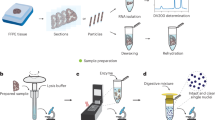
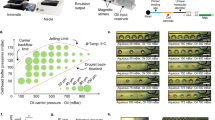
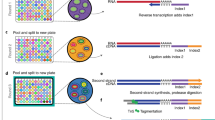
Abstract
Single-cell genomics technologies have accelerated our understanding of cell-state heterogeneity in diverse contexts. Although single-cell RNA sequencing identifies rare populations that express specific marker transcript combinations, traditional flow sorting requires cell surface markers with high-fidelity antibodies, limiting our ability to interrogate these populations. In addition, many single-cell studies require the isolation of nuclei from tissue, eliminating the ability to enrich learned rare cell states based on extranuclear protein markers. In the present report, we addressed these limitations by developing Programmable Enrichment via RNA FlowFISH by sequencing (PERFF-seq), a scalable assay that enables scRNA-seq profiling of subpopulations defined by the abundance of specific RNA transcripts. Across immune populations (n = 184,126 cells) and fresh-frozen and formalin-fixed, paraffin-embedded brain tissue (n = 33,145 nuclei), we demonstrated that programmable sorting logic via RNA-based cytometry can isolate rare cell populations and uncover phenotypic heterogeneity via downstream, high-throughput, single-cell genomics analyses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Sequencing data associated with this work are available on the Gene Expression Omnibus (GEO) at accession no. GSE262355. A full step-by-step protocol for PERFF-seq is available on protocols.io (https://doi.org/10.17504/protocols.io.14egn3k6ql5d/v1 and https://doi.org/10.17504/protocols.io.8epv5x8r4g1b/v1).
Code availability
Customized code and intermediate code files to reproduce all analyses supporting this manuscript are available at https://github.com/clareaulab/perffseq_reproducibility and via Zenodo at https://doi.org/10.5281/zenodo.14089656 (ref. 52).
References
Montoro, D. T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018).
Drokhlyansky, E. et al. The human and mouse enteric nervous system at single-cell resolution. Cell 182, 1606–1622.e23 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Lareau, C. A. et al. Latent human herpesvirus 6 is reactivated in CAR T cells. Nature 623, 608–615 (2023).
Chung, H. et al. Joint single-cell measurements of nuclear proteins and RNA in vivo. Nat. Methods 18, 1204–1212 (2021).
Chen, A. & Koehler, A. N. Transcription factor inhibition: lessons learned and emerging targets. Trends Mol. Med. 26, 508–518 (2020).
Evers, D. L., Fowler, C. B., Cunningham, B. R., Mason, J. T. & O’Leary, T. J. The effect of formaldehyde fixation on RNA: optimization of formaldehyde adduct removal. J. Mol. Diagn. 13, 282–288 (2011).
Janesick, A. et al. High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nat. Commun. 14, 8353 (2023).
Wang, Y. et al. EASI-FISH for thick tissue defines lateral hypothalamus spatio-molecular organization. Cell 184, 6361–6377.e24 (2021).
Choi, H. M. T. et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018).
Choi, H. M. T., Beck, V. A. & Pierce, N. A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano. 8, 4284–4294 (2014).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Reilly, S. K. et al. Direct characterization of cis-regulatory elements and functional dissection of complex genetic associations using HCR-FlowFISH. Nat. Genet. 53, 1166–1176 (2021).
Alon, S. et al. Expansion sequencing: spatially precise in situ transcriptomics in intact biological systems. Science 371, eaax2656 (2021).
Marshall, J. L. et al. HyPR-seq: single-cell quantification of chosen RNAs via hybridization and sequencing of DNA probes. Proc. Natl Acad. Sci. USA 117, 33404–33413 (2020).
Lubeck, E., Coskun, A. F., Zhiyentayev, T., Ahmad, M. & Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 11, 360–361 (2014).
Cano-Gamez, E. et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nat. Commun. 11, 1801 (2020).
Yu, Y. et al. Bcl11a is essential for lymphoid development and negatively regulates p53. J. Exp. Med. 209, 2467–2483 (2012).
Orkin, S. H. & Zon, L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008).
Choi, J. et al. Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res. 47, D780–D785 (2018).
Guo, X. et al. Mosaic loss of human Y chromosome: what, how and why. Hum. Genet. 139, 421–446 (2020).
Bruhn-Olszewska, B. et al. Loss of Y in leukocytes as a risk factor for critical COVID-19 in men. Genome Med. 14, 139 (2022).
Mattisson, J. et al. Loss of chromosome Y in regulatory T cells. BMC Genom. 25, 243 (2024).
Lleo, A. et al. Y chromosome loss in male patients with primary biliary cirrhosis. J. Autoimmun. 41, 87–91 (2013).
Kozareva, V. et al. A transcriptomic atlas of mouse cerebellar cortex comprehensively defines cell types. Nature 598, 214–219 (2021).
Siokas, V. et al. Myelin-associated oligodendrocyte basic protein rs616147 polymorphism as a risk factor for Parkinson’s disease. Acta Neurol. Scand. 145, 223–228 (2022).
Irwin, D. J. et al. Myelin oligodendrocyte basic protein and prognosis in behavioral-variant frontotemporal dementia. Neurology 83, 502–509 (2014).
Kon, T. et al. Immunoreactivity of myelin-associated oligodendrocytic basic protein in Lewy bodies. Neuropathology 39, 279–285 (2019).
Patel, K. S. et al. Decorin expression is associated with predictive diffusion MR phenotypes of anti-VEGF efficacy in glioblastoma. Sci. Rep. 10, 14819 (2020).
Serres, E. et al. Fibronectin expression in glioblastomas promotes cell cohesion, collective invasion of basement membrane in vitro and orthotopic tumor growth in mice. Oncogene 33, 3451–3462 (2014).
Mojiri, A. et al. Functional assessment of von Willebrand factor expression by cancer cells of non-endothelial origin. Oncotarget 8, 13015–13029 (2017).
Sung, H.-Y. et al. Down-regulation of interleukin-33 expression in oligodendrocyte precursor cells impairs oligodendrocyte lineage progression. J. Neurochem. 150, 691–708 (2019).
Huang, H.-T. & Tzeng, S.-F. Interleukin-33 has the protective effect on oligodendrocytes against impairment induced by cuprizone intoxication. Neurochem. Int. 172, 105645 (2024).
Floriddia, E. M. et al. Distinct oligodendrocyte populations have spatial preference and different responses to spinal cord injury. Nat. Commun. 11, 5860 (2020).
Langlieb, J. et al. The molecular cytoarchitecture of the adult mouse brain. Nature 624, 333–342 (2023).
Du, J. et al. S100B is selectively expressed by gray matter protoplasmic astrocytes and myelinating oligodendrocytes in the developing CNS. Mol. Brain 14, 154 (2021).
Auguste, Y. S. S. et al. Oligodendrocyte precursor cells engulf synapses during circuit remodeling in mice. Nat. Neurosci. 25, 1273–1278 (2022).
Morel, L. et al. Intracortical astrocyte subpopulations defined by astrocyte reporter mice in the adult brain. Glia 67, 171–181 (2019).
Xie, Y. et al. Key molecular alterations in endothelial cells in human glioblastoma uncovered through single-cell RNA sequencing. JCI insight 6, e150861 (2021).
Wälchli, T. et al. Single-cell atlas of the human brain vasculature across development, adulthood and disease. Nature 632, 603–613 (2024).
Mei, Y. et al. Osteoglycin promotes meningioma development through downregulation of NF2 and activation of mTOR signaling. Cell Commun. Signal. 15, 34 (2017).
Amamoto, R. et al. Probe-Seq enables transcriptional profiling of specific cell types from heterogeneous tissue by RNA-based isolation. eLife 8, e51452 (2019).
Clark, I. C. et al. Identification of astrocyte regulators by nucleic acid cytometry. Nature 614, 326–333 (2023).
Ippolito, G. C. et al. Dendritic cell fate is determined by BCL11A. Proc. Natl Acad. Sci. USA 111, E998–E1006 (2014).
Warren, C. J. et al. Quantification of virus-infected cells using RNA FISH-Flow. STAR Protoc. 4, 102291 (2023).
Antony, C., Somers, P., Gray, E. M., Pimkin, M. & Paralkar, V. R. FISH-Flow to quantify nascent and mature ribosomal RNA in mouse and human cells. STAR Protoc. 4, 102463 (2023).
González-Vasconcellos, I., Cobos-Fernández, M. A., Atkinson, M. J., Fernandez-Piqueras, J. & Santos, J. Quantifying telomeric lncRNAs using PNA-labelled RNA-Flow FISH (RNA-Flow). Commun. Biol. 5, 513 (2022).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Rouillard, A. D. et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, baw100 (2016).
Lareau, C. A. PERFF-seq reproducibility. Zenodo https://doi.org/10.5281/zenodo.14089656 (2024).
Acknowledgements
We thank the Satpathy Lab, Lareau Lab, Gladstone Flow Cytometry Core and Single-cell Analytics Innovation Lab members for helpful discussions. We acknowledge a helpful blog post from 10x Genomics describing the singlet unligated probe set. We thank T. Nawy for helpful feedback on the manuscript, A. Chow for assistance with flow cytometry and N. Pereira with S. Jovanovich of S2 Genomics for support with nuclei isolation from FFPE tissue. This work was supported by National Insitutes of Health grants (nos. P30CA008748, R00HG012579 and U01AT012984 to C.A.L. and UM1HG012076 to A.T.S. and L.S.L.) and a Gates Foundation seed award. A.T.S. is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists, the Parker Institute for Cancer Immunotherapy, a Pew-Stewart Scholars for Cancer Research Award, a Cancer Research Institute Lloyd J. Old STAR Award, a Scholar Award from the American Society of Hematology and a Baxter Foundation Faculty Scholar Award. Y.H.H. is supported by a PhD fellowship from the Hector Fellow Academy. L.S.L. is supported by the Hector Fellow Academy, the Paul Ehrlich Foundation, the European Molecular Biology Organization Young Investigator Programme, an Emmy Noether fellowship (grant no. LU 2336/2-1) and grants by the German Research Foundation (Dnos. LU 2336/3-1, LU 2336/6-1, STA 1586/5-1, TRR241 and SFB1588, and Heinz Maier-Leibnitz Award). The Single-cell Analytics Innovation Lab is supported by the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center at Memorial Sloan Kettering. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
T.A., R.R.S., M.T.T., R.C., A.T.S. and C.A.L. conceived and designed this work. T.A. and R.R.S. led assay development with input from A.T.S. and C.A.L. T.A., R.R.S., M.T.T., B.N.N., Y.-H.H., S.H. and C.S. performed experiments. K.K.H.Y., Z.A.-M., V.T., B.E.H. and L.S.L. aided in the interpretation of data analyses. R.R.S., R.C., A.T.S. and C.A.L. supervised the work. C.A.L. led bioinformatics analyses with input from T.A. and R.R.S. T.A. led the development of the protocol with input from R.R.S. and M.T.T. T.A., R.R.S., A.T.S. and C.A.L. drafted the manuscript with input from the other authors.
Corresponding authors
Ethics declarations
Competing interests
A.T.S. is a founder of Immunai, Cartography Biosciences, Santa Ana Bio and Prox Biosciences, an advisor to Wing Venture Capital and receives research funding from Astellas and Merck Research Laboratories. R.R.S., L.S.L. and C.A.L. are consultants to Cartography Biosciences. R.C. is a consultant for Sanavia Oncology, S2 Genomics and LevitasBio. The other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Dominic Gruen, Sydney Shaffer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analyses supporting PERFF-seq development.
(a) Schematic overview of Flex workflow, including probe hybridization to transcript fragments in cells upstream of Chromium and bead oligo extension of the ligation product. (b) Representative Bioanalyzer (Agilent Technologies) trace outlining complete versus incomplete sequencing molecules. (c) Graphical summary of probe capture sequence (pCS1) bead oligo capture sequence (left) and percent of reads with pCS1 detected in first 25 bases. (d) Comparison of FlowFISH signal using either unstained cells or the hairpin only comparing the sorted CD3E+ population and/or stripped via formamide. (e) Same as in (d) but using dsDNase for stripping. Note: quantifying fluorescence after sorting/stripping (d, purple; e, blue) is not standard for the PERFF-seq protocol but shown here as part of assay development. (f) Bioanalyzer traces for representative libraries from panels in Fig. 1, highlighting half- and fully-mapped probes. (g) Bioanalyzer traces of library preparation where the full PERFF-seq workflow was completed except omiting the dsDNAse stripping step.
Extended Data Fig. 2 Supporting analyses of assay benchmarking.
(a) HCR FISH staining and quantification of EPCAM across cell lines. Mean fluorescence intensity (MFI) and bulk RNA-seq transcripts per million (TPM from49) are noted for each condition. (b) Replication of cell line mixing experiment and staining for XIST. (c) Reduced dimensionality embedding of all cells in the four-plex benchmarking experiment. PERFF-seq was enriched for CD3E+ cells. No probe, no sort (NPNS) and Yes probe, no sort (YPNS) profile all PBMC subpopulations with minor modifications to the reaction. (d) Marker genes supporting annotation of key populations. (e) Same as (c) but stratified by library. Boxes indicate B cell and monocyte populations that are depleted from the PERFF-seq library. (f) Percent of T cells from each library with at least 1 UMI for CD3D or CD3E. (g) log2 counts per million (CPM) of CD3D and CD3E across different library conditions. (h) Differentially expressed genes between different facets of PERFF-seq compared to Flex. Two genes were differentially expressed, including CD3E in both the YPNS and PERFF-seq conditions. ∅ means empty or no genes detected.
Extended Data Fig. 3 Supporting analyses of combinatorial PBMC cell states.
(a) Additional marker genes for distinct populations from PBMC cell type analyses. Arrows indicate markers for rare populations expected from PBMC profiling. (b) Relative enrichment of cell populations (colours) for each PERFF-seq library compared to the Flex PBMC library. (c) Subclustering of CD3E+/CD4+ cells, highlighting rare subclusters marked by relevant genes. (d) Summary of CD4 HCR FISH signal, stratified by CD3E populations. (e) Bulk RNA-seq expression of CD4 from FACS-isolated populations21. Replicates for e are all libraries from Haemopedia21 with no statistical test. (f) Design and results of antibody and HCR FISH co-staining to evaluate CD4 RNA and protein expression.
Extended Data Fig. 4 Supporting analyses of unconventional enrichments of rare cell states.
(a) Azimuth Violin plots for BCL11A and SPI1 RNA expression across well-annotated populations in peripheral blood mononuclear cell types. (b) Summary of FACS populations, including unsorted, BCL11A+, and SPI1+ populations. (c) Additional marker genes supporting cell type annotations. (d) Empirical cumulative distribution plot of scaled expression of BCL11A (left) and SPI1 (right) stratified by the captured PERFF-seq library. (e) Bulk RNA-seq of sorted populations of BCL11A21. (f) Design (left) and results (right) of cytometry analysis of PBMCs co-stained with BCL11A mRNA (via HCR-FISH) and CD19 and CD123 protein (via antibodies). Mean fluorescence intensity (MFI) for BCL11A of each population is quantified. (g) Comparison of B cells from BCL11A+ FlowFISH or negative/SPI1+ populations for BCL11A expression or BCL11A target gene module scores. Uncorrected p-value for the two-sided Wilcoxon rank-sum test is shown. (h) Additional violin plots of marker genes, stratified by the FlowFISH library. All genes were significantly differentially expressed at a false discovery rate (FDR) < 0.01. (i) Summary of IL3RA+ FACS sort and population characterized with PERFF-seq. (j) Proportion of cell types from the Azimuth L1 reference for IL3RA+/- PERFF-seq libraries. (k) Reduced dimensionality representation of IL3RA+ PERFF-seq library, highlighting profiled AS DCs. (l) Gene-gene Spearman correlations of all AS DCs from the IL3RA+ sort. Genes match those in Fig. 4k. (m) Summary of CD123 expression from antibody-derived tags (ADT) of PBMC CITE-seq. (n) Bulk RNA-seq of sorted populations of IL3RA21. Replicates for e and n are all libraries from Haemopedia21 with no statistical test.
Extended Data Fig. 5 Supporting analyses for mosaic loss of Y chromosome.
(a) FlowFISH cytometry gating scheme, including control (no probe, left) and four male (XY) donors of different ages. The percent of cells corresponding to MSY+ (red) and MSY- (blue) in each donor gate are shown as numeric values. (b) Empirical cumulative distribution plot of the percent of Y chromosome UMIs stratified by the captured PERFF-seq library. Percentages of cells with 0 MSY UMIs from the scRNA-seq library are noted. P-values are from a two-sided Kolmogorov–Smirnov test comparing the distributions of the positive and negative samples.
Extended Data Fig. 6 Supporting analyses of nuclei enrichment from fresh and fixed tissues.
(a) UMAP of adult mouse cerebellum atlas26, including cell types (top) and Mobp expression (bottom). The Mobp+ oligodendrocytes and oligodendrocyte precursors are circled with their frequency noted. (b) Reduced dimensionality representation of public GBM FFPE Flex data showing 17 clusters. (c) Annotation of marker genes for cluster 10, the population highlighted by the arrow in (b). (d) Supporting marker genes annotating other subpopulations from the PERFF-seq experiment, including the primary cluster of granule cells. (e) Additional marker genes from Mobp+ cells were profiled with PERFF-seq. Atp-associated genes (Atp1b1, Aqp4) supporting rare subclsuters are highlighted in the boxes as well as marker genes highly expressed in all cells. (f) Empirical cumulative distribution plot of the raw UMI counts for each of the three genes enriched via FlowFISH, stratified by the captured PERFF-seq library. (g) Additional marker genes showing heterogeneity defining subclusters of endothelial cells and pericytes.
Supplementary information
Supplementary Information
Supplementary Note on the design of PERFF-seq experiments.
Supplementary Tables
Supplementary Tables 1–6.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Abay, T., Stickels, R.R., Takizawa, M.T. et al. Transcript-specific enrichment enables profiling of rare cell states via single-cell RNA sequencing. Nat Genet 57, 451–460 (2025). https://doi.org/10.1038/s41588-024-02036-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-024-02036-7
This article is cited by
-
PURE-seq integrates FACS and PIP-seq for single-cell genomics of ultra-rare cells
Nature Communications (2026)
-
Unveiling heterogeneity in rare cells by combining RNA-based sorting and scRNA-seq
Nature Genetics (2025)