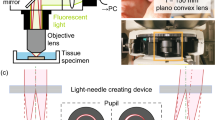
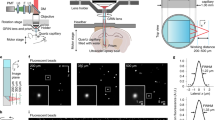
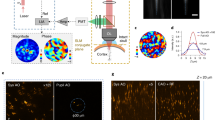
Abstract
Advanced brain-wide mapping is critical for addressing complex questions in neuroscience. However, current imaging methods are limited by throughput, resolution and signal-to-noise ratio, constraining their broader applicability. Here, we present confocal Airy beam integrated with single-photon oblique light-sheet tomography (CAB-OLST): a system that integrates single-photon excitation with a scanned Airy beam light sheet, virtual slit detection and automated mechanical sectioning. CAB-OLST enables high-throughput, high-resolution and high-signal-to-noise ratio volumetric imaging, achieving an optical resolution of 0.77 μm × 0.49 μm × 2.61 μm. This allows for mouse brain-wide cell type distribution mapping at a voxel size of 0.37 μm × 0.37 μm × 1.77 μm in 10 h and single-neuron projectome imaging with a voxel size of 0.26 μm × 0.26 μm × 1.06 μm over 58 h. Compared to existing light-sheet and point-scanning systems, CAB-OLST provides a scalable and robust platform for comprehensive neuronal morphology reconstruction and high-precision cell atlas generation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Processed, downsampled image data sufficient to reproduce all figures and analyses are publicly available at Zenodo (https://doi.org/10.5281/zenodo.17191164)59. Due to their substantial size (4–60 TB per dataset), the raw, high-resolution datasets have been deposited in the specialized Brain Image Library (https://www.brainimagelibrary.org/) archive as part of the Brain Initiative Cell Census Network.
Code availability
All custom code developed for this study is released under the MIT License and is archived on Zenodo to ensure long-term availability and reproducibility. The archived versions correspond to the exact code used for the analyses in this paper. The specific packages are as follows: cell type distribution analysis, https://doi.org/10.5281/zenodo.17170411 (ref. 60) and https://github.com/coreyelowsky/OLSTv2; stripe removal, provided as Supplementary Code 1, https://github.com/Vaa3D/vaa3d_tools/tree/master/hackathon/mBrainAligner; 3D U-Net for cellular segmentation, https://doi.org/10.5281/zenodo.17172303 (ref. 61) and https://github.com/rmunozca/CAB-OLST_Analysis; SmartStitcher, https://doi.org/10.5281/zenodo.17178922 (ref. 62) and https://github.com/polya1998/SmartStitcher.git. The publicly available software Vaa3D (version 4.001), including Vaa3D-TeraVR and Vaa3D-TeraFly, is accessible at http://www.vaa3d.org/.
Change history
19 November 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41592-025-02982-y
References
Li, A. Micro-optical sectioning tomography. Science 1404, 1404–1408 (2010).
Ragan, T. et al. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 9, 255–258 (2012).
Economo, M. N. et al. A platform for brain-wide imaging and reconstruction of individual neurons. eLife 5, e10566 (2016).
Ueda, H. R. et al. Whole-brain profiling of cells and circuits in mammals by tissue clearing and light-sheet microscopy. Neuron 106, 369–387 (2020).
Jiang, T., Gong, H. & Yuan, J. Whole-brain optical imaging: a powerful tool for precise brain mapping at the mesoscopic level. Neurosci. Bull. 39, 1840–1858 (2023).
Oh, S. W. et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014).
Gong, H. et al. Continuously tracing brain-wide long-distance axonal projections in mice at a one-micron voxel resolution. Neuroimage 74, 87–98 (2013).
Qi, X. et al. Fluorescence micro-optical sectioning tomography using acousto-optical deflector-based confocal scheme. Neurophotonics 2, 041406 (2015).
Qi, X. et al. Improved detectability of neuronal connectivity on mechanical sectioning setup by using confocal detection. J. Biomed. Opt. 18, 050506 (2013).
Gong, H. et al. High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 7, 12142 (2016).
Wang, X. et al. Chemical sectioning fluorescence tomography: high-throughput, high-contrast, multicolor, whole-brain imaging at subcellular resolution. Cell Rep. 34, 108709 (2021).
Zhong, Q. et al. High-definition imaging using line-illumination modulation microscopy. Nat. Methods 18, 309–315 (2021).
Chen, H. et al. Sparse imaging and reconstruction tomography for high-speed high-resolution whole-brain imaging. Cell Rep. Methods 1, 100089 (2021).
Narasimhan, A. et al. Oblique light-sheet tomography: fast and high resolution volumetric imaging of mouse brains. Preprint at bioRxiv https://doi.org/10.1101/132423 (2016).
Seiriki, K. et al. High-speed and scalable whole-brain imaging in rodents and primates. Neuron 94, 1085–1100 (2017).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. K. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Dodt, H.-U. et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331–336 (2007).
Engelbrecht, C. J. & Stelzer, E. H. Resolution enhancement in a light-sheet-based microscope (SPIM). Opt. Lett. 31, 1477–1479 (2006).
Power, R. M. & Huisken, J. A guide to light-sheet fluorescence microscopy for multiscale imaging. Nat. Methods 14, 360–373 (2017).
Voie, A. H., Burns, D. H. & Spelman, F. A. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J. Microsc. 170, 229–236 (1993).
Silvestri, L., Bria, A., Sacconi, L., Iannello, G. & Pavone, F. S. Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain. Opt. Express 20, 20582–20598 (2012).
Mertz, J. & Kim, J. Scanning light-sheet microscopy in the whole mouse brain with HiLo background rejection. J. Biomed. Opt. 15, 016027 (2010).
Dean, K. M., Roudot, P., Welf, E. S., Danuser, G. & Fiolka, R. Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys. J. 108, 2807–2815 (2015).
Chakraborty, T. et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nat. Methods 16, 1109–1113 (2019).
Voigt, F. F. et al. The mesoSPIM initiative: open-source light-sheet microscopes for imaging cleared tissue. Nat. Methods 16, 1105–1108 (2019).
Vladimirov, N. et al. Benchtop mesoSPIM: a next-generation open-source light-sheet microscope for cleared samples. Nat. Commun. 15, 2679 (2024).
Tang, L. et al. Curved light sheet microscopy for centimetre-scale cleared tissue imaging. Nat. Photonics 19, 577–584 (2025).
Glaser, A. et al. Expansion-assisted selective plane illumination microscopy for nanoscale imaging of centimeter-scale tissues. eLife 12, RP91979 (2024).
Tomer, R., Khairy, K., Amat, F. & Keller, P. J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–763 (2012).
Krzic, U., Gunther, S., Saunders, T. E., Streichan, S. J. & Hufnagel, L. Multiview light-sheet microscope for rapid in toto imaging. Nat. Methods 9, 730–733 (2012).
Kim, B. et al. Open-top axially swept light-sheet microscopy. Biomed. Opt. Express 12, 2328–2338 (2021).
Glaser, A. K. et al. A hybrid open-top light-sheet microscope for versatile multi-scale imaging of cleared tissues. Nat. Methods 19, 613–619 (2022).
Yang, B. et al. DaXi—high-resolution, large imaging volume and multi-view single-objective light-sheet microscopy. Nat. Methods 19, 461–469 (2022).
Wu, Y. et al. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 108, 17708–17713 (2011).
Hu, B., Bolus, D. & Brown, J. Q. Improved contrast in inverted selective plane illumination microscopy of thick tissues using confocal detection and structured illumination. Biomed. Opt. Express 8, 5546–5559 (2017).
Dunsby, C. Optically sectioned imaging by oblique plane microscopy. Opt. Express 16, 20306–20316 (2008).
Bouchard, M. B. et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nat. Photonics 9, 113–119 (2015).
Voleti, V. et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat. Methods 16, 1054–1062 (2019).
Vettenburg, T. et al. Light-sheet microscopy using an Airy beam. Nat. Methods 11, 541–544 (2014).
Siviloglou, G. A. & Christodoulides, D. N. Accelerating finite energy Airy beams. Opt. Lett. 32, 979–981 (2007).
Hosny, N. A. et al. Planar Airy beam light-sheet for two-photon microscopy. Biomed. Opt. Express 11, 3927–3935 (2020).
Gong, S. et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 (2003).
Taniguchi, H. et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Wang, D. et al. Color liquid crystal grating based color holographic 3D display system with large viewing angle. Light Sci. Appl. 13, 16 (2024).
Matho, K. S. et al. Genetic dissection of the glutamatergic neuron system in cerebral cortex. Nature 598, 182–187 (2021).
Kim, Y. et al. Brain-wide maps reveal stereotyped cell-type-based cortical architecture and subcortical sexual dimorphism. Cell 171, 456–469 (2017).
Muñoz-Castañeda, R. et al. A comprehensive atlas of cell type density patterns and their role in brain organization. Preprint at bioRxiv https://doi.org/10.1101/2024.10.02.615922 (2024).
Ronneberger, O., Fischer, P., & Brox, T. U-net: convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention — MICCAI 2015: 18th international conference (eds Navab, N. et al.) 234–241 (Springer, 2015).
Li, H. et al. Single-neuron projectomes of mouse paraventricular hypothalamic nucleus oxytocin neurons reveal mutually exclusive projection patterns. Neuron 112, 1081–1099 (2024).
Pfau, S. J. et al. Characteristics of blood–brain barrier heterogeneity between brain regions revealed by profiling vascular and perivascular cells. Nat. Neurosci. 27, 1892–1903 (2024).
Aakhte, M., Akhlaghi, E. A. & Müller, H.-A. J. SSPIM: a beam shaping toolbox for structured selective plane illumination microscopy. Sci. Rep. 8, 10067 (2018).
Zhang, Q. et al. High axial resolution imaging system for large volume tissues using combination of inclined selective plane illumination and mechanical sectioning. Biomed. Opt. Express 8, 5767–5775 (2017).
Kumar, A. et al. Using stage-and slit-scanning to improve contrast and optical sectioning in dual-view inverted light sheet microscopy (diSPIM). Biol. Bull. 231, 26–39 (2016).
Qu, L. et al. Cross-modal coherent registration of whole mouse brains. Nat. Methods 19, 111–118 (2022).
Peng, H., Ruan, Z., Long, F., Simpson, J. H. & Myers, E. W. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 28, 348–353 (2010).
Peng, H., Bria, A., Zhou, Z., Iannello, G. & Long, F. Extensible visualization and analysis for multidimensional images using Vaa3D. Nat. Protoc. 9, 193–208 (2014).
Wang, Y. et al. TeraVR empowers precise reconstruction of complete 3-D neuronal morphology in the whole brain. Nat. Commun. 10, 3474 (2019).
Qi, X. Confocal Airy beam oblique light-sheet tomography. Zenodo https://doi.org/10.5281/zenodo.17191164 (2025).
jdp-cshl & Elowsky, C. coreyelowsky/OLSTv2: publication. Zenodo https://doi.org/10.5281/zenodo.17170411 (2025).
rmunozca. rmunozca/CAB-OLST_Analysis: v1. Zenodo https://doi.org/10.5281/zenodo.17172303 (2025).
Li, Y. & Ding, L. SmartSticher. Zenodo https://doi.org/10.5281/zenodo.17178922 (2025).
Acknowledgements
This work was mainly supported by National Institutes of Health grants U01MH114824 (to P.O.) and U19MH114821 (to Z. Josh Huang).H.P. is a New Cornerstone Investigator and a Shanghai Academy of Natural Sciences Senior Investigator. Additional support was provided by National Natural Science Foundation of China grants 62401009 (to Y.L.) and 62271003 (to L.Q.), in part by the Sci-Tech Innovation 2030 Agenda (2022ZD0205200 and 2022ZD0205204). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper. We thank J. Daniels (ASI), F. Albeanu (Cold Spring Harbor Laboratory) and J. Mizrachi (Cold Spring Harbor Laboratory) for their discussions during microscopy development. We also thank R. Eifert (Cold Spring Harbor Laboratory) for the custom fabrication of microscopy components and R. Campbell (Advanced Microscopy Facility, Sainsbury Wellcome Centre, UCL) for providing the vibratome design specifications.
Author information
Authors and Affiliations
Contributions
P.O. and H.P. supervised and coordinated the project. X.Q. conceptualized and led the project under the supervision of P.O. and H.P. X.Q. designed and constructed the microscope with input from A.N. and performed data acquisition. X.Q. and Y.Y. drafted the manuscript with guidance from P.O. and H.P. S.X. developed the Airy deconvolution algorithm and reviewed the optical design. H.P., R.M.-C. and X.Q. jointly supervised image processing. Z.W. and W.W. contributed to sample preparation, including optical clearing and immunostaining. R.D. and J.S. performed animal surgeries including virus injections and brain embedding. C.E., J.P., L.Q. and Y.L. developed computational tools for imaging analysis. L.D. and X.C. conducted single-cell neuronal tracing under the supervision of H.P. J.W. performed data visualization for Fig. 5c,d. All authors participated in the interpretation of results and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
P.O. is a cofounder and the CEO of Theracast. The company had no role in or financial gain from this work. All other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CAB-OLST configurations.
(a-b) Configuration for imaging 5 mm-thick tissue using long working-distance (WD) objectives. (c) Configuration showing laser illumination of a cleared mouse brain. (d) Full view of CAB-OLST prototype, with a single camera, and objectives immersed in U.Clear buffer.
Extended Data Fig. 2 Comparative analysis of Airy, deconvolved Airy, and confocal Airy microscopy.
a, Representative neurite images. b, Normalized intensity profiles of a. c, Quantitative comparison of image contrast across the three microscopy modes. Data are presented as mean ± s.e.m. (n = 5 neurites). Statistical analysis was performed using a one-way ANOVA with Tukey’s post-hoc test. Significant differences were found between the standard Airy (Control) and both Deconvolved (P = 0.0013) and Confocal (P = 0.0019) modes. No significant difference was observed between the Deconvolved and Confocal modes (P = 0.976). On the graph, asterisks denote significance levels: **P < 0.01.
Extended Data Fig. 3 XZ and YZ projections of dual-channel cell distribution data.
Representative XY, YZ, and XZ projections of a dual-channel dataset. The top row shows Channel 1 (CH1) and the bottom row shows Channel 2 (CH2). Scale bars, 80 μm.
Extended Data Fig. 4 Axial resolution and detection efficiency across the FOV.
(a) Maximum intensity projection of fluorescent microspheres across the full FOV. (b) Representative images of isolated microspheres at different Z’ positions (1: 250 μm, 2: 131 μm, 3: 491 μm). (c, e) Full-width at half maximum (FWHM) in X, Y, and Z for microspheres imaged at the original full FOV (c) and with Z-stitching (e). Original FOV (median ± s.d.): X: 0.78 ± 0.09 μm; Y: 0.46 ± 0.08 μm; Z: 2.95 ± 0.89 μm; Stitching FOV (median ± s.d.): X: 0.77 ± 0.10 μm; Y: 0.49 ± 0.08 μm; Z: 2.61 ± 0.79 μm. (d) Schematic of the Z-stitching strategy. (f) Background image from a median-filtered AVP brain. (g) Intensity profiles of microspheres versus distance from the FOV center (Z’). Data are presented as median± s.d.
Extended Data Fig. 5 Quantitative comparison of Slit Confocal Airy LS and standard Airy LS.
(a, b) Quantification of image contrast (or signal-to-noise ratio, SNR) for somata (a) and neurites (b) imaged with either Slit Confocal Airy LS or standard Airy LS. For this comparison, laser powers were adjusted to achieve the same maximum noise intensity across modalities.
Extended Data Fig. 6 Quantitative comparison with Slit Confocal Airy LS and standard Airy LS in different signal locations.
a,b, Representative images (a) and quantification of image contrast (b) for somata. c,d, Representative images (c) and quantification of image contrast (d) for neurites. All images were acquired with the same laser power to ensure equivalent maximum signal intensity. Data are presented as mean ± s.e.m. (n = 5 for both). Statistical significance was determined by a two-tailed, paired Student’s t-test (P = 0.0011 for somata; P = 0.0001 for neurites). On the graph, asterisks denote significance levels: **P < 0.01, ***P < 0.001.
Extended Data Fig. 7 Imaging comparison with and without mechanical sectioning (MS).
Representative images and corresponding intensity profiles of four somata (a) and four neurites (b), imaged with and without MS under identical conditions.
Extended Data Fig. 8 Imaging performance comparison using Depth LS and Sectional LS.
(a) Representative images comparing Depth LS (left) and Sectional LS (right) imaging of neuronal structures. Insets are magnified views. Scale bar, 50 μm. (b) Corresponding intensity profiles along the lines indicated in (a), for Depth LS (black) and Sectional LS (red).
Extended Data Fig. 9 3D U-Net segmentation workflow.
a) Overview of ground truth annotation generation process. An unsupervised segmentation generates initial cell masks, which are then refined to create three-class ground truth labels (background, cell boundary, intracellular). A representative cell (right panels) is shown at different processing stages (indicated by green arrows). b) Example of final segmentation results. Left: Raw image of clustered cell bodies with varying intensities. Middle: The segmented output, with cell boundaries shown in green and intracellular regions in red. Right: Detected cell centroids (red dots) overlaid on the original image.
Supplementary information
Supplementary Information
Supplementary Tables 1–4
Supplementary Code 1
Stripe removal code.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qi, X., Muñoz-Castañeda, R., Yue, Y. et al. Confocal Airy beam oblique light-sheet tomography for brain-wide cell type distribution and morphology. Nat Methods 22, 2622–2630 (2025). https://doi.org/10.1038/s41592-025-02888-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02888-9