignment markers and corner spots with the H&E images through affine transformation. Finally, spots covered by the identified tissue masks were recognized based on the transformed coordinates.
Image registration
To register the images, the original high-resolution H&E images were first reduced to low resolution to expedite the registration process. Subsequently, binarized images were generated using the OTSU algorithm45. The image registration process was initiated by anchoring the first image’s position. The second image underwent affine transformations, including translation, rotation and scaling. After the transformation, the registration loss was computed by measuring the difference between the transformed image and the first image. The registration matrix with the lowest loss value was ultimately selected. This process was sequentially applied to align each subsequent image with the preceding one. Finally, a set of transformation matrices was derived to register all the H&E images.
Three-dimensional transcriptome reconstruction and visualization
Using the obtained H&E registration matrices, we aligned and positioned all H&E images within the same 3D coordinate system. The open3d (v0.17.0) package46 was used to generate 3D point clouds from these coordinates. Next, the Spateo (v1.0.2) package47 constructed 3D meshes based on the Marching Cubes algorithm for visualization. The registration matrices were then used to project the spatial spot coordinates into the 3D coordinate system. Finally, the K3D (v2.16.1) and pyvista (v0.41.1) packages48 enabled the 3D visualization of the point cloud, 3D mesh and transcriptomic data. In summary, the image registration provided the necessary alignment for 3D reconstruction, point clouds were instrumental in mesh generation and coordinate mapping localized spots within the 3D space, allowing for the integrated visualization of both imaging and omics data.
Generation of gene expression matrix
The seqkit49 (v2.0.0) tool was used to extract spatial coordinates and UMI sequences from read 1 of the original sequencing data. The STARsolo pipeline of STAR (v2.7.10b) software50 was then used to process the spatial transcriptomic data by aligning to the mouse reference genome (mm10), obtaining the raw gene expression matrix for ST. The rows represented the spatial coordinate sequences and the columns represented the aligned genes. The spatial coordinate sequences were then aligned with the spatial coordinates of the fluorescence chip images, finally yielding the gene expression matrix with spatial coordinate information for downstream analysis.
Comparison of MAGIC-seq with published methods
To compare MAGIC-seq, 10x Visium and other microfluidic technologies (DBiT-seq and Decoder-seq), we used the same type of samples and resolution. We also normalized the raw sequencing data by sampling according to the tissue area, ensuring that each spot had the same number of raw reads. Subsequently, we processed the sequencing data using the same workflow and parameters.
Data preprocessing and integration
Spots that were not covered by tissue and spots with fewer than 500 detected genes were excluded. Genes expressed in fewer than 10 spots were also filtered out. Subsequently, the filtered gene expression data were normalized to 10,000 counts per spot by using the ‘normalize total’ function in SCANPY51 (v1.9.3), ensuring that the counts were made comparable across all spots. The normalized expression matrix was then subjected to logarithmic transformation for the identification of spatially variable genes. Harmony52 (v0.0.9) was used to integrate data from multiple samples.
Dimension reduction, clustering and marker identification
Dimensionality reduction was performed using principal component analysis (PCA). Clustering was performed by constructing a shared nearest-neighbor graph based on the transcriptomic data using established components and clusters identified through the Louvain53 or Leiden algorithm54. For spatially constrained clustering, we built a spatial k-nearest-neighbor graph using Squidpy55 (v1.3.0) and combined it with transcriptomic data. A 2D UMAP or tSNE embedding was constructed from the established top principal components for each tissue region. The marker genes for each cluster and differential expression for different groups were determined by the Wilcoxon rank-sum test.
Cell-type deconvolution and neighborhood enrichment analysis
To spatially map cell types of mouse brain, we used the snRNA-seq dataset of mouse whole brain by 10x Genomics Chromium platform. and performed deconvolution for each section by the cell2location (v0.1.3). We computed the neighborhood enrichment score using the nhood_enrichment function in Squidpy55. If the spot distance is close, the score will be high, otherwise it will be low. To investigate the spatial organization of various cell types, we computed the Ripley’s L statistic using the Ripley function in Squidpy to evaluate whether specific cell types exhibited dispersion, clustering or random distribution.
Vector fields and developmental trajectory calculation
Morphogenesis vector fields were calculated by spateo. First, we extracted spots of DPall and MPall, and placed spots from adult and embryo mouse brain in the same coordinate system based on gene expression information. Then, we used aligned coordinates and optimal transport couplings to learn a morphometric vector field using sparseVFC56 and predicted the cell developmental trajectory based on the vector field.
Spatial transcriptomic interpolation and exception handling
For the missing gaps and anomalous channel data of splicing-grid chips, we applied nearest-neighbor interpolation57, which is commonly used in image scaling algorithms, to fill and replace the data. Specifically, we located the coordinate of spots to be interpolated or replaced based on the image information and the original gene expression data, and used the nearest spots data for interpolation or replacement.
Pseudotime analysis
To elucidate the pseudotime dynamics in mouse cerebellum and cortex development, we used Monocle3 (ref. 58) to construct pseudotime trajectories for each region. We used the learn_graph function of py_monocle to learn the principal graph for the UMAP embeddings of the gene expression matrix. We then assigned pseudotime to individual projected points based on their distance to a root point within an undirected acyclic graph.
RNA velocity analysis
The unspliced and spliced RNA data matrix was preprocessed using scVelo59 (v0.3.2), which included normalization, selection of characteristic genes and PCA dimension reduction. Default parameters were used to estimate the dynamic parameters and gene-wise RNA velocity vectors. These velocity vectors were projected onto the UMAP space for visualization and analysis. We calculated a gene-shared latent time using the dynamical model to recover the underlying cellular processes.
GO enrichment analysis
GO enrichment analysis for selected genes was performed using GSEApy60 (v1.1.1). The significant P values of GO terms were calculated by Fisher’s exact test and adjusted by false discovery rate.
Statistics and reproducibility
No statistical method was used to predetermine the sample size. No data were excluded from the analyses. All boxplots consist of upper and lower limits (first and third quartiles), middle line (median), and whiskers extending to the largest/smallest values within 1.5× the interquartile range from the limits of each box. P value <0.05 was considered statistically significant unless otherwise stated. Statistical analysis and related plots were carried out using Python Jupyter Note with numpy (v1.23.3) and pandas (v1.5.0), matplotlib (v3.6.3), and seaborn (v0.12.2). All codes to replicate the analysis are available as part of code availability.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The MAGIC-seq and 10x Visium sequencing data reported in this study are available from the Genome Sequence Archive (GSA) in the National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences under the accession code PRJCA026358. In detail, the raw sequencing data have been deposited under the accession code CRA016614. DBiT-seq sequencing data were obtained from NCBI’s Gene Expression Omnibus archive under accession GSE137986 and Decoder-seq sequencing data were obtained from NCBI under accession GSE235896. The snRNA-seq data used for adult mouse brain deconvolution were accessed in ArrayExpress under accession E-MTAB-11115. The snRNA-seq data used for developing mouse brain deconvolution were accessed in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under accession PRJNA637987. The processed MERFISH data were accessed in the Allen Brain Cell Atlas. All the image and spatial barcode files used for MAGIC-seq are available at Zenodo (https://doi.org/10.5281/zenodo.11243023)61. Source data are provided at GitHub (https://github.com/bioinfo-biols/MAGIC-seq/tree/main/data/source_data) and Zenodo (https://doi.org/10.5281/zenodo.13235505)62.
Code availability
All the codes for sequencing processing and data analysis are available at GitHub (https://github.com/bioinfo-biols/MAGIC-seq) and Zenodo (https://doi.org/10.5281/zenodo.13235505)62.
References
Chen, A. et al. Single-cell spatial transcriptome reveals cell-type organization in the macaque cortex. Cell 186, 3726–3743 (2023).
Qu, F. et al. Three-dimensional molecular architecture of mouse organogenesis. Nat. Commun. 14, 4599 (2023).
Wang, M. et al. High-resolution 3D spatiotemporal transcriptomic maps of developing Drosophila embryos and larvae. Dev. Cell 57, 1271–1283 (2022).
Ortiz, C. et al. Molecular atlas of the adult mouse brain. Sci. Adv. 6, eabb3446 (2020).
Yao, Z. et al. A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332 (2023).
Langlieb, J. et al. The molecular cytoarchitecture of the adult mouse brain. Nature 624, 333–342 (2023).
Zhang, M. et al. Molecularly defined and spatially resolved cell atlas of the whole mouse brain. Nature 624, 343–354 (2023).
Arutyunyan, A. et al. Spatial multiomics map of trophoblast development in early pregnancy. Nature 616, 143–151 (2023).
Cassier, P. A. et al. Netrin-1 blockade inhibits tumour growth and EMT features in endometrial cancer. Nature 620, 409–416 (2023).
Kanemaru, K. et al. Spatially resolved multiomics of human cardiac niches. Nature 619, 801–810 (2023).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Wei, X. et al. Single-cell Stereo-seq reveals induced progenitor cells involved in axolotl brain regeneration. Science 377, eabp9444 (2022).
Yang, M. et al. Spatiotemporal insight into early pregnancy governed by immune-featured stromal cells. Cell 186, 4271–4288 (2023).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Fu, X. et al. Polony gels enable amplifiable DNA stamping and spatial transcriptomics of chronic pain. Cell 185, 4621–4633 (2022).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681 (2020).
Zhang, D. et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Liu, Y. et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41, 1405–1409 (2023).
Jiang, F. et al. Simultaneous profiling of spatial gene expression and chromatin accessibility during mouse brain development. Nat. Methods 20, 1048–1057 (2023).
Wirth, J. et al. Spatial transcriptomics using multiplexed deterministic barcoding in tissue. Nat. Commun. 14, 1523 (2023).
Cao, J. et al. Decoder-seq enhances mRNA capture efficiency in spatial RNA sequencing. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-02086-y (2024).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Islam, A. et al. Fatty acid binding protein 3 is involved in n-3 and n-6 PUFA transport in mouse trophoblasts. J. Nutr. 144, 1509–1516 (2014).
Tomasi, T. B. Structure and function of alpha-fetoprotein. Annu. Rev. Med. 28, 453–465 (1977).
Nie, G., Li, Y., He, H., Findlay, J. K. & Salamonsen, L. A. HtrA3, a serine protease possessing an IGF-binding domain, is selectively expressed at the maternal-fetal interface during placentation in the mouse. Placenta 27, 491–501 (2006).
Simmons, D. G., Rawn, S., Davies, A., Hughes, M. & Cross, J. C. Spatial and temporal expression of the 23 murine prolactin/placental lactogen-related genes is not associated with their position in the locus. BMC Genomics 9, 352 (2008).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Liang, Y. et al. Temporal analyses of postnatal liver development and maturation by single-cell transcriptomics. Dev. Cell 57, 398–414.e5 (2022).
Guo, M. et al. Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nat. Commun. 10, 37 (2019).
Whitsett, J. A., Wert, S. E. & Weaver, T. E. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu. Rev. Med. 61, 105–119 (2010).
La Manno, G. et al. Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021).
Marzban, H. et al. Cellular commitment in the developing cerebellum. Front. Cell Neurosci. 8, 450 (2014).
Sepp, M. et al. Cellular development and evolution of the mammalian cerebellum. Nature 625, 788–796 (2024).
Araujo, A. P. B., Carpi-Santos, R. & Gomes, F. C. A. The role of astrocytes in the development of the cerebellum. Cerebellum 18, 1017–1035 (2019).
Asp, M. et al. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 179, 1647–1660 (2019).
Sampath Kumar, A. et al. Spatiotemporal transcriptomic maps of whole mouse embryos at the onset of organogenesis. Nat. Genet. 55, 1176–1185 (2023).
Vickovic, S. et al. Three-dimensional spatial transcriptomics uncovers cell type localizations in the human rheumatoid arthritis synovium. Commun. Biol. 5, 129 (2022).
Zhang, M. et al. Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137–143 (2021).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Cho, C.-S. et al. Microscopic examination of spatial transcriptome using Seq-Scope. Cell 184, 3559–3572 (2021).
Shi, H. et al. Spatial atlas of the mouse central nervous system at molecular resolution. Nature 622, 552–561 (2023).
McKellar, D. W. et al. Spatial mapping of the total transcriptome by in situ polyadenylation. Nat. Biotechnol. 41, 513–520 (2023).
Ben-Chetrit, N. et al. Integration of whole transcriptome spatial profiling with protein markers. Nat. Biotechnol. 41, 788–793 (2023).
Llorens-Bobadilla, E. et al. Solid-phase capture and profiling of open chromatin by spatial ATAC. Nat. Biotechnol. 41, 1085–1088 (2023).
Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man. Cybern. 9, 62–66 (1979).
Zhou, Q., Park, J. & Koltun, V. Open3D: A modern library for 3D data processing. Preprint at https://arxiv.org/abs/1801.09847 (2018).
Qiu, X. et al. Spateo: multidimensional spatiotemporal modeling of single-cell spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2022.12.07.519417 (2022).
Sullivan, C. B. & Kaszynski, A. A. PyVista: 3D plotting and mesh analysis through a streamlined interface for the Visualization Toolkit (VTK). J. Open Source Softw. 4, 1450 (2019).
Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 11, e0163962 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods 19, 171–178 (2022).
Ma, J., Zhao, J., Tian, J., Bai, X. & Tu, Z. Regularized vector field learning with sparse approximation for mismatch removal. Pattern Recognit. 46, 3519–3532 (2013).
Lehmann, T. M., Gönner, C. & Spitzer, K. Survey: interpolation methods in medical image processing. IEEE Trans. Med. Imaging 18, 1049–1075 (1999).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
Fang, Z., Liu, X. & Peltz, G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 39, btac757 (2023).
Kun, P. Image and barcode files for MAGIC-seq. Zenodo https://doi.org/10.5281/zenodo.11243023 (2024).
Zhu, J. et al. Custom microfluidic chip design enables cost-effective three-dimensional spatiotemporal transcriptomics with a wide field of view. Zenodo https://doi.org/10.5281/zenodo.13235505 (2024).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32025009 to F.Z.), Natural Science Foundation of Beijing, China (Z230007 to F.Z.) and National Key R&D Project (2022YFA1303900, 2021YFA1300500 and 2022YFC2703200 to F.Z.).
Author information
Authors and Affiliations
Contributions
F.Z. conceived the study. J.Z., B.H., N. W. and Z. J. performed the experiments. J.Z. and B.H. generated the data. K.P., J.Z., R.H. and P.J. analyzed the data. J.Z., K.P., B.H. and F.Z. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Bo Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Schematic representation and AutoCAD designs of the polydimethylsiloxane (PDMS) chip for MAGIC-seq.
a. Design of triple-grid chips X and Y; each chip possesses 70 channels with a channel width of 50 μm. b. Design of nine-grid chips X and Y; each chip possesses 70 channels with a channel width of 50 μm. c. Design of splicing-grid chips X, Y and Z; chips X and Y possess 70 channels with a channel width of 50 μm. d. Design of splicing-grid chips X, Y and Z; chips X and Y possess 70 channels with a channel width of 15 μm.
Extended Data Fig. 2 Data analysis and comparison of MAGIC-seq with other methods.
a. Sequencing and data analysis of MAGIC-seq. After staining, permeabilization and reverse transcription (RT), sequencing libraries were prepared and sequenced by paired-end reads. The images of spots and H&E staining were aligned using AFIR algorithm. The sequencing reads were transformed into a spot-gene count matrix which was then remapped to the spots on the tissue sections. AFIR, affine transformation and image registration. b. Sample preparation device and fluorescence images for a triple-grid chip. The sample preparation device (top) consists of an aluminum alloy clamp, a three-chamber silicone adaptor and a barcoded slide with tissue stained. After assembly with screws in the corner, enzymatic reactions and washing steps could be performed through the well on the top. Representative fluorescence images of barcoded arrays in triple-grid chip were shown (bottom left). The fluorescence of each spot in each region was measured and plotted (bottom right). Boxes, interquartile range. Centerlines, median. Whiskers, values within 1.5× interquartile range of the top and bottom quartiles. c. Violin plot showing the number of UMIs per spot captured by MAGIC-seq in comparison with DBiT-seq, Decoder-seq and our sequenced 10x Visium datasets. d. Violin plot showing the number of genes and UMIs per spot captured by MAGIC-seq in comparison with Decoder-seq for mouse olfactory bulb (MOB) at different resolutions. For c and d, boxes, interquartile range. Centerlines, median. Whiskers, values within 1.5× interquartile range of the top and bottom quartiles. e. Saturation curves of median UMI counts per spot using MAGIC-seq and other spatial transcriptomic methods under different sequencing depths. f Saturation curves of median gene (left) and UMI (right) counts per spot for mouse olfactory bulb using MAGIC-seq and Decoder-seq under different sequencing depths.
Extended Data Fig. 3 Clustering analysis and comparison between MAGIC-seq and 10x Visium.
a. UMAP plots of MAGIC-seq and 10x Visium samples based on gene expression. The colored dots represented spots from different samples (top) and clusters (bottom). b. Spatial distribution of clusters across different mouse brain samples from MAGIC-seq and 10x Visium. Samples starting with ‘grid’ were processed by MAGIC-seq at 50 μm resolution and samples starting with ‘Visium’ were processed by 10x Visium for comparison. c. Correlation analysis of gene expression among different samples. R, Spearman correlation coefficient. d. Mean expression of top 5 marker genes for each cluster across different samples. Each row represents a cluster from a sample, and each column represents a gene.
Extended Data Fig. 4 Spatial expression of region-specific marker genes in the mouse brain.
Samples starting with ‘grid’ were processed by MAGIC-seq at 50 μm resolution and samples starting with ‘Visium’ were processed by 10x Visium.
Extended Data Fig. 5 MAGIC-seq enables spatial transcriptomic profiling of diverse tissue types at high throughput.
a. Fluorescence intensity of barcoded arrays in the nine-grid chip. Cy3-labeled linker was used to mark the position of all spots and the average fluorescence of each spot was calculated and summarized for each region. Boxes, interquartile range. Center lines, median. Whiskers, values within 1.5× interquartile range of the top and bottom quartiles. b. Evaluation of MAGIC-seq’s performance among different regions on the same slide. Three types of tissues (olfactory bulb, cerebellum and colon) were sectioned at a thickness of 10 μm and mounted to the same nine-grid chip. The sections on the left or right side for each tissue type were selected as replicates for downstream sample preparation and sequencing analysis. Unsupervised clustering and spatial mapping of all spots from two replicates of each tissue type were performed. The correlation between two replicates was calculated based on gene expression of all spots. c. Correlation analysis of gene expression among four brain sections, related to Fig. 1h. R, Spearman correlation coefficient. d. UMAP plots of spots from four brain sections. The colored dots represented spots from different sections (left) and clusters (right), respectively, related to Fig. 1h. e. Spatial distribution of clusters across different brain sections. f. Distribution of the number of UMI (left) and gene (right) counts per spot for the nine tissue types. Boxes, interquartile range. Center lines, median. Whiskers, values within 1.5× interquartile range of the top and bottom quartiles. g. Unsupervised clustering of spots from nine tissue types based on gene expression. h. Mean expression of marker genes for each cluster in the nine tissue types. i. Spatial mapping of representative marker genes for each cluster in the placenta. Exp., expression.
Extended Data Fig. 6 MAGIC-seq enables high-definition profiling of the mouse brain.
a. Unsupervised clustering and spatial mapping of all spots from mouse brain section using the splicing-screen chip. b. Unsupervised clustering and spatial mapping of all spots from mouse brain section using the single-grid chip. c. Measurement of cell type clustering/dispersion using Ripley’s L statistics with increasing distance.
Extended Data Fig. 7 Comparison between the single and splicing grid chip.
a. Spatial expression of region-specific marker genes in mouse brain for the splicing grid chip at 15 μm resolution. b. Neighborhood enrichment analysis of cell types in the single (left) and splicing (right) grid chip.
Extended Data Fig. 8 Spatially resolved transcriptomic atlas of mouse organogenesis.
Related to Fig. 4. a. Spatial visualization of the captured gene counts across sections from E17.5, P0 and P4 mice. b. Spatial visualization of tissue-specific marker genes across sections from E17.5, P0 and P4 mice. c. Dot plot showing the expression levels of the differentially expressed genes in the developing cerebellum. d. RNA velocity streamline plot (left) and pseudotime trajectory analysis (right and middle, respectively) of the cerebellums from E17.5, P0 and P4 mice. Spots are colored by the developmental stages (left), cerebellum regions (middle) or pseudotime (right). e. Spatial visualization of the dynamically changed genes across E17.5, P0 and P4 cerebellums. Exp., expression.
Extended Data Fig. 9 Spatial distribution of region-specific marker genes and neurotransmitter-related genes in 3D brain atlas.
a. Three-dimensional visualization of spots from the nine typical brain regions throughout the whole brain. b. Heatmap showing the expression of top 3 expressed genes enriched in individual brain regions (colored by bars). c. The Allen Brain Atlas in situ hybridization (ISH) validation of spatial expression of representative region-specific marker genes, related to Fig. 5d. d. Spatial expression of representative region-specific marker genes in the 3D space and validation by the Allen Brain Atlas in situ hybridization (ISH) results. e. Heatmap showing the scaled expression pattern of genes identified along the latent time of DPall. f. Fraction of non-neuronal cells in different brain regions. g. Cell abundance of non-neuronal cells in different brain regions. h. Spatial expression of Slc17a7 and Slc17a6 in the E18.5 and adult mouse brain and validation of E18.5 brain from the Allen Brain Atlas. i. Spatial distribution of serotoninergic and dopaminergic neurons in the 3D model. Four slides, slide 113, 276, 444, and 597 were selected for illustration. Exp., expression.
Supplementary information
Supplementary Information
Supplementary Note (MAGIC-seq protocol).
Supplementary Table 1
PCR oligos and barcodes used in this study.
Supplementary Data 1
CAD file for microfluidic chips used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, J., Pang, K., Hu, B. et al. Custom microfluidic chip design enables cost-effective three-dimensional spatiotemporal transcriptomics with a wide field of view. Nat Genet 56, 2259–2270 (2024). https://doi.org/10.1038/s41588-024-01906-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-024-01906-4
This article is cited by
-
Spatial omics in 3D culture model systems: decoding cellular positioning mechanisms and microenvironmental dynamics
Journal of Translational Medicine (2025)
-
From morphology to single-cell molecules: high-resolution 3D histology in biomedicine
Molecular Cancer (2025)
-
Targeting leukemic stem cell biomechanics suppresses stemness and enhances NK cell-mediated immunotherapy
Nature Communications (2025)
-
Bridging the dimensional gap from planar spatial transcriptomics to 3D cell atlases
Nature Methods (2025)
-
Circular RNA discovery with emerging sequencing and deep learning technologies
Nature Genetics (2025)