Abstract
Single-particle cryogenic electron microscopy (cryo-EM) enables reconstruction of atomic-resolution 3D maps of proteins by visualizing thousands to millions of purified protein particles embedded in vitreous ice. This corresponds to picograms of purified protein, which can potentially be isolated from a few thousand cells. Hence, cryo-EM holds the potential of a very sensitive analytical method for delivering high-resolution protein structure as a readout. In practice, millions of times more starting biological material is required to prepare cryo-EM grids. Here we show that using a micro isolation (MISO) method, which combines microfluidics-based protein purification with cryo-EM grid preparation, cryo-EM structures of soluble bacterial and eukaryotic membrane proteins can be solved starting from less than 1 µg of a target protein and progressing from cells to cryo-EM grids within a few hours. This scales down the amount of starting biological material hundreds to thousands of times, opening possibilities for the structural characterization of hitherto inaccessible proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The protein models with the PDB accession codes 1F4A, 6P46 and 6UZ8 were used in this study. The newly generated cryo-EM-refined atomic models, density maps and raw data were deposited in the PDB, EMDB and EMPIAR databases under accession codes: βG from 20 μg 9HPL, EMD-52333, EMPIAR-12734; βG from 1 μg 9HPM, EMD-52334, EMPIAR-12740; btTMEM206 conventional 9HQN, EMD-52344, EMPIAR-12773; btTMEM206–YFP MISO 9HQO, EMD-52345, EMPIAR-12769; mTMEM16F–YFP 9HQP, EMD-52346, EMPIAR-12757; TRPC6 EMD-52486, EMD-52487, EMPIAR-12770. Source data are provided with this paper.
Code availability
The LabVIEW code to run MISO device is available at https://github.com/EfremovLab/MISO.
References
de la Cruz, M. J. & Eng, E. T. Scaling up cryo-EM for biology and chemistry: the journey from niche technology to mainstream method. Structure 31, 1487–1498 (2023).
Zhu, K.-F. et al. Applications and prospects of cryo-EM in drug discovery. Mil. Med. Res. 10, 10 (2023).
Henderson, R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 28, 171–193 (1995).
Vinothkumar, K. R. & Henderson, R. Single particle electron cryomicroscopy: trends, issues and future perspective. Q. Rev. Biophys. 49, e13 (2016).
Grigorieff, N. Direct detection pays off for electron cryo-microscopy. eLife 2, e00573 (2013).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Bouvette, J. et al. Automated systematic evaluation of cryo-EM specimens with SmartScope. eLife 11, e80047 (2022).
Cheng, A. et al. Fully automated multi-grid cryoEM screening using Smart Leginon. IUCrJ 10, 77–89 (2023).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Gómez-Blanco, J. et al. Using Scipion for stream image processing at cryo-EM facilities. J. Struct. Biol. 204, 457–463 (2018).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 478, 4169–4185 (2021).
Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature 628, 450–457 (2024).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018).
Zwart, P. H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008).
Terwilliger, T. C. et al. Improved AlphaFold modeling with implicit experimental information. Nat. Methods 19, 1376–1382 (2022).
McMullan, G. et al. Structure determination by cryoEM at 100 keV. Proc. Natl Acad. Sci. USA 120, e2312905120 (2023).
Karia, D. et al. Sub-3 Å resolution protein structure determination by single-particle cryo-EM at 100 keV. Structure 33, 1717–1727.e4 (2025).
D'Imprima, E. et al. Protein denaturation at the air-water interface and how to prevent it. eLife 8, e42747 (2019).
Noble, A. J. et al. Reducing effects of particle adsorption to the air–water interface in cryo-EM. Nat. Methods 15, 793–795 (2018).
Noble, A. J. et al. Routine single particle cryoEM sample and grid characterization by tomography. eLife 7, e34257 (2018).
Zheng, L. et al. Uniform thin ice on ultraflat graphene for high-resolution cryo-EM. Nat. Methods 20, 123–130 (2023).
Russo, C. J. & Passmore, L. A. Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014).
Naydenova, K., Jia, P. & Russo, C. J. Cryo-EM with sub–1 Å specimen movement. Science 370, 223–226 (2020).
Huber, S. T. et al. Nanofluidic chips for cryo-EM structure determination from picoliter sample volumes. eLife 11, e72629 (2022).
Naydenova, K., Peet, M. J. & Russo, C. J. Multifunctional graphene supports for electron cryomicroscopy. Proc. Natl Acad. Sci. USA 116, 11718–11724 (2019).
Pantelic, R. S., Meyer, J. C., Kaiser, U., Baumeister, W. & Plitzko, J. M. Graphene oxide: a substrate for optimizing preparations of frozen-hydrated samples. J. Struct. Biol. 170, 152–156 (2010).
Wang, F. et al. General and robust covalently linked graphene oxide affinity grids for high-resolution cryo-EM. Proc. Natl Acad. Sci. USA 117, 24269–24273 (2020).
Sader, K., Stopps, M., Calder, L. J. & Rosenthal, P. B. Cryomicroscopy of radiation sensitive specimens on unmodified graphene sheets: reduction of electron-optical effects of charging. J. Struct. Biol. 183, 531–536 (2013).
Jain, T., Sheehan, P., Crum, J., Carragher, B. & Potter, C. S. Spotiton: a prototype for an integrated inkjet dispense and vitrification system for cryo-TEM. J. Struct. Biol. 179, 68–75 (2012).
Rubinstein, J. L. et al. Shake-it-off: a simple ultrasonic cryo-EM specimen-preparation device. Acta Crystallogr. D 75, 1063–1070 (2019).
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988).
Henderikx, R. J. M. et al. VitroJet: new features and case studies. Acta Crystallogr. D 80, 232–246 (2024).
Ravelli, R. B. G. et al. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nat. Commun. 11, 2563 (2020).
Arnold, S. A. et al. Total sample conditioning and preparation of nanoliter volumes for electron microscopy. ACS Nano 10, 4981–4988 (2016).
Arnold, S. A. et al. Blotting-free and lossless cryo-electron microscopy grid preparation from nanoliter-sized protein samples and single-cell extracts. J. Struct. Biol. 197, 220–226 (2016).
Schmidli, C. et al. Microfluidic protein isolation and sample preparation for high-resolution cryo-EM. Proc. Natl Acad. Sci. USA 116, 15007–15012 (2019).
Kelly, D. F., Abeyrathne, P. D., Dukovski, D. & Walz, T. The affinity grid: a pre-fabricated EM grid for monolayer purification. J. Mol. Biol. 382, 423–433 (2008).
Yu, G., Li, K. & Jiang, W. Antibody-based affinity cryo-EM grid. Methods 100, 16–24 (2016).
Arnold, S. A. et al. Miniaturizing EM sample preparation: opportunities, challenges and “visual proteomics”. Proteomics 18, 1700120–1700176 (2018).
Duffy, D. C., McDonald, J. C., Schueller, O. J. A. & Whitesides, G. M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984 (1998).
Weerappuli, P., Kojima, T., Takayama, S. & Basu, A. Novel monolithic "slightly-open doormat" (SOD) valve enables efficient fabrication of highly-scalable microfluidic gas-on-gas multiplexer. Sens Actuators B 297, 126776 (2019).
Datta, S. & Ghosal, S. Characterizing dispersion in microfluidic channels. Lab Chip 9, 2537 (2009).
Bartesaghi, A. et al. 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science 348, 1147–1151 (2015).
Torino, S., Dhurandhar, M., Stroobants, A., Claessens, R. & Efremov, R. G. Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting. Nat. Methods 20, 1400–1408 (2023).
Wetzl, B. K., Yarmoluk, S. M., Craig, D. B. & Wolfbeis, O. S. Chameleon labels for staining and quantifying proteins. Angew. Chem. Int. Ed. 43, 5400–5402 (2004).
Juers, D. H. et al. High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation. Protein Sci. 9, 1685–1699 (2000).
Mira, P., Yeh, P. & Hall, B. G. Estimating microbial population data from optical density. PLoS ONE 17, e0276040 (2022).
Ruan, Z., Osei-Owusu, J., Du, J., Qiu, Z. & Lü, W. Structures and pH-sensing mechanism of the proton-activated chloride channel. Nature 588, 350–354 (2020).
Deng, Z. et al. Cryo-EM structure of a proton-activated chloride channel TMEM206. Sci. Adv. 7, eabe5983 (2021).
Wang, C., Polovitskaya, M. M., Delgado, B. D., Jentsch, T. J. & Long, S. B. Gating choreography and mechanism of the human proton-activated chloride channel ASOR. Sci. Adv. 8, 3942 (2022).
Ullrich, F. et al. Identification of TMEM206 proteins as pore of PAORAC/ASOR acid-sensitive chloride channels. eLife 8, e49187 (2019).
Suzuki, J., Umeda, M., Sims, P. J. & Nagata, S. Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 (2010).
Lhermusier, T., Chap, H. & Payrastre, B. Platelet membrane phospholipid asymmetry: from the characterization of a scramblase activity to the identification of an essential protein mutated in Scott syndrome. J. Thromb. Haemost. 9, 1883–1891 (2011).
Braga, L. et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature 594, 88–93 (2021).
Alvadia, C. et al. Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. eLife 8, e44365 (2019).
Brunner, J. D., Lim, N. K., Schenck, S., Duerst, A. & Dutzler, R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212 (2014).
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A. & Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121 (2013).
Giss, D., Kemmerling, S., Dandey, V., Stahlberg, H. & Braun, T. Exploring the interactome: microfluidic isolation of proteins and interacting partners for quantitative analysis by electron microscopy. Anal. Chem. 86, 4680–4687 (2014).
Kastner, B. et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2007).
Hauer, F. et al. GraDeR: membrane protein complex preparation for single-particle cryo-EM. Structure 23, 1769–1775 (2015).
Lomeli, G. & Herr, A. E. Reducing cathodic drift during isoelectric focusing using microscale immobilized pH gradient gels. Anal. Chem. 96, 8648–8656 (2024).
Sharma, H. et al. A scalable high-throughput isoelectric fractionation platform for extracellular nanocarriers: comprehensive and bias-free isolation of ribonucleoproteins from plasma, urine, and saliva. ACS Nano 17, 9388–9404 (2023).
Bhattacharjee, S. et al. Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling. Cell 187, 782–796 (2024).
Rashid, S., Ward-Bond, J., Krupin, O. & Berini, P. Non-specific adsorption of protein to microfluidic materials. Colloids Surf. B 208, 112138 (2021).
Ettre, L. S. Nomenclature for chromatography (IUPAC Recommendations 1993). Pure Appl. Chem. 65, 819–872 (1993).
Meier, R. J., Steiner, M.-S., Duerkop, A. & Wolfbeis, O. S. SDS–PAGE of proteins using a chameleon-type of fluorescent prestain. Anal. Chem. 80, 6274–6279 (2008).
Geertsma, E. R. & Dutzler, R. A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry 50, 3272–3278 (2011).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29, 1069–1078 (2020).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015).
Feng, S. et al. Cryo-EM studies of TMEM16F calcium-activated ion channel suggest features important for lipid scrambling. Cell Rep. 28, 567–579 (2019).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Bai, Y. et al. Structural basis for pharmacological modulation of the TRPC6 channel. eLife 9, e53311 (2020).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Acknowledgements
We thank P. Kolata for assistance with the assembly of the MISO device, R. Claessens for advising on the design of the light detector and M. Fislage for assistance with cryo-EM data collection. We acknowledge the funding provided by Vlaams Instituut Voor Biotechnologie, Fonds Wetenschappelijk Onderzoek (grant nos. G0H5916N, G054617N to R.G.E.), and by the European Research Council (grant no. 726436 to R.G.E.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
G.E. developed the microfluidic chip, instrument and software. G.E. and R.G.E. designed and constructed the plunger device. G.E. characterized and optimized the operation of MISO chips and the plunger. G.E. and S.D.G. characterized the MISO chip. A.S. fabricated MISO chips and prepared E. coli cells expressing βG. S.S. and J.D.B. designed the constructs for TMEM206 and TMEM16F, generated the stable cell lines, and established purification conditions for btTMEM206 and mTMEM16F. B.S. and P.E. provided cells and purification protocols for TRPC6. S.S. and J.D.B. purified and plunged btTMEM206 using the conventional approach. G.E. and S.D.G. optimized MISO purification protocols. G.E. performed MISO experiments with β-galactosidase, btTMEM206 and TMEM16F. S.D.G. performed MISO experiments with TRPC6. G.E., S.D.G. and J.D.B. collected and processed cryo-EM data. S.D.G. built, refined and validated atomic models. R.G.E. prepared the original paper draft. R.G.E., G.E., S.D.G., S.S. and J.D.B. prepared figures and reviewed and edited the paper. R.G.E. conceived, managed and supervised the project, and acquired funding.
Corresponding author
Ethics declarations
Competing interests
G.E. and R.G.E. are inventors on the patent application WO2023/232662/A1 disclosing the MISO instrument and chip design filed by VIB and VUB. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Thomas Braun and Arjen Jakobi for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Arunima Singh, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Experimental setup.
a, An overview of the experimental setup. b, Close-up of the MISO chip and plunger module. Key elements are labeled.
Extended Data Fig. 2 Layout of MISO chips.
a, b, Chip photograph and schematics of chip elements for a 2-column MISO chip. c, d, Chip photograph and schematics of chip elements for a 1-column MISO chip. Three single-column microfluidics circuits with columns of 0.5, 3 and 10 μl were designed on a single chip. The valves are shown as red circles, and pneumatic channels are not shown in the schematics of the b and d panels. The following abbreviations are used: IN – inlet, WO – waste outlet, CF- column filling port, DZ – detection zone. In a and c, long graduations of the ruler correspond to 1 cm. The chip designs are provided as Supplementary Data 1.
Extended Data Fig. 3 Processing of β-galactosidase data for cryo-EM sample prepared starting from 20 μg βG containing cytoplasmic extract.
a, A micrograph; representative from n > 3000 micrographs. b, 2D class averages. The box size is 278 Å. c, Image processing scheme. d, Fourier Shell Correlation curves for unmasked and masked half-maps and between model and map. e, Heat map shows the distribution of particle orientations. f, 3D map surface colored by local resolution.
Extended Data Fig. 4 Processing of β-galactosidase data for cryo-EM sample prepared starting from 1 μg βG containing cytoplasmic extract.
a, A micrograph; representative from n > 5000 micrographs. b, 2D class averages. The box size is 280 Å. c, Image processing scheme. d, Fourier Shell Correlation curves for unmasked and masked half-maps and between model and map. e, Heat map shows distribution of particle orientations. f, 3D map surface colored by local resolution.
Extended Data Fig. 5 Processing of TMEM206 purified and plunged using the conventional approach.
a, A micrograph; representative from n > 5000 micrographs. b, Selected 2D class averages. The box size is 304 Å. c, Image processing scheme. d, Fourier Shell Correlation curves for unmasked and masked half-maps and between model and map. e, The heat map shows distribution of particle orientations. f, 3D map surface colored by local resolution.
Extended Data Fig. 6 Processing of TMEM206-YFP purified and plunged using MISO.
a, A micrograph; representative from n > 7000 micrographs. b, Selected 2D class averages. The box size is 278 Å. c, Image processing scheme. d, Fourier Shell Correlation curves for unmasked and masked half-maps and between model and map. e, Heat map shows the distribution of particle orientations. f, 3D map surface colored by local resolution.
Extended Data Fig. 7 Processing of TMEM16F-YFP purified and plunged using MISO.
a, A micrograph; representative from n > 12000 micrographs. b, Selected 2D class averages. The box size is 278 Å. c, Image processing scheme. d, Fourier Shell Correlation curves for unmasked and masked half-maps and between model and map. e, Heat map shows distribution of particle orientations. f, 3D map surface colored by local resolution.
Extended Data Fig. 8 Processing of TRPC6.
a, A cryo-EM micrograph (representative from n > 4000 micrographs) and, b 2D class averages. The box size is 288 Å. c, Image processing scheme. d, g, Heat maps show distribution of particle orientations for soluble domain and complete trans-membrane complex, respectively. e, h Fourier Shell Correlation curves for unmasked and masked half-maps. f, i, 3D map surfaces colored by local resolution.
Extended Data Fig. 9 Identification of the TRPC6 by sequencing cryo-EM map.
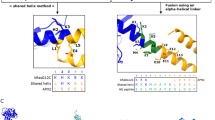
a, ModelAngelo modelled a sequence of 151 HHM segments into a reconstructed cryo-EM map of the protein of interest. TRPC6 was identified as the highest likelihood target upon HHM profiles searched against the HMMER v3.3 reference proteome database. hTRPC6 was refined into the density (pink) and superimposed onto hTRPC6 (PDB 6UZ8, grey). b, Examples of density map regions with ModelAngelo sculptured fragments. c, Examples of HMMR search results for 2 sequences against the database.
Supplementary information
Supplementary Information
Supplementary Note, Figs. 1–9 and uncropped scans of gels.
Supplementary Video 1
Blotless deposition of protein solution on EM grid through capillary from MISO chip.
Supplementary Video 2
MISO protein deposition on EM grid, blotting and plunging.
Supplementary Data 1
AutoCAD files in.dwg format containing MISO chip designs.
Supplementary Data
Numerical source data for Supplementary Figs. 1 and 6.
Source data
Source Data Fig. 1
Chromatograms and plots.
Source Data Fig. 1
Unprocessed gels.
Source Data Fig. 2
Chromatogram.
Source Data Fig. 2
Unprocessed gels.
Source Data Fig. 3
Chromatogram.
Source Data Fig. 3
Unprocessed gels.
Source Data Fig. 4
Chromatogram.
Source Data Fig. 4
Unprocessed gels.
Source Data Fig. 5
Chromatogram.
Source Data Fig. 5
Unprocessed gels.
Source Data Fig. 6
Chromatogram.
Source Data Fig. 6
Unprocessed gels.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Eluru, G., De Gieter, S., Schenck, S. et al. MISO: microfluidic protein isolation enables single-particle cryo-EM structure determination from a single cell colony. Nat Methods 22, 2563–2573 (2025). https://doi.org/10.1038/s41592-025-02894-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02894-x