Abstract
GABA (γ-aminobutyric acid) is the primary inhibitory neurotransmitter in the mammalian central nervous system. GABAergic neuronal types play important roles in neural processing and the etiology of neurological disorders; however, there is no comprehensive understanding of their functional diversity. Here we perform two-photon imaging of GABA release in the inner plexiform layer of male and female mice retinae (8–16 weeks old) using the GABA sensor iGABASnFR2. By applying varied light stimuli to isolated retinae, we reveal over 40 different GABA-releasing neuron types. Individual types show layer-specific visual encoding within inner plexiform layer sublayers. Synaptic input and output sites are aligned along specific retinal orientations. The combination of cell type-specific spatial structure and unique release kinetics enables inhibitory neurons to sculpt excitatory signals in response to a wide range of behaviorally relevant motion structures. Our findings emphasize the importance of functional diversity and intricate specialization of GABAergic neurons in the central nervous system.
Similar content being viewed by others
Main
Neurons communicate between one another primarily through neurotransmitter release and reception. GABA is the predominant inhibitory neurotransmitter in the vertebrate brain. Although less abundant than excitatory neurons, GABAergic neurons are essential for increasing and diversifying the computational power of circuits by gating excitatory signals from principal neurons1. Accordingly, dysfunction of GABAergic signaling results in diverse neurodevelopmental disorders, including autism spectrum disorders2, cognitive disorders3, mood disorders4, dystonia5 and congenital visual disorders6.
In the vertebrate retina, an accessible part of the central nervous system, GABAergic neurons play critical roles in visual processing. Light is detected by photoreceptors, which form excitatory synapses with glutamatergic interneurons called bipolar cells. Bipolar cells transmit this signal to more than 30 types of retinal ganglion cell, each encoding different visual features and transmitting them to the brain in parallel7. The axon terminals of bipolar cells and dendrites of retinal ganglion cells are suppressed by neurotransmitters, such as GABA and glycine, released from amacrine cells, a class of retinal interneuron comprising diverse types8,9,10. A recent study identified 63 molecular clusters of amacrine cells, ~70% of them GABAergic11, indicating that GABA provides varied inhibitory modulation in the inner retina.
Despite this understanding of basic circuit connectivity and neurotransmitter usage, the specific functional properties of a wide array of GABAergic signaling remain uncharacterized, due to large cellular diversity and a lack of tools to genetically target and study these neurons. Physiological and morphological studies can identify some specific amacrine cell types, including wide-field A17 cells12 and direction-selective starburst amacrine cells (SACs)13,14,15, from their distinctive features; however, a comprehensive cataloging of the functional properties of all amacrine cell types has not been provided. It is even more challenging for cortical interneurons given their complexity and diversity16.
Here we combine two-photon imaging of the recently developed GABA indicator iGABASnFR2 (ref. 17), unsupervised clustering of release types, mapping of receptive and projective fields, and computational modeling to determine the functional diversity of GABA signaling in the retina of both male and female mice (8–16 weeks old). The GABA response profiles reveal >40 cell types, each with unique synaptic release kinetics. We discovered spatiotemporally ordered relationships between receptive and projection fields for many amacrine cell types, which would allow encoding of diverse features of visual inputs.
Functionally divergent GABA signal in the inner retina
To investigate the diversity of GABA signaling in the retina, we labeled amacrine and retinal ganglion cells by intravitreal injection of AAV2/9-hSyn1 encoding iGABASnFR2 (Fig. 1a,b and Extended Data Fig. 1a–c) and performed two-photon imaging of extracellular GABA signals released from amacrine cells. The iGABASnFR2 likely reports either GABA released from amacrine cells or received by ganglion and amacrine cells. We imaged ~7,100 regions of interest (ROIs) on their dendrites throughout the inner plexiform layer (IPL) during light stimulation. By using a ChAT-IRES-Cre mouse line crossed with ROSA26-STOP-tdTomato mice, SACs were co-labeled with the red fluorophore tdTomato, which allowed separation of the IPL into nine sublayers (L1–L9) based on SAC process stratification depth (Extended Data Fig. 1d). Three types of visual stimuli were used to characterize the functional properties of GABA signals (Extended Data Figs. 1e–i and 2a–d): a static spot of modulating light intensity to characterize response polarity, kinetics, and modulation by temporal frequency and contrast; a moving spot to measure direction and orientation selectivity; and dense noise to estimate receptive field properties.
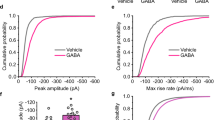
a, Left: schematic of retinal neurons. AC, amacrine cell; BC, bipolar cell; SAC, starburst AC; RGC, retinal ganglion cell. Right: two-photon cross-sectional images of iGABASnFR2 and ChAT-IRES signals. L4 and L6 denote depths of OFF and ON ChAT processes, respectively. b, iGABASnFR2 (green) and ChAT-IRES (magenta) signals (7,098 ROIs, 11 retinae) in different imaging planes. The results were replicated 11 times independently. c, Dendrogram sorting of 49 identified GABA signal groups by direction/orientation selectivity and temporal dynamics (labels) (7,098 ROIs, 11 retinae). DS, direction selective (magenta); OS, orientation selective (blue); Sust., sustained; Trans., transient. d, Heatmap of ON response index (ON resp.), OFF response index (OFF resp.), bi-response index (Bi-resp.), transience index (Trans.) and latency index. e, Average static flash stimulus-evoked (left) and motion stimulus-evoked (right) signal for individual groups. Responses to a motion direction that produced maximum responses were temporally aligned relative to peak response timing and averaged. Gray shading indicates the s.d. Black line shows the average GABA signal for ROIs assigned to each group. f, Histogram of receptive field (RF) diameter for individual groups (color) and all groups (gray), each normalized to respective peaks.
Our stimulation protocols revealed that GABA signals in the inner retina show functional divergence depending on the sublayer being imaged (Extended Data Fig. 2e). Analysis of GABA signals using sparse principal component analysis (PCA) and Gaussian mixture modeling uncovered 49 groups7,18 (Fig. 1 and Extended Data Fig. 2f). The identified groups were further assigned to direction-selective or orientation-selective groups, according to their motion responses, with 14.3% being direction-selective and 20.4% being orientation-selective. We then categorized the 49 groups according to their response to light onset (ON) and offset (OFF). Overall, 21 were classified as ON, 12 as OFF and 16 as ON–OFF (Fig. 1c–e). Six of the ON groups were subclassified as delayed-ON due to their noncanonical long response latency.
To investigate the physiological properties of individual groups, we performed k-means clustering, which successfully classified 89.8% of ROIs according to their temporal dynamics (Extended Data Fig. 3a–c). Cluster analysis of receptive field size subsequently revealed four categories: small, small–medium (s–medium), large–medium (l–medium) and large (Fig. 1f and Extended Data Fig. 3d). These receptive field categories were then assigned to the 42 groups (85.7%) that were dominated by a single receptive field size (Extended Data Fig. 3e). Of 42 groups, 30.9% were also classified as orientationally biased (Extended Data Fig. 3f,g). Because wide-field cells (medium- and large-field in our classification) include polyaxonal cell types, characterized by long axon-like dendritic processes and Na+ spikes19, we performed imaging during pharmacological block of voltage-gated sodium channels (NaV) by tetrodotoxin (TTX). Of the 49 groups, 34% were TTX-sensitive: 28% inhibited and 6% disinhibited (Extended Data Fig. 4a,b). Medium- and large-field-receptive field groups inhibited by NaV block likely correspond to known amacrine cell types with thick axon-like processes (Supplementary Table 1).
Together, the 49 groups encompass many essential components of visual space representation, including receptive field size, contrast sensitivity, temporal frequency sensitivity (Fig. 2a and Extended Data Fig. 3h–k) and motion response properties (Fig. 2b). A positive correlation was apparent between response modulation by contrast and temporal frequency, as previously observed in retinal ganglion cells7 and visual cortex neurons20, suggesting that this is a common property across the mouse visual system. Mapping individual groups to the sublayers where they were observed revealed group-specific distribution among layers, together populating the entire IPL (Fig. 2c). This distribution is consistent with previous reports that the termination patterns of dendritic processes differ between cell types8,21 and underscores the diversity of GABA signaling within the retina.
a, Left: relationship between RF size and ON–OFF index for 49 groups (color coded). Circle size denotes fraction of recorded population. Right: relationship between contrast modulation and temporal frequency modulation indexes. b, Motion response properties: direction-selective index (DSI), orientation-selective index (OSI), motion/flash preference and speed tuning. dON, delayed ON. c, Heatmap, distribution of observed ROIs for 49 groups in layers between L2 and L8. Bars show the layer dominance index for each group (color coded). There were 7,098 ROIs and 11 retinae. Boxes represent 25th to 75th percentiles and median; whiskers represent 1.5 × interquartile range.
The mouse retina has 44 distinct functional GABAergic groups
Different axon terminals from a single neuron share intrinsic biological noise patterns22,23,24. We examined this idea by computing noise correlation in ROIs from individual imaging planes (Extended Data Fig. 5a). First, we confirmed that noise correlation allowed us to assign activity in ROIs to individual cells. We targeted SAC processes by injecting AAV encoding floxed iGABASnFR2 into ChAT-IRES-Cre mice, and patching and filling a single SAC with Alexa 594 dye to visualize its dendritic processes (Extended Data Fig. 5b). As predicted, GABA signals in a single cell constituted a group (assembly) with higher noise correlation than with those in processes of nearby SACs (Extended Data Fig. 5c–e). Therefore, high intrinsic neural noise correlation between different signals is indicative of assignment of ROIs to individual cells.
Next, we used noise correlation to assign ROIs from each field of view (FOV) to different assemblies (Fig. 3a–c) and examined the response identity of the assigned ROIs. Receptive fields of ROIs assigned to the same assembly were highly overlapping, suggesting that the assembled ROIs belong to the same cells (Fig. 3d,e). Indeed, ROIs representing the same response group had higher noise correlation than those of different groups (Fig. 3f). Specifically, 41 groups were typically observed in the same assemblies (Fig. 3g,h), indicating that the ROIs contained ≥41 functionally distinct cell types, each characterized by a single release group. Of the remaining eight groups, noise correlation analysis revealed three assemblies comprising multiple response groups: G2/G5, G12/G19/G21 and G35/G42/G47 (Extended Data Fig. 5f). Because the connections were not random, but fixed between specific groups in the same layers (Extended Data Fig. 5g), they likely reflect multiplexed output properties of a single cell type. While response temporal dynamics varied between individual groups in multigroup assemblies, receptive fields were similar (Extended Data Fig. 5h). These data therefore suggest that the imaged ROIs contain a total of 44 functional amacrine cell types.
a, Example ROIs of two different GABA signal groups (magenta and cyan). b, Noise correlation in four example pairs. c, Correlation matrix denoting three assemblies with intrinsic noise. d, RFs of ROIs in a using same color scheme. e, Comparison of distances between RF centers in assembled ROI pairs (cyan; 36 pairs, 1 retina) and others (gray; 192 pairs, 1 retina) used in a. P = 3.42 × 10−14; two-sided Mann–Whitney–Wilcoxon test. f, Relationship between ROI-to-ROI distances and noise correlation in ROI pairs in the same (blue; 4,373 pairs) and different (gray; 15,199 pairs) groups. There were 11 retinae. g, Matrix denoting frequency of GABA signal groups sharing significant neural noise. h, Dominance index for noise coincidence within the same group. Orange, groups with heterogeneous connections. ***P < 0.001; NS, not significant. Boxes represent 25th to 75th percentiles and median; whiskers represent 1.5 × interquartile range.
Direction-selective cell types
We found three ON groups (G7, G10 and G20), one ON–OFF group (G31) and three OFF groups (G37, G38 and G45) with direction selectivity (Fig. 4a,b). Because the only genetically defined direction-selective GABAergic amacrine cells are ON and OFF SACs, we labeled SAC processes by injecting floxed-AAV encoding iGABASnFR2 into the eyes of ChAT-IRES-Cre mice and later imaging GABA signaling in the IPL (Extended Data Fig. 4c). As expected, responses showed smaller variance than recordings from nonspecific amacrine cell types (Extended Data Fig. 4d). ON and OFF SAC signals resembled groups G10 and G37, respectively (Extended Data Fig. 4e,f). The dominance of a single receptive field category (Extended Data Fig. 3e) and stratification in the ChAT layer (ON and OFF SACs) depths (Fig. 4f) supports a cellular assignment for G10 and G37. Moreover, this close correspondence demonstrates the reliability of our clustering method for identifying distinct functional cell types.
a, DSI of ON (dark gray; 88 ROIs, P = 3.18 × 10−34) and OFF (light gray; 62 ROIs, P = 3.58 × 10−35) ChAT, non-DS groups (gray; 6,180 ROIs) and DS groups (color coded; 189 G10 ROIs, P = 1.74 × 10−82; 148 G37, P = 2.64 × 10−58; 133 G7, P = 6.38 × 10−57; 128 G20, P = 2.11 × 10−68; 109 G31, P = 3.71 × 10−77; 129 G38, P = 3.14 × 10−81; 141 G45, P = 9.16 × 10-47). ON and OFF ChAT targeted imaging, four retinae. Nontargeted imaging, 11 retinae. Kruskal–Wallis test, Bonferroni correction for multiple comparisons. b,c, Distributions of preferred directions in individual DS groups (b) and all TTX-sensitive DS groups (c). D, dorsal; T, temporal; V, ventral; N, nasal. d, Multidimensional features projected along the principal axes in datasets pooling functionally labeled (G10, G37, G7, G20, G31, G38 and G45) and genetically labeled (ON and OFF ChAT) DS groups (gray dots). Color-coded circles, average of each group. e, Top: similarity between functionally labeled and genetically labeled groups (11 retinae). Bottom: RF sizes (compared to G10: G7, P = 9.40 × 10−43; G20, P = 3.22 × 10−79; G31, P = 5.01 × 10−104; G38, P = 5.53 × 10−23; G45, P = 2.32 × 10−78; compared to G37: G7, P = 2.88 × 10−21; G20, P = 2.90 × 10-47; G31, P = 1.41 × 10−67; G38, P = 7.99 × 10−9; G45, P = 9.28 × 10−46). Kruskal–Wallis test, Bonferroni correction for multiple comparisons. f, Layer distribution of DS groups. g, Top: population of TTX-sensitive groups (left) and DS groups among them (right) in the functional clustering. Bottom: population of NaV-expressing groups (left) and NaV and acetylcholine receptor (AChR) coexpressing groups among them (right) in the molecular clustering. h, Expression of key neurotransmitter receptors in NaV-expressing molecular groups. Groups expressing AChRs highlighted (pink, GABAergic; green, glycinergic). mGluR, metabotropic glutamate receptors; iGluR, ionotropic glutamate receptors; GlyR, glycine receptors; GABAR, GABA receptor; Markers, known amacrine cell type markers. Gene counts and metadata from Yan et al.11 (g,h). ***P < 0.001; boxes represent 25th to 75th percentiles and median; whiskers represent 1.5 × interquartile range.
The other five direction-selective groups (G7, G20, G31, G38 and G45) showed no correlation with SAC signals (Extended Data Fig. 4g,h). Notably, the five direction-selective groups had significantly larger receptive fields than others (Fig. 4e and Extended Data Fig. 3e), and their activities decreased under pharmacological block of NaV (Extended Data Fig. 3b), suggesting that they correspond to TTX-sensitive polyaxonal wide-field amacrine cells. While the tuning directions of the TTX-insensitive G10 and G37 groups (SACs) are widely distributed14,22, those of each TTX-sensitive group cluster along one or multiple cardinal directions (Fig. 4b,c). The preferred directions of all TTX-sensitive groups cluster along the four cardinal directions, as do those of direction-selective ganglion cells25, although the preferred direction of many amacrine cells was predominantly horizontal. Multidimensional analysis of response features revealed functional segregation of TTX-sensitive and -insensitive groups (Fig. 4d,e and Extended Data Fig. 4i).
Our data indicate a greater functional diversity of wide-field amacrine cell types than previously appreciated. To relate physiologically to molecularly identified types, we analyzed published single-cell amacrine cell transcriptomes11. The frequency of TTX-sensitive response groups (29%) resembled that of molecular groups expressing TTX-sensitive NaV channels (32%, 20 of 62 groups) (Fig. 4g and Extended Data Fig. 4k). The 20 NaV-expressing molecular groups showed diverse expression patterns of neurotransmitter receptors (Fig. 4h), consistent with the large functional diversity we observe in TTX-sensitive response groups. We found that 35% of NaV-expressing molecular groups also expressed acetylcholine receptors (AChRs; Fig. 4g,h). As acetylcholine release from SACs is direction-selective26, these AChR-expressing groups might establish direction selectivity. Indeed, TTX-sensitive direction-selective signals tended to stratify in the same depth as SACs (ChAT layers; Fig. 4f) and the direction selectivity decreased during pharmacological blockade of AChRs (Extended Data Fig. 4j). Recent electron microscopy studies in mouse and primate retinae identified synapses from SACs onto wide-field amacrine cells22,27. Collectively, these suggest the involvement of TTX-sensitive polyaxonal wide-field amacrine cell types in direction-selectivity circuits22,27. We posit that AChR/NaV-coexpressing molecular groups correspond to the identified direction-selective, TTX-sensitive wide-field cells (29%, 7 of 20 clusters; Fig. 4g). On the other hand, G31, the ON–OFF TTX-sensitive direction-selective group, was an exception to this trend, terminating broadly in middle-to-inner layers (L6–L8), rather than either ON or OFF ChAT layer depths (Fig. 4f). This resembles the morphological features of wide-field direction-selective A1 amacrine cells, a displaced polyaxonal amacrine cell type in the primate retina27.
Visual features are encoded in specific IPLs
We sought to examine, in an unbiased manner, how our observed functional groups encode visual information by characterizing prominent features of visual stimulus encoding using PCA (Extended Data Fig. 6a–d). We used direction selectivity, orientation selectivity, motion/flash preference, speed tuning, response modulation by contrast and response modulation by temporal frequency as visual features for our analysis. A subset of 30 of the 49 groups (61%) was sufficient to robustly encode all six visual features (Fig. 5a,b and Extended Data Fig. 6e,f). The 30 groups were sorted according to their information score (an estimate of the extent to which they encode visual features; Fig. 5c). We observed redundant encoding of motion/flash preference and response modulation by contrast (70% of the top ten informative groups encoded one or both). In general, feature encoding was distributed across the groups without obvious bias, indicating that multiple amacrine cell types process each visual feature.
a, Population of functionally characterized groups based on flash and motion responses. b, Fraction of significantly informative visual features encoded by specific groups. TF, temporal frequency. c, Profiles of visual encoding in 30 informative groups (Grps) sorted by Hotelling’s T2 score. White diamonds show significantly informative features. d, Left: schematic of retinal layers. IPL sublayers denoted by light and dark gray. PR, photoreceptor; OPL, outer plexiform layer; INL, inner nuclear layer; GCL, ganglion cell layer. Right: fraction of significantly informative visual features for each IPL sublayer. Largest and second-largest features in each layer marked by pink and light pink, respectively; 4,149 ROIs, 6 retinae.
Nevertheless, assigning feature sensitivity to individual sublayers revealed laminar differences (Fig. 5d). For example, outer layers (L2 and L3) are more sensitive to changes in temporal frequency and contrast. ROIs in the inner layers (L7 and L8) were enriched in those showing motion/flash and orientation selectivity. Intermediate layers had different profiles: L4 and L6 are exclusively direction-selective, and L4 and L5 specialize in speed tuning. These results highlight that individual amacrine cells, differentially distributed across the IPL, encode specific aspects of visual stimuli.
GABA signaling is differentially compartmentalized
Many amacrine cells lack axons per se, and the transmitter is released from the same dendritic processes that receive synaptic inputs10. Some amacrine cell types, however (including polyaxonal wide-field cells19 and SACs13,14,28), have processes compartmentalized into synaptic input and output sites. Thus, dendritic segmentation of synaptic inputs and outputs can be used as an indicator of cell type. We investigated compartmentalization of inputs and outputs along the nasotemporal and dorsoventral axes of the retina (Extended Data Fig. 7a,b). For this, the receptive field locations of individual GABA signals were mapped and ROI locations were aligned with each receptive field center. The area occupied by the ROI locations for each cell was defined as the projective field, indicating the spatial extent of the output GABA signal (Extended Data Fig. 7c,d).
We found that spatial relationships between receptive fields and projective fields differed across response groups (Fig. 6a–c). For example, projective fields of the delayed-ON G6 group lay inside their respective receptive fields. On the other hand, projective fields of two other delayed-ON groups (G2 and G4) were biased to the temporal and ventral sides of each receptive field, respectively. Furthermore, receptive fields of group G4 displayed spatial offset from the receptive field, suggestive of anatomical compartmentalization. The projective fields of TTX-sensitive wide-field cells tended to be spatially offset, suggesting that Na+ action potentials drive transmitter release at distant sites along the long axon-like processes (Extended Data Fig. 7g,h). Overall, the projective fields of response groups varied in size, shape and extent of overlap with their corresponding receptive fields (Fig. 6d–f). Thus, amacrine cell processes show different compartmentalization of synaptic input and output sites, depending on cell type.
a–c, Projective fields of three example groups. Left: ROI location mapping (cyan dots, release sites) relative to RFs (purple). Black indicates the RF envelope. Middle left: density of release sites. Middle right: histogram denoting orientation of ROI locations (140 G6, 254 G2 and 147 G4 ROIs; 17 retinae). Right: estimated RFs and projective fields (PFs). Dots indicate centroids. d, Relationship between RF area and PF area for 49 groups (color coded). Gray circles indicate RFs and PFs of glutamatergic groups (six ON and six OFF). Inset shows the ratio of PF area to RF area for amacrine cells (green; 49 groups and 17 retinae) and glutamatergic cells (gray; 12 groups and 8 retinae). e, Relationship between directional and orientational bias indexes (DBI and OBI, respectively) for projective fields. Gray circles show bipolar cells. Gray thick and dotted lines show 95% CIs. Inset shows example directionally (top, G15) and orientationally (bottom, G13) biased projective fields. f, Orientation relative to RF (angle) and extent of overlap between RF and PF. g, Relationship between overlap index (OLI) and size change index (SCI). Gray circles indicate glutamatergic cells. Inset shows RFs and PFs for three example groups. h, Angular tunings of projective fields for example orientationally (top) and directionally (bottom) biased groups. i, Top: angular tunings of projective fields for orientationally (left) and directionally (right) biased groups. Arrows represent tuning in each group. Bottom: histogram of preferred angles. j, Clustering of projective field properties. Magenta and yellow squares, directionally and orientationally biased groups, respectively. Boxes represent 25th to 75th percentiles and median; whiskers represent 1.5 × interquartile range.
Notably, projective fields with orientational or directional bias were aligned along the retinal cardinal axes (Fig. 6h,i and Extended Data Fig. 7d,i), as previously shown for the preferred directions of direction-selective ganglion cells25, with orientationally biased projective fields only observed along the horizontal axes. Remarkably, this variety of receptive–projective field relationships was not found for glutamatergic signals mediated by mainly bipolar cell types, which we monitored by imaging glutamate on the dendrites of ON–OFF direction-selective ganglion cells (Extended Data Fig. 7e,f) using the iGluSnFR sensor18,22. These results agree with a previous study of the tiger salamander retina showing a single bipolar cell transferring its signal exclusively vertically29. Thus, GABA seems to diversify signal processing pathways more than glutamate in the mammalian retina.
Amacrine cells heterogeneously filter visual motion
Finally, we asked how individual response groups modulate post-synaptic cells. The efficacy by which amacrine cells modulate the activity of post-synaptic cells (modulation efficacy) depends, in part, on the latency between amacrine cells detecting and transmitting signal. Thus, any offset between the receptive field and projective field (where post-synaptic cells are located) could directly affect modulation efficacy according to the speed of activity propagation along processes (Extended Data Figs. 8a and 9).
We built a computational model to simulate light-evoked responses (Extended Data Fig. 8b), assuming that amacrine cells modulate post-synaptic bipolar cells at release sites. The time courses of modulation were greatly affected by both speed of motion stimuli and functional properties of amacrine cells (Extended Data Fig. 8c); thus, individual cell groups were tuned to different motion speeds (Extended Data Fig. 8d,e). Half the groups were tuned to faster motion speed due to short response latency (Extended Data Fig. 8e). Of note, displaced release sites and long response latencies in delayed-ON groups like G6 gave rise to slow-motion tuning, relevant for visual stimuli induced during slow drift of head and/or eye. Tuning speeds are spread over a wide range, covering behavioral contexts from slow slips associated with eye drift (~10 ° s−1) to fast saccadic eye movements (~100 ° s−1).
Discussion
This work shows that there are 49 functionally different types of GABA signal associated with amacrine cells in the mouse retina (Supplementary Table 1). Noise correlation analysis suggests that 44 distinct amacrine cell types give rise to 49 groups: 41 cell types with single waveforms and 3 types showing diverse waveforms at distinct IPL depths. The number of functional cell types is very similar to the 43 molecular groups identified in a previous study11.
Cell type identification
Our results reveal previously undescribed amacrine cell types, including those with noncanonical long latencies (delayed-ON cells), non-SAC direction-selective cells, and cell types with multiplexed outputs. A direction-selective cell (G31) transmits signals to broad IPLs other than the ON and OFF SAC layers (Fig. 4f). Furthermore, although GABAergic amacrine cells have been assumed to be solely wide-field cells8,23, our results show that they range from small-field (comparable to known narrow-field glycinergic amacrine cells8) to wide-field types (Extended Data Fig. 3d). We also found that TTX-sensitive wide-field cells segregated into different types. This physiological variation was reflected in broad diversity in expression of neurotransmitter receptors among NaV-expressing amacrine cells (Fig. 4h).
We confirmed that ON and OFF SAC types correspond to G10 and G37, respectively, by genetically targeted recordings of SACs (Fig. 4), suggesting that our designations of other groups are likely correct, as well. Furthermore, assuming that (1) axon-like processes provide sensitivity to NaV block (Extended Data Fig. 4b) and (2) dendritic stratification and arborization in the IPL correlate with observed sublayer depth and layer dominance (Fig. 2c), we predict cellular identities for the following groups: G3, SST-1 cells; G14, CRH-1 cells; G18, A17 (CCK-2) cells30,31; G22, TH2-cells32; G30, nNOS-2 cells; G31, CRH-2 (A1, nNOS-1) cells; G32, VIP-1 cells (Supplementary Tables 1 and 2). CRH-1 cells morphologically resemble Gbx2+ sublamina 5-targeting cells (S5-Gbx2+), which were genetically identified by expression of Maf and Lhx9 (refs. 11,33). The ON–OFF transient spiking and direction selectivity of G31 are a good match for A1 amacrine cells27. Further studies involving recordings from different genetically labeled amacrine cell types are required to confirm our assignments.
Directional and orientational specificity of GABA release
We identified five direction-selective amacrine cell types in addition to SACs. Given their TTX sensitivity (Fig. 4f), they should molecularly correspond to NaV-expressing wide-field cell types. Our gene expression analysis found NaV/AChR co-positive molecular groups (Fig. 4h). NaV/AChR positive groups may receive directionally tuned cholinergic inputs from SACs to generate direction selectivity26 (Extended Data Fig. 10a). Indeed, synapses from SACs onto the axons of wide-field amacrine cells have been identified by electron microscopy22. Thus, the direction-selective amacrine cells we observed might correspond to NaV/AChR positive groups.
Alternatively, three other mechanisms could establish direction selectivity in NaV-expressing cells. The first is the indirect effect mediated by bipolar cells. Direction-selective glutamatergic inputs from type 2 and 7 bipolar cells22 could promote direction selectivity in the post-synaptic wide-field cells. The second is spatiotemporal organization of excitatory synaptic activity. Spatially structured excitatory inputs with different temporal kinetics along local dendritic segments could generate direction-selective activity, as shown in SACs34,35 (Extended Data Fig. 10b). The third is spatially segregated input and output. Previous modeling studies suggested that segregated input and output synapses (with inputs restricted to proximal dendrites) could generate direction-selective activity at distal SAC dendrites14,34. Consistent with this, we found that projective fields of direction-selective types tend to be shifted (Extended Data Fig. 7g), with the angles of offsets matching the preferred directions of motion responses (Extended Data Fig. 7i). These hypotheses will be tested in future experiments, such as genetic manipulation of SACs, anatomical tracing of pre-synaptic cells, and electrophysiological analysis of synaptic inputs from individual NaV-expressing cells. It should be noted that projective field mapping is limited to the imaging FOV size. The complete mapping of wide-field, polyaxonal types, which have processes > 1 mm, would require macroscale imaging from genetically targeted cell types.
Our spatial projective and projective field mapping revealed that ~60% of response groups had laterally offset and/or orientational bias, suggesting compartmentalization of dendrites. For example, dendrites of SACs are functionally segregated into proximal regions for bipolar cell inputs (giving rise to their receptive field) and distal regions for neurotransmitter release (giving rise to their projective field)13,14. Previous electrophysiological measurements of synaptic inputs to polyaxonal wide-field cells showed hotspots of synaptic inputs on proximal processes36. It is also known that cell types with wide dendritic arbors have proximal dendrite-like and distal axon-like processes19,27,31,36.
Orientational bias suggests that dendrites are asymmetrically directed along specific retinal orientations31. For example, the directional projective field of G14 resembles the asymmetrically aligned dendrites of CRH-1. Such orientationally aligned tuning might explain the cardinal alignment observed for projective fields. Indeed, some orientation-selective response groups (G30, G34 and G44) have horizontally elongated projective fields.
Functional contribution of GABA signal in visual processing
The information processing of GABA signal groups displayed layer specificity (Fig. 5d). While OFF-dominant layers (L2 and L3) were tuned to static features (contrast and temporal frequency), ON-dominant layers (L7 and L8) were tuned to image motion features (orientation selectivity, speed tuning). Of note, human psychophysics studies have shown a greater contrast gain in the OFF system37 and better motion speed tuning in the ON system38. The functional lamination of GABA signals might explain known asymmetries in ON and OFF pathways.
The spatial offset of projective fields relative to receptive fields will generate diverse configurations of lateral inhibition (Extended Data Fig. 7j). We found a horizontal bias in the offsets (Fig. 6h), indicating that lateral inhibition is more enhanced along the horizontal axis. Indeed, psychophysics studies in human have suggested that visual feature detection is enhanced in horizontal over vertical orientations, resulting in better visual attention and cognitive tasks39. The horizontally biased lateral inhibition in the retina might contribute to this spatial asymmetry in visual processing.
The identified direction-selective amacrine cell types showed strong horizontal preference (Fig. 4c). It is known that spontaneous rapid shifts of eye positions to capture an object (saccades) occur more frequently in the horizontal than in the vertical axis in head-restrained mice40,41. Freely moving mice make the conjugate horizontal eye movements, with coupled head rotation, to produce a saccade and fixate gaze shifts41. Thus, the retina receives rapid horizontal flows during natural behaviors. The horizontally tuned GABAergic inhibition may allow selective gating of retinal activity, such as saccadic suppression, to effectively stabilize visual perception. Of the identified response groups, three (G7, G38 and G45) correspond to such gating cell types. Indeed, previous physiological studies on isolated retinae showed that fast, global optic flow, simulating saccadic eye movements, suppressed retinal ganglion cells via far-surround inhibition mediated by TTX-sensitive polyaxonal amacrine cells42, although direction selectivity of the amacrine cell types was not examined.
Furthermore, selectivity to the nasal direction (corresponding to posterior optical flow) was overrepresented in G20 and G31 (Fig. 4b). Given their middle-speed range tuning (Extended Data Fig. 8f), it is plausible that GABAergic suppression by G20 and G31 contributes to the processing of retinal image during forward locomotion. Notably, the posterior motion preference is also dominant in retinal ganglion cells and cortical direction-selective cells43,44, indicating that G20 and G31 would not suppress those direction-selective cells. One plausible function of these groups may be to prevent the activation of local object motion detectors45 in response to global optic flow. Indeed, G20 and G31 would be activated by global optic flow, but not local object motion, because of their wide-field receptive fields. It would be possible that retinal circuits in the mouse retina are built to differentiate horizontal-global versus horizontal-local motion, depending on behavioral context.
Technical limitations
We labeled amacrine and ganglion cell processes with iGABASnFR2 using the pan-neuronal synapsin-1 promoter, which will express in both GABAergic and glycinergic amacrine cells. Further, iGABASnFR2 is expressed continuously on the plasma membrane, and as such detects both released and received GABA17,46. The first limitation of our study is that iGABASnFR2 might be picking up physiologically irrelevant GABA signal such as spillover to extra-synaptic space where there are no GABA receptors expressed on the post-synaptic structures; however, GABAergic synapses in the inner retina are tight and ‘wraparound’26,47, likely shortening any delays between pre-synaptic release and post-synaptic reception and preventing detectable spillover. If there are non-negligible spillovers, there might be artificial GABA signals reflecting mixes of the spillovers at the extra-synaptic spaces. Instead, the GABA signal from the genetically labeled SAC processes showed slight variance in the response measurements and directional tunings (Fig. 4a and Extended Data Fig. 5d), indicating that iGABASnFR2 picks the local GABA signals that are separated at the synapses.
The second limitation of our study is that we performed imaging, and thus noise correlation, at single layers in the IPL. Thus, we might miss the noise correlation across layers, resulting in an overestimation of the number of cell types. Given our finding of multiplexed types, it is possible that GABA signal groups across different layers, which are currently assigned into different types, are involved in the same single cell, sharing noise correlation. Indeed, it is known that a glycinergic/glutamatergic amacrine cell type, the vGluT3 cells, has different response profiles across layers48. Cholinergic and GABAergic pre-synaptic inputs mediate the multiplexed direction selectivity in type 7 bipolar cells22. The subcellular compartmentalization, as shown in AII amacrine cell12 and type 6 bipolar cells49, would be a plausible mechanism underlying the multiplexity. On the other hand, common shared inputs to different amacrine cells from pre-synaptic bipolar cells could mediate noise correlation. The noise correlation among ROIs on the same single SAC was higher than ROIs on different SACs (Extended Data Fig. 5e). Given that SACs share common inputs from the same bipolar cells, this result indicates that such common inputs alone might not be enough to drive strong noise correlation. Based on the noise correlation mapping, the distances between coupled GABA signal ROIs ranged from 100–400 µm (Fig. 3). Considering that the axonal arbors of single bipolar cells extend 10–50 µm50, it is unlikely that a single bipolar cell alone can mediate the noise correlation. Future studies should employ volumetric imaging to capture responses at multiple depths near-simultaneously and yield reliable characterization of the full GABA signal diversity in the IPL. Studies combining functional imaging with post hoc molecular profiling would also help delineate specific cell types.
Methods
Animals
Wild-type mice (C57BL/6J) were obtained from Janvier Labs. ChAT-IRES-Cre (strain Chattm2(cre)Lowl/MwarJ; The Jackson Laboratory stock 028861) were used to label starburst cell processes. ChAT-IRES-Cre crossed with ROSA26-STOP-tdTomato (Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J; The Jackson Laboratory stock 007905) were used for GABA imaging. Oxtr-T2A-Cre (strain Cg-Oxtrtm1.1(cre)Hze/J; The Jackson Laboratory stock 031303) was used for glutamate imaging from direction-selective ganglion cell dendrites. The mice were purchased from The Jackson Laboratory and maintained in a C57BL/6J background. We used 8–16-week-old mice of either sex. Mice were group-housed throughout and maintained in a 12-h light–dark cycle with ad libitum access to food and water. There were no significant differences in the functional clustering among different retinal locations (temporal, dorsal, nasal and ventral), male or female, ages and individual mice (Supplementary Fig. 3). All animal experiments were performed according to standard ethical guidelines and were approved by the Danish National Animal Experiment Committee (permission no. 2015−15−0201−00541 and 2020-15-0201-00452) and the National Institute of Genetics (R3-23).
Retinal preparation
Retinae were isolated from the left eye of mice dark-adapted for 1 h before experiments. The isolated retina was mounted on a small piece of filter paper (MF-membrane, Millipore), in which a 2 × 2-mm window had been cut, with the ganglion cell side up. During the procedure, the retina was illuminated by dim red light (KL 1600 LED, Olympus) filtered with a 650 ± 45-nm bandpass optical filter (ET650/45×, Chroma) and bathed in Ringer’s medium (in mM): 110 NaCl, 2.5 KCl, 1 CaCl2, 1.6 MgCl2, 10 d-glucose and 22 NaHCO3 bubbled with 5% CO2 and 95% O2. The retina was kept at 35–36 °C and continuously superfused with oxygenated Ringer’s medium during recordings.
For pharmacological experiments, we used tetrodotoxin (1 μM, Tocris) to block Na+ channels by bath application (Extended Data Fig. 4), methyl lycaconitine (100 nM, Tocris) and hexamethonium (100 μM, Tocris) to block nicotinic receptors, and atropine (3 μM, Sigma) to block muscarinic receptors.
To visualize dendritic morphology, AlexaFluor 594 (Thermo Fisher) was added in intracellular solution (Extended Data Fig. 5) (in mM): 112.5 CsCH3SO3, 1 MgSO4, 7.8 × 10−3 CaCl2, 0.5 BAPTA, 10 HEPES, 4 ATP-Na2, 0.5 QX314-Br and 7.5 neurobiotin chloride. The solution was loaded through borosilicate glass micropipettes pulled by a micropipette puller (P-97, Sutter Instrument).
AAV production
Plasmids pGP-AAV-hSyn1-flex-iGABASnFR2 (var 610.4409) and pGP-AAV-hSyn1-iGABASnFR2-WPRE (var 609.4409) and the virus AAV2/9-hSyn1-iGABASnFR2-WPRE (var 609.4409) (3.21 × 1013 vg per ml) were designed and provided by the Genetically Encoded Indicator and Effector (GENIE) Project, Janelia Research Campus, HHMI. Virus was produced by the Janelia Viral Shared Resource Facility. A part of the AAVs used in this study were produced by the Zurich Viral Vector Core (ssAAV-9/2-hSyn1-dlox-iGABASnFR2(rev)-dlox-WPRE-SV40p(A) (1.9 × 1013 vg per ml) and ssAAV-9/2-hSyn1-iGABASnFR2(var 609.4409)-WPRE-SV40p(A) (1.4 × 1013 vg per ml)) based on the same plasmids provided by the GENIE project. The imaging datasets obtained by AAVs from the GENIE Project and the Zurich Viral Vector Core were pooled because there were no significant differences among the signals in the datasets. AAV9-hSyn1-Flex-SF-iGluSnFR.WPRE.SV40 for direction-selective ganglion cell imaging was obtained from Penn Vector Core (98931; 7.73 × 1013 GC per ml)51.
Viral injections
Mice were anesthetized with an i.p. injection of fentanyl (0.05 mg kg−1 body weight; Actavis), midazolam (5.0 mg kg−1 body weight; Dormicum, Roche) and medetomidine (0.5 mg kg−1 body weight; Domitor, Orion) mixture dissolved in saline. We made a small hole at the border between the sclera and the cornea with a 30-gauge needle. The AAV was delivered through a pulled borosilicate glass micropipette (30-µm tip diameter). All pressure injections were performed using a Picospritzer III (Parker) under a stereomicroscope (SZ61; Olympus). We pressure-injected 1–2 µl AAV into the vitreous of the left eye. Mice were returned to their home cage after anesthesia was antagonized by an i.p. injection of flumazenil (0.5 mg kg−1 body weight; Anexate, Roche) and atipamezole (2.5 mg kg−1 body weight; Antisedan, Orion Pharma) mixture dissolved in saline and, after recovering, were placed on a heating pad for 1 h.
Two-photon imaging
Three to four weeks after virus injection, we performed two-photon GABA and glutamate imaging. The isolated retina was placed under a microscope (SliceScope, Scientifica) equipped with a galvo-galvo scanning mirror system, a mode-locked Ti: Sapphire laser tuned to 940 nm (MaiTai DeepSee, Spectra-Physics; average excitation power, 20–30 mW), and an Olympus ×60 (1.0 NA) or Olympus ×25 (1.05 NA) objective, as described previously22. The microscope stage was manipulated by LinLab2 (Scientifica). The size of FOVs varied from 5,000 to 150,000 μm2. The retina was superfused with oxygenated Ringer’s medium. Emitted iGABASnFR2 or SF-iGluSnFR signals were passed through a set of optical filters (ET525/50m, Chroma; lp GG495, Schott) and collected with a GaAsP detector. Images were acquired at 8–12 Hz using custom software developed by Zoltan Raics (SENS Software). Temporal information about scan timings was recorded by transistor–transistor logic signals generated at the end of each scan, and the scan timing and visual stimulus timing were subsequently aligned during off-line analysis. We imaged GABA signals from different retinal areas (nasal, ventral, temporal, and dorsal parts) except for a very central part including the optic disc. We acquired GABA signals throughout the inner retinal layers during experiments, and the imaging depths were projected in the normalized IPL axis, which was computed based on the ON and OFF ChAT signals obtained by tdTomato signals (Extended Data Fig. 1d). The orientation of retinal samples did not affect the responses (Supplementary Fig. 1). We acquired glutamate signals from the genetically labeled dendrites of direction-selective ganglion cells22.
Visual stimulation
Visual stimulation was generated via custom-made software (Python and LabVIEW) developed by Zoltan Raics (SENS Software). For electrophysiological recordings, the stimulus was projected through a DLP projector (NP-V311X, NEC). The stimulus was focused on the photoreceptor layer of the mounted retina through a condenser (WI-DICD, Olympus). The intensity was measured using a photodiode power meter (Thorlabs), and the power of the spectrum was measured using a spectrometer (Ocean Optics). The calculated photoisomerization rate ranged from 0.0025–0.01 × 107 photons absorbed per rod per second (R* s−1) both for electrophysiological recordings and two-photon imaging. For two-photon imaging, the stimulus was projected using a DLP projector (LightCrafter Fiber E4500 MKII, EKB Technologies) coupled via a liquid light guide to an LED source (4-Wavelength High-Power LED Source, Thorlabs) with a 400 nm LED (LZ4-00UA00, LED Engin) through a bandpass optical filter (ET405/40×, Chroma). The stimuli were exclusively presented during the fly-back period of the horizontal scanning mirror18. The contrast of visual stimulus (\({C}_{\rm{s}}\)) was calculated as:
in which \({L}_{\rm{s}}\) and \({L}_{\rm{b}}\) indicate luminance intensity in stimulus and background, respectively.
Region of interest detection
ROIs for GABA signals were determined by customized programs in MATLAB. First, acquired individual images were spatially aligned based on spatial cross-correlograms. The stack of adjusted images was filtered with a Gaussian filter (3 × 3 pixels), and then each image was downsampled to 0.8 of the original using the MATLAB imresize function. The signal in each pixel was resampled using the MATLAB interp function with a rate of 2 and smoothed temporally by a moving average filter with a window size of 2× the signal time-bin. The parameters for the filter processing provided better signal-to-noise ratio across the ranges (window size in Gaussian filter, 1 × 1, 3 × 3, 5 × 5; scale factor for downsampling, 0.8, 0.6 and 0.4; time-bin of moving average, 1, 2, 4, 6 and 8) (Extended Data Fig. 1f–i). Next, we computed the temporal correlation among the pixels within 10 µm of each other located during static flash stimulus based on a raw cross-correlation (\({\rm{Crr}}_{p,q}^{\rm{r}}\)):
in which \({F}_{p}\) and \({F}_{q}\) indicate GABA signals during the term between \({T}_{i}\) and \({T}_{\!j}\) in pixel \(p\) and \(q\), respectively. The noise correlation (\({\rm{nc}}_{p,q}\)) was then given by a subtraction:
in which \({\rm{Crr}}^{\rm{s}}\) indicates trial-shuffled cross-correlation. The noise correlation was normalized as a score (\({\rm{NC}}_{p,q}\)):
in which 〈 〉 and \({\rm{Var}}\) [ ] indicate mean and variance, respectively. We set a threshold of the correlation score at 0 time-lag as 0.5 to determine which pixels were to be grouped as a single ROI. To exclude signal from somata of displaced cell types, we set a threshold for ROI size at <10 µm. The majority of ROI sizes were smaller than 3.6 µm (Extended Data Fig. 1e). Then the response of each ROI (\(\Delta F(t)\)) was determined as:
where \(F(t)\) is the fluorescence signal in arbitrary units, and \({F}_{0}\) is the baseline fluorescence measured as the average fluorescence in a 1-second window before stimulus presentation. After processing, responsive pixels were detected based on a response index (RI):
where \({R}^{i}\) is a peak response amplitude during motion stimulus to direction i in ROI \(P\), and \({r}_{P}^{\,i}\) indicates GABA signals before the stimulus (1 s period). ROIs with RI higher than 0.6 were deemed responsive.
To evaluate reliability in responses, we computed the response quality index (QI)7:
where C is a matrix constructed by response \(\Delta F(t)\) in all stimulus trials, and 〈 〉x and Var[ ]x denote the mean and variance across the indicated dimension \(x\). If all responses are identical in all stimulus trials, QI is equal to 1. Responses with QI > 0.6 were deemed reliable7 and used for the following analysis of response measures. The ROI locations were not spatially biased on individual FOVs (Supplementary Fig. 2).
Response measures
To evaluate sensitivity to luminance increments (ON) or decrements (OFF), we used static, flashing spots (300 µm in diameter, 2 s in duration, 100% positive contrast). To evaluate release kinetics, we used modulating spots7,18,23. The stimulus (300 µm in diameter) had four phases: static flashing spot of 100% contrast, one of 50% contrast, one with increasing temporal frequency from 0.5–8 Hz and one with increasing contrast from 5–80%. To quantify the sensitivity to luminance ON and OFF, we computed ON (OFF) RI:
where \({R}_{\rm{stim}}\) denotes response amplitudes in stimulus (luminance increments for ON and decrements for OFF) and \({R}_{\rm{base}}\) denotes baseline activity. To compute bi-response index, ON and OFF response amplitudes were used for calculation. To quantify the response transience, we computed transience index23,35:
where \({R}_{{\rm{plateau}}+{t}_{a}}\) denotes response amplitude \(a\) ms (400 ms) after the timing of peak response, \({R}_{\rm{plateau}}\). A cell showing sustained responses shows an index of 0. To quantify the response modulation by frequency and contrast, we calculated the mean response strength before the start of each phase (1 s) as the baseline strength, and the peak response amplitude during the modulating phases was divided by the baseline strength. The indices represent response modulation to high versus low temporal frequency and contrast7.
To measure directional tuning and motion speed preference, we used a spot (300 µm in diameter, 100% positive contrast) moving in eight directions (0–315°, ∆45°) at 150, 300, 800, 1,200 and 2,400 µm s−1. To quantify direction tuning, the direction-selective index (DSI) and preferred direction were defined as the length and angle of the sum of eight vectors divided by the sum of the lengths of eight vectors, respectively:
where \({\bf{v}}_{D}\) are response vectors in the motion direction \(D\) and \({\bf{r}}_{D}\) are length. The preferred direction was defined as the direction that elicited the maximum response, and the null direction was the opposite. The DSI ranged from 0 to 1, with 0 indicating a perfectly symmetrical response, and 1 indicating a response only in the preferred direction. To quantify response amplitudes, response to one of eight motion directions with the nearest distance from the preferred direction was measured. Response amplitudes to the null direction were measured as the response to the opposite direction. To quantify orientational tuning, the orientation-selective index (OSI) was defined as:
where \({R}_{\rm{P}}\) indicates response in the preferred axis, and \({R}_{\rm{O}}\) indicates response in the orthogonal axis. To precisely quantify the directional biases in motion responses, an angle of preferred direction (\(\theta\)) was defined by the vector sum:
where i denotes the motion direction and Ri denotes the response amplitude.
To evaluate network activity, ROIs with noise correlation higher than 0.5 during static light stimulus were assigned to the same assembly (Fig. 3). If the assigned ROIs had less than 80% overlap in their receptive fields, those ROIs were removed from the assembly. To evaluate the statistical significance in the network activity, we shuffled pairs of ROIs so that all ROI pairs were not from simultaneous recordings and computed noise correlation. We performed bootstrapping to compute P values by using the shuffled noise correlation. To compute the number of coincidences of ROIs on a FOV, first, we computed the noise correlation of each ROI pair on each FOV. Next, we counted the number of coincidences among the 49 groups and made a fraction matrix for individual FOVs. Then, the fraction matrix was averaged across different FOVs. We computed dominance index (DI), representing the extent to which each group was dominated by a congruent group (for example, G1 ROIs were coincident with G1 ROIs) (Fig. 3h). \(\rm{DI}\) for group i was computed as follows:
where \({\rm{freq}}_{{\rm{c}}^{i}}\) denotes the maximum frequency of the coincidences within the same group i, and \(\rm{{freq}}_{\left\langle c\right\rangle }\) denotes an expected frequency, assuming no bias exists among the groups. If ROIs of a group were perfectly coincident with their own group, the DI was 1.
To evaluate the subgroups in the response measures, we performed k-means clustering using MATLAB k-means function (Extended Data Fig. 3). For receptive field subgroups, as we did not have a priori group numbers, we calculated silhouette scores (\({\rm{SC}}_{i}\)) to estimate optimal cluster numbers52:
where \({a}_{i}\) is an average distance between i and all other points included in the same cluster. \({b}_{i}\) is the smallest average distance between i and all other points. A silhouette score of 1 indicates that all data points are perfectly clustered. For other response measures, we performed k-means clustering under the assumptions of group numbers. We computed \(\rm{DI}\) for response time course (Extended Data Fig. 3b), kinetics (Extended Data Fig. 3c), receptive field size (Extended Data Fig. 3e) and receptive field orientation bias (Extended Data Fig. 3g).
To examine the functional segregation of response properties among direction-selective groups, we performed multidimensional analysis (Fig. 4d). We first formed a decomposed response matrix, consisting of response transience, latency, receptive field size, direction selectivity, orientation selectivity and sensitivity to TTX (Extended Data Fig. 3b), by using PCA (Fig. 4d). We computed a modulation index as:
where \({r}_{\rm{ttx}}\) and \({r}_{\rm{c}}\) denote response amplitudes in control and after application of TTX. The sensitivity to TTX was statistically evaluated by comparisons of average response amplitudes between control and TTX conditions by using the Wilcoxon signed-rank test (Extended Data Fig. 3b). Next the response matrix pooled the ROIs of direction-selective groups (ROIs in G7, G10, G20, G31, G37, G38 and G45 groups) and starburst cell signals (ROIs in ON and OFF ChAT), and was projected onto the principal-component axes. The similarity between GABA signals in ON (or OFF) ChAT groups and TTX-sensitive DS groups (Fig. 4e) was quantified by an average of the similarity index:
where \({v}_{i}^{j}\) denotes an average feature i of a group j, and \({v}_{\rm{ON}}^{\;j}\) denotes an average feature i of ON ChAT group. To compute the similarity index for OFF ChAT, \({v}_{\rm{OFF}}^{\;j}\) was used.
To quantify the contrast sensitivity in responses, we applied static flashes with different contrast conditions (5, 10, 20, 40 and 80% contrasts, 2 s duration; Extended Data Fig. 3h–j). Based on linear regression for contrast response tuning curve in each group, we quantified two measurements: (1) a contrast sensitivity index:
where \({r}_{\mathrm{10,20}}\) and \({r}_{\rm{base}}\) denote mean responses to 10% and 20% contrasts and mean responses before visual stimulus application, respectively (a higher index of an ROI indicates that it is activated by the lower contrast); and (2) a nonlinearity index, residual sum between observed tuning curves (Extended Data Fig. 3i) and a fitted linear model (Extended Data Fig. 3i). Higher index of a tuning curve reflects nonlinear modulation. Furthermore, we divided each group into two contrast sensitivity groups (higher versus lower) and two nonlinearity groups (linear versus nonlinear) by k-means clustering with a cluster number of 2 (Extended Data Fig. 3i). We computed the DI to examine whether each group was dominated by a single contrast sensitivity (high threshold versus low threshold) and nonlinearity (nonlinear versus linear) (Extended Data Fig. 3k).
Screening of motion direction selectivity
To statistically evaluate direction/orientation selectivity by the statistical methods, we first performed a shuffling test for directional tuning22. For individual ROIs, shuffled tunings (\({T}_{\rm{s}}\)) were generated by random shuffling among eight directions with noise following a normal distribution:
where \({T}_{\rm{s}}^{\,d}\) denotes the generated tuning value to direction \(d\), and \(n\) denotes a noise value taken from a normal distribution with the same mean (\({M}_{\rm{r}}\)) and s.d. (\({\rm{SD}}_{\rm{r}}\)) as the raw original tunings. To estimate a false-positive probability (\({P}_{{\rm{False}}\; {\rm{Positive}}}\)), we tested whether the shuffled DSI and OSI were higher than the original indexes with bootstrapping (20,000 replications). ROIs with \({P}_{{\rm{False}}\; {\rm{Positive}}}\) < 0.05 were identified as directionally or orientationally tuned ROIs.
Clustering
Clustering of GABA signals was based on the temporal kinetics of responses to the modulating spot, as described previously7,18,23. We used sparse PCA to extract temporal features in response to a modulating flash using the SpaSM toolbox on MATLAB53. Next, we fitted a Gaussian mixture model based on the expectation-maximization algorithm using the MATLAB gmdistribution function to fit the dataset of detected sparse features. To determine the optimal number of clusters in the model, we calculated the Bayesian information criterion score54:
in which \(L\) is the log-likelihood of the model, \(k\) is the number of dimensions in the model and \(n\) is the number of datasets. To separate ON, OFF and ON–OFF input groups, we first performed clustering using responses to a static flash (the first phase of the modulating spot), then we repeated the clustering for the dissected individual groups using responses to the entire stimulus phase of the modulating spot. Clusters were further segregated into subclusters based on motion direction selectivity (direction/orientation-selective groups). If individual clusters had significant variance (for example, kinetics and response measurements), they were further divided manually into subgroups by k-means. The variance within the clusters was evaluated by a shuffling test. We sorted detected clusters by similarity calculated with hierarchical clustering analysis using a standard linkage algorithm in the MATLAB linkage function. The similarity was represented by ON RI, OFF RI, bi-response index, transience index and response latency (Fig. 1d). After the clustering, we computed the layer DI for individual assigned groups (Fig. 2c) as:
where \({\rm{freq}}_{{\rm{L}}^{i}}\) denotes maximum frequency at the layer i and \({\rm{{freq}}}_{\left\langle {\rm{L}}\right\rangle }\) denotes an expected frequency assuming no bias exists among the layers. A DI of 1 indicates a dominance of signals in a specific layer depth.
Visual encoding
To characterize the visual encoding by individual GABA signal groups, the six visual features were used: direction selectivity, orientation selectivity, motion/flash preference, speed modulation, response modulation by contrast and response modulation by temporal frequency (Extended Data Fig. 6a–c). The four measurements, motion/flash preference, speed modulation, contrast modulation and temporal frequency modulation, were computed as indexes:
where \({f}_{{\rm{C}}^{\max }}\) and \({f}_{{\rm{C}}^{\min }}\) denote responses in a stimulus condition inducing maximum and minimum responses, respectively. We obtained barcodes of the six features for each GABA signal group and performed PCA to decompose the feature weights (Extended Data Fig. 6d–f). The significance of the weights was evaluated by shuffling test. The information weights in each GABA signal group were sorted by an information score (Hotelling’s T2 score) to denote variations in measures within individual groups. To evaluate the statistical significance of the Hotelling T2 score, we performed bootstrapping by using shuffled datasets. To evaluate DS and OS, significantly positive values were adopted. In the cell type characterization (Supplementary Table 1), we labeled DS and OS types based on the result of shuffling test, even if the Hotelling’s T2 score was not significant.
Mapping projective fields
To estimate the spatial representation of GABA release from individual amacrine cells, we mapped projective fields (Extended Data Fig. 7). First, we mapped ROIs onto the space relative to the receptive field center while registering retinal orientations (Extended Data Fig. 7a). This ROI remapping was performed for ROIs assigned to each GABA signal group (Extended Data Fig. 7b). Based on the remapped ROI distribution, we computed a density map and convex hull representing the spatial projective field. We also computed orientational angles of ROI locations relative to the receptive field center. The directional bias of the ROI angles was computed from a vector sum of the polar histogram. To quantify angular tuning along the cardinal axes, we separately generated density maps for temporal, dorsal, nasal, and ventral areas, respectively (Extended Data Fig. 7d). The angles and distances from the center provided angular tunings along the cardinal axes. To compare receptive field and projective field profiles, we defined the SCI as:
in which \({D}_{\rm{PF}}\) and \({D}_{\rm{RF}}\) denote the size of the projective field and receptive field, respectively. An overlap index was computed as the fraction of the projective field area included in the receptive field.
Modeling of amacrine cell modulation
To simulate spatiotemporal filtering by GABAergic amacrine cells, we modeled synaptic transmission from amacrine cells to post-synaptic cells (bipolar cells) during visual stimulation. The response time course in each circuit component was represented by a spatiotemporal receptive field, estimated from a dense noise stimulus (Extended Data Fig. 8). The dense noise was constructed from black and white pixels (40 µm in length, 50 × 50 matrix; 15 µm in length, 50 × 50 matrix for bipolar cells), each flickering randomly at 20 Hz (ref. 18):
in which \(F\left(x,y,\tau \right)\) is the receptive field at a location \(\left(x,y\right)\) at delay \(\tau\), \(r(t)\) is the response to dense noise, and \(S(x,y,t+\tau )\) is the stimulus input at the location. For amacrine cell models, an average receptive field was computed for individual GABA signal groups. For the bipolar cell model, an average receptive field was computed using the dataset of glutamate imaging from ON–OFF direction-selective ganglion cell dendrites. The model response was described by the spatiotemporal convolution of the stimulus input18,55:
in which \(r\left(x,y,\tau \right)\) is the model response at location \((x,y)\), \(s(x,y,t-\tau )\) is the stimulus input, and \(F(x,y,\tau )\) is the receptive field. The decay in outputs of the linear filter was modulated by an exponential function with double-decay constants18. The stimulus input was a square (300 × 300 µm) moving at 50, 100, 200, 400, 800, 1,600, 2,400 or 3,200 µm s−1.
Based on the mapped projective fields (Fig. 6), the release sites where the post-synaptic bipolar cell receives GABA (bipolar cell receptive field) were displaced from receptive fields of amacrine cells (Extended Data Fig. 8a). The modeled bipolar cell responses were modulated by amacrine cell inputs under the given spatiotemporal profiles of the moving stimulus (Extended Data Fig. 8c,d). The modulation efficacy was quantified as the difference between the original post-synaptic response and the response after amacrine cell modulation.
Mapping to molecularly identified amacrine cell types
The raw gene counts matrix and associated metadata from Yan et. al.11 were downloaded from the Gene Expression Omnibus (GSE149715). The counts matrix was loaded into R (R v.4.2.1 GUI 1.79)56 and a Seurat object created with CreateSeuratObject() from the Seurat package (Seurat v.4.3.0.1; https://CRAN.R-project.org/package=SeuratObject)57. Cluster Identities were assigned from the Yan et. al. metadata file ‘MouseAC_metafile.csv’ using the Idents() function. Cluster AC_16 was removed due to the presence of cell doublets before normalization with ‘logNormalize’. Cells within each cluster were then filtered for expression of the following GABA gene markers, where expression is defined as a normalized value ≥0.5: GABA-synthesizing enzymes (Gad1 or Gad2), vesicular GABA transporter (Slc32a1), GABA receptors (any one of Gabbr1, Gabbr2, Gabra1, Gabra2, Gabra3, Gabra4, Gabra5, Gabrb1, Gabrb2, Gabrb3, Gabrg1, Gabrg2, Gabrg3, Gabrd, Gabre, Gabrq, Gabrr1, Gabrr or Gabrr3). Expression of genes encoding TTX-sensitive voltage-gated sodium channel subunits in the filtered clusters was assessed by the RidgePlot() function and Scn1a and Scn3a chosen for further analysis. Hierarchical clustering of the amacrine cell clusters was performed using the Euclidian distance metric and the complete linkage clustering method using the Clustered_DotPlot() function of scCustomize (scCustomize v.1.1.2; https://doi.org/10.5281/zenodo.5706430, RRID:SCR_024675)58 to obtain a dendrogram for identification of the top TTX-sensitive clusters. The cutoff for top clusters based on the dendrogram was 60% of cells within the cluster expressing either Scan1a or Scn3a at an average scaled normalized expression of ≥1.5. Two clusters, AC_30 and AC_36 met the expression but not the cell percentage cutoff (20%) but were included because of their location within the dendrogram of other top TTX-sensitive clusters. TTX-sensitive clusters coexpressing acetylcholine receptors were identified by re-clustering of clusters and features based on expression of the receptors (Chrna2, Chrna3, Chrna4, Chrna5, Chrna6, Chrnb2, Chrnb3 and Chrm1) using the cutoff of ≥50% of cells expressing any of these genes at an average scaled normalized expression ≥1.5. Hierarchical clustering of the top TTX-sensitive clusters was then performed using the Euclidian distance metric and the complete linkage clustering method with the Seurat DotPlot() function and ggplot2 (ggplot2 v.3.4.2)59 customization based on genes of interest, which included the TTX-sensitive voltage-gated sodium channel subunits identified previously in the mouse retina: Scn1a, Scn3a; acetylcholine receptors: Chrm1, Chrna2, Chrna3, Chrna4, Chrna5, Chrna6, Chrnb2 and Chrnb3; glutamate receptors: Grm1, Grm2, Grm3, Grm4, Grm5, Grm7, Grm8, Gria1, Gria2, Gria3, Gria4, Grid2, Grin1, Grin2a and Grin2b; glycine receptors: Glra1, Glra2, Glra3 and Glrb; and GABA receptors: Gabbr1, Gabbr2, Gabra1, Gabra2, Gabra3, Gabrb1, Gabrb3, Gabrg2, Gabrg3 and Gabrd. Marker genes for specific amacrine cell clusters of interest were also included in the dot plot but not used for clustering (Sst, Penk, Nos1, Crh, Cck and Chat).
Statistics and reproducibility
Statistical tests were not conducted to predetermine sample sizes. Data distribution was assumed to be normal, but this was not formally tested. All experiments were replicated more than three times independently. Our experimental results and analysis were consistent across animals, retinal locations, ages and either sex (Supplementary Fig. 3). For all experiments, mice were randomly allocated. Experimenters were blind for the sex. Imaging data processing, analysis and clustering analysis were performed blind to the experimental conditions. Data collection was not carried out blind to experimental conditions. The data that did not meet the criteria of response validation were excluded.
All analyses and statistical tests were performed in MATLAB 2017b (MathWorks) unless otherwise noted. Population data are shown as mean ± s.d. The box in the box-and-whisker plots marks the median and 25th and 75th percentiles. The whiskers are set to 1.5 × interquartile range. To compare the differences in paired conditions, a Wilcoxon signed-rank test was used. To compare the differences in different groups, a Mann–Whitney–Wilcoxon test and Kruskal–Wallis test were used. The variance within groups was evaluated by a shuffling test. The Bonferroni correction was used when multiple comparisons were performed. No statistical tests were performed to predetermine sample sizes, but the sample sizes in this study were similar or larger than those in previous publications6,13,23,25,26.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All relevant data are available from GitHub at https://github.com/AkiatNIG/Matsumoto_et_al_GABA2024 and Zenodo at https://doi.org/10.5281/zenodo.14854477 (ref. 60). Source data are provided with this paper.
Code availability
Our custom scripts and relevant datasets are available from GitHub at https://github.com/AkiatNIG/Matsumoto_et_al_GABA2024 and Zenodo at https://doi.org/10.5281/zenodo.14854477(ref. 60).
References
Tremblay, R., Lee, S. & Rudy, B. GABAergic interneurons in the neocortex: from cellular properties to circuits. Neuron 91, 260–292 (2016).
Tang, X., Jaenisch, R. & Sur, M. The role of GABAergic signalling in neurodevelopmental disorders. Nat. Rev. Neurosci. 22, 290–307 (2021).
Jiménez-Balado, J. & Eich, T. S. GABAergic dysfunction, neural network hyperactivity and memory impairments in human aging and Alzheimer’s disease. Semin. Cell Dev. Biol. 116, 146–159 (2021).
Brambilla, P., Perez, J., Barale, F., Schettini, G. & Soares, J. C. GABAergic dysfunction in mood disorders. Mol. Psychiatry 8, 721–37, 715 (2003).
Levy, L. M. & Hallett, M. Impaired brain GABA in focal dystonia. Ann. Neurol. 51, 93–101 (2002).
Yonehara, K. et al. Congenital nystagmus gene FRMD7 is necessary for establishing a neuronal circuit asymmetry for direction selectivity. Neuron 89, 177–193 (2016).
Baden, T. et al. The functional diversity of retinal ganglion cells in the mouse. Nature 529, 345–350 (2016).
Werblin, F. S. The retinal hypercircuit: a repeating synaptic interactive motif underlying visual function. J. Physiol. 589, 3691–3702 (2011).
Helmstaedter, M. et al. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500, 168–174 (2013).
Diamond, J. S. Inhibitory interneurons in the retina: types, circuitry, and function. Annu. Rev. Vis. Sci. 3, 1–24 (2017).
Yan, W. et al. Mouse retinal cell atlas: molecular identification of over sixty amacrine cell types. J. Neurosci. 40, 5177–5195 (2020).
Grimes, W. N., Zhang, J., Graydon, C. W., Kachar, B. & Diamond, J. S. Retinal parallel processors: more than 100 independent microcircuits operate within a single interneuron. Neuron 65, 873–885 (2010).
Vlasits, A. L. et al. A role for synaptic input distribution in a dendritic computation of motion direction in the retina. Neuron 89, 1317–1330 (2016).
Ding, H., Smith, R. G., Poleg-Polsky, A., Diamond, J. S. & Briggman, K. L. Species-specific wiring for direction selectivity in the mammalian retina. Nature 535, 105–110 (2016).
Euler, T., Detwiler, P. B. & Denk, W. Directionally selective calcium signals in dendrites of starburst amacrine cells. Nature 418, 845–852 (2002).
Tasic, B. et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346 (2016).
Kolb, I. et al. iGABASnFR2: Improved genetically encoded protein sensors of GABA. Preprint at bioRxiv https://doi.org/10.1101/2025.03.25.644953 (2025).
Matsumoto, A., Briggman, K. L. & Yonehara, K. Spatiotemporally asymmetric excitation supports mammalian retinal motion sensitivity. Curr. Biol. 29, 3277–3288.e5 (2019).
Lin, B. & Masland, R. H. Populations of wide-field amacrine cells in the mouse retina. J. Comp. Neurol. 499, 797–809 (2006).
Camillo, D., Ahmadlou, M. & Heimel, J. A. Contrast-dependence of temporal frequency tuning in mouse V1. Front. Neurosci. 14, 868 (2020).
Masland, R. H. The tasks of amacrine cells. Vis. Neurosci. 29, 3–9 (2012).
Matsumoto, A. et al. Direction selectivity in retinal bipolar cell axon terminals. Neuron 109, 2928–2942.e8 (2021).
Franke, K. et al. Inhibition decorrelates visual feature representations in the inner retina. Nature 542, 439–444 (2017).
Liang, L. et al. A fine-scale functional logic to convergence from retina to thalamus. Cell 173, 1343–1355.e24 (2018).
Sabbah, S. et al. A retinal code for motion along the gravitational and body axes. Nature 546, 492–497 (2017).
Sethuramanujam, S. et al. Rapid multi-directed cholinergic transmission in the central nervous system. Nat. Commun. 12, 1374 (2021).
Kim, Y. J. et al. Origins of direction selectivity in the primate retina. Nat. Commun. 13, 2862 (2022).
Poleg-Polsky, A., Ding, H. & Diamond, J. S. Functional compartmentalization within starburst amacrine cell dendrites in the retina. Cell Rep. 22, 2898–2908 (2018).
Asari, H. & Meister, M. The projective field of retinal bipolar cells and its modulation by visual context. Neuron 81, 641–652 (2014).
Badea, T. C. & Nathans, J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J. Comp. Neurol. 480, 331–351 (2004).
Zhu, Y., Xu, J., Hauswirth, W. W. & DeVries, S. H. Genetically targeted binary labeling of retinal neurons. J. Neurosci. 34, 7845–7861 (2014).
Knop, G. C., Feigenspan, A., Weiler, R. & Dedek, K. Inputs underlying the ON-OFF light responses of type 2 wide-field amacrine cells in TH::GFP mice. J. Neurosci. 31, 4780–4791 (2011).
Kerstein, P. C., Leffler, J., Sivyer, B., Taylor, W. R. & Wright, K. M. Gbx2 identifies two amacrine cell subtypes with distinct molecular, morphological, and physiological properties. Cell Rep. 33, 108382 (2020).
Kim, J. S. et al. Space-time wiring specificity supports direction selectivity in the retina. Nature 509, 331–336 (2014).
Srivastava, P. et al. Spatiotemporal properties of glutamate input support direction selectivity in the dendrites of retinal starburst amacrine cells. eLife 11, e81533 (2022).
Murphy-Baum, B. L. & Taylor, W. R. The synaptic and morphological basis of orientation selectivity in a polyaxonal amacrine cell of the rabbit retina. J. Neurosci. 35, 13336–13350 (2015).
Zemon, V., Gordon, J. & Welch, J. Asymmetries in ON and OFF visual pathways of humans revealed using contrast-evoked cortical potentials. Vis. Neurosci. 1, 145–150 (1988).
Luo-Li, G., Mazade, R., Zaidi, Q., Alonso, J.-M. & Freeman, A. W. Motion changes response balance between ON and OFF visual pathways. Commun. Biol. 1, 60 (2018).
Chakravarthi, R., Papadaki, D. & Krajnik, J. Visual field asymmetries in numerosity processing. Atten. Percept. Psychophys. 84, 2607–2622 (2022).
Sakatani, T. & Isa, T. Quantitative analysis of spontaneous saccade-like rapid eye movements in C57BL/6 mice. Neurosci. Res. 58, 324–331 (2007).
Meyer, A. F., O’Keefe, J. & Poort, J. Two distinct types of eye-head coupling in freely moving mice. Curr. Biol. 30, 2116–2130.e6 (2020).
Roska, B. & Werblin, F. Rapid global shifts in natural scenes block spiking in specific ganglion cell types. Nat. Neurosci. 6, 600–608 (2003).
Rasmussen, R., Matsumoto, A., Dahlstrup Sietam, M. & Yonehara, K. A segregated cortical stream for retinal direction selectivity. Nat. Commun. 11, 831 (2020).
Hillier, D. et al. Causal evidence for retina-dependent and -independent visual motion computations in mouse cortex. Nat. Neurosci. 20, 960–968 (2017).
Olveczky, B. P., Baccus, S. A. & Meister, M. Segregation of object and background motion in the retina. Nature 423, 401–408 (2003).
Marvin, J. S. et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods 16, 763–770 (2019).
Dowling, J. E., Boycott, B. B. & Wells, G. P. Organization of the primate retina: electron microscopy. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 166, 80–111 (1997).
Chen, M., Lee, S. & Zhou, Z. J. Local synaptic integration enables ON-OFF asymmetric and layer-specific visual information processing in vGluT3 amacrine cell dendrites. Proc. Natl Acad. Sci. USA 114, 11518–11523 (2017).
Swygart, D., Yu, W.-Q., Takeuchi, S., Wong, R. O. L. & Schwartz, G. W. A presynaptic source drives differing levels of surround suppression in two mouse retinal ganglion cell types. Nat. Commun. 15, 599 (2024).
Tsukamoto, Y. & Omi, N. Classification of mouse retinal bipolar cells: type-specific connectivity with special reference to rod-driven aii amacrine pathways. Front. Neuroanat. 11, 92 (2017).
Marvin, J. S. et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10, 162–170 (2013).
Bos, R., Gainer, C. & Feller, M. B. Role for visual experience in the development of direction-selective circuits. Curr. Biol. 26, 1367–1375 (2016).
Zou, H., Hastie, T. & Tibshirani, R. Sparse principal component analysis. J. Comput. Graph. Stat. 15, 265–286 (2006).
Fraley, C. & Raftery, A. E. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 97, 611–631 (2002).
Baccus, S. A., Olveczky, B. P., Manu, M. & Meister, M. A retinal circuit that computes object motion. J. Neurosci. 28, 6807–6817 (2008).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2022).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Wickham, H. Ggplot2 (Springer, 2016).
Matsumoto, A. et al. AkiatNIG/Matsumoto_et_al_GABA2024: Functional clustering of GABA signal. Zenodo https://doi.org/10.5281/zenodo.14854477 (2025).
Wallace, D. J. et al. Rats maintain an overhead binocular field at the expense of constant fusion. Nature 498, 65–69 (2013).
Martinez-Conde, S. & Macknik, S. L. Fixational eye movements across vertebrates: comparative dynamics, physiology, and perception. J. Vis. 8, 28.1–16 (2008).
Michaiel, A. M., Abe, E. T. & Niell, C. M. Dynamics of gaze control during prey capture in freely moving mice. eLife 9, e57458 (2020).
Payne, H. L. & Raymond, J. L. Magnetic eye tracking in mice. eLife 6, e29222 (2017).
Harrod, C. G. & Baker, J. F. The vestibulo ocular reflex (VOR) in otoconia deficient head tilt (het) mutant mice versus wild type C57BL/6 mice. Brain Res. 972, 75–83 (2003).
Migliaccio, A. A., Meierhofer, R. & Della Santina, C. C. Characterization of the 3D angular vestibulo-ocular reflex in C57BL6 mice. Exp. Brain Res. 210, 489–501 (2011).
Acknowledgements
We thank Z. Raics for developing our visual stimulation and imaging system and B. Thomsen, M. Yonehara, C. Thiesen and E. Helga for technical assistance. iGABASnFR2 plasmids and AAVs were a gift from the GENIE Project, Janelia Research Campus, Howard Hughes Medical Institute. We also thank L. Anson for English proofreading and providing comments on the paper. This work was supported by grants from VELUX FONDEN Postdoctoral Ophthalmology Research Fellowship (27786), KAKENHI (23K19412), JST PRESTO (JPMJPR2489), The Food Science Institute Foundation (Ryoushoku-kenkyukai) (2023A12) and Narishige Neuroscience Research Foundation to A.M.; and the Lundbeck Foundation (DANDRITE-R248-2016-2518; R344-2020-300), Novo Nordisk Foundation (NNF15OC0017252; NNF20OC0064395), European Research Council Starting Grant (638730), KAKENHI (20K23377; 22K21353; 23H04241; 24H02311), Chugai Foundation for Innovative Drug Discovery Science, Daiichi Sankyo Foundation of Life Science, Mochida Memorial Foundation for Medical and Pharmaceutical Research, Mitsubishi Foundation, Toray Science Foundation and Naito Foundation to K.Y.
Author information
Authors and Affiliations
Contributions
A.M. and K.Y. conceived and designed the experiments and analyses. A.M. performed all experiments and analyzed all physiology data. L.L.L. and J.M. analyzed gene expression data and edited and advised on the paper. A.M. and K.Y. interpreted the data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Laura Busse, Gregory Schwartz, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Two-photon GABA imaging in the inner retina.
(a) Schematic of retinal neurons. Different amacrine cell types (cyan) stratify their processes in the IPL. (b and c) ChAT (b) and iGABASnFRs2 (c) signals in the inner retina. (d) Left, ChAT signal intensity in IPL. Based on the distance between ON (L6) and OFF (L4) peaks of ChAT signal, IPL was divided into nine layers (L1–L9). Since L1 and L9 approach the somatic layers (INL and GCL, respectively), we used seven layers (L2-L8) for analysis. Right, histogram denoting locations of imaging planes relative to ON (depth = 0) and OFF (depth = 1) ChAT depths (circles, individual recordings) and fraction of the 49 groups at each depth (cyan line). 177 imaging planes, 11 retinae. (e) Histogram of ROI size and a fitted exponential curve to estimate a length constant (red line). (f) Light stimulus-evoked GABA signal before (“raw”) after Gaussian filter processing (1 × 1, 3 × 3, 5 × 5 pixels). 529 ROIs in a retina. (g) Changes of signal-to-noise ratio after Gaussian filtering (1 × 1, 3 × 3, 5 × 5 pixels) and down-sampling (scale factors, 0.8, 0.6, 0.4) from raw signal. Signal-to-noise ratio was computed as the difference in response amplitudes compared with signal variance before stimulation. *, parameters giving the best signal-to-noise ratio. (h) Light stimulus-evoked GABA signal before (“raw”) and after moving average filter processing (2, 4, 6 time-bin). 529 ROIs in a retina. (i) Changes of signal-to-noise ratio after moving average processing from raw signal (529 ROIs, 1 retina). Error bars, s.e.m.
Extended Data Fig. 2 Light stimulus-induced GABA signal.
(a and b) Light-evoked GABA signals in response to modulating spot (a) and motion stimuli (b). Left, locations of regions of interest (ROI; yellow) on each imaging field of-view (FOV). Gray and black lines, each trial and an average. Right radar plots on (b), directional tuning curves with a vector sum (arrow). (c) Relationship between response magnitudes in the first and last trials (left) and example FOVs with/without light stimulus (right). 7098 ROIs, 11 retinae. (d) Left, estimated receptive field of an example ROI. Right, receptive fields (cyan) of two example ROIs (coded by magenta pixels) imaged on a FOV. (e) GABA signals in different IPL depths (L2-L8) during modulating spots. 689 ROIs in a retina. (f) Left and right, sorted by determined GABA signal groups (G1-G49) and example signals (average and SD, black and gray, respectively). 689 ROIs in a retina. Error bars, s.e.m.
Extended Data Fig. 3 Statistical determination of response groups.
(a) Histograms of transience index for delayed-ON, ON, ON–OFF, and OFF types. Based on k-means clustering with a given cluster number (2 clusters), ROIs were grouped into two kinetics groups: sustained (smaller transience index) and transient. After labeling kinetics for each ROI, we computed a dominance of the response kinetics for each group and statistically examined if each group was dominated by a single kinetics type (“Slow” or “Fast”). If a group was not dominated by a single kinetics type, the group was defined as “Undefined”. In the end, all groups were separated into three categories: sustained, transient, and undefined. The same clustering procedures were performed for response latency. 689 ROIs in a retina. (b and c) Population of response-determined kinetics groups for time course (b) and transience (c). (d) Histogram of receptive field sizes of GABA signals. Based on k-means clustering and silhouette score, we determined four receptive field types: small, small-medium (s-medium), large-medium (l-medium), and large. 7098 ROIs, 11 retinae. (e) Top, population of receptive field categories in individual GABA signal groups. Bottom, dominance of receptive field categories. +, groups significantly dominated by a single receptive field category. (f) Histogram of orientation bias index (OBI) in individual receptive fields. Inset, two example receptive fields with different OBI. Based on k-means clustering, ROIs were labeled as not biased (darker blue) or biased (light blue). (g) Relative frequency of orientation bias labels in those GABA signal groups. Colors are denoted in (f). (h) Responses to static flashes with different contrasts (2 s duration, 500 μm spot) in example ROIs. (i) Relationship between contrast sensitivity and non-linearity in contrast response tuning curves. Circles represent the average for each group (color-coded 49 groups). Individual groups were divided into 2 contrast sensitivity (higher vs lower) and two non-linearity (linear vs non-linear) by k-means clustering with cluster number, 2 (Extended Data Fig. 2i, dotted lines). (j) Contrast tuning curves of groups denoted in (h). G28, linear type. G33, non-linear type. G14, lower contrast sensitive type. (k) Left, distribution of contrast sensitivity categories (gray, high threshold; dark gray, low threshold) (top) and dominance of each category (bottom). +, groups significantly dominated by a single category. Right, distribution of non-linearity categories (gray, linear; dark gray, linear). Error bars, s.e.m.
Extended Data Fig. 4 Characterization of DS groups.
(a) Changes of light-evoked responses of example ROIs in control (gray) and TTX application (orange). (b) Modulation index denoting changes of responses by TTX application. Purples and blues, direction- and orientation-selective groups. 7098 ROIs, 11 retinae. (c) Left, schematic of targeted expressions of iGABASnFR2 in starburst cells (SACs; green). Middle, field of view for imaging from SAC processes in ChAT-IRES-Cre mice. Right, light-evoked responses in example ROIs (#1 and #2 in left) and an average response of group G10. (d) Comparisons of response kinetics, receptive field size, DSI, OSI, and sensitivity to NaV block in untargeted imaging (gray; 7098 ROIs, 11 retinae) and SAC -targeted imaging from ON ChAT layer depth (red; 88 ROIs, 4 retinae). (e) Comparisons of response measures between G10 (orange; 189 ROIs, 11 retinae) and ON ChAT layer imaging (red; 88 ROIs, 4 retinae). (f) Comparisons of response measures between G37 (blue; 113 ROIs, 11 retinae) and OFF ChAT layer imaging (red; 62 ROIs, 4 retinae). (g) Left, correlation of SAC signal obtained by targeted imaging at ON ChAT layer depth with G10 (with the highest correlation; dark gray; 189 ROIs, 11 retinae), with G12 (second best match; gray; 139 ROIs, 11 retinae, P = 9.19 \(\times\) 10-34, Kruskal–Wallis test, Bonferroni correction for multiple comparisons), and with all other groups (light gray; P = 2.33 \(\times\) 10-44). Comparison of receptive field size between G10 and G12 (P < 0.00001, Z = 9.56, Two-sided Mann–Whitney-Wilcoxon test). Right, correlation of targeted imaging at OFF ChAT layer depth with G37 (with the highest correlation; dark gray; 113 ROIs, 11 retinae), with G42 (second best match; gray; 137 ROIs, 11 retinae, P = 5.05 \(\times\) 10-36, Kruskal–Wallis test, Bonferroni correction for multiple comparisons), and with all other groups (light gray, P = 1.68 \(\times\) 10-46). Comparison of receptive field size between G37 and G42 (P < 0.00001, Z = 14.75, Two-sided Mann–Whitney-Wilcoxon test). Although the second best match groups (G12, G42) were also highly correlated with GABA signal in ChAT layer depths, the receptive field size did not correspond to those of SACs. (h) Frequency of SACs (G10, G37) and the newly identified DS groups. (i) Multidimensional features projected along the principal axes. ROIs in each group were denoted by different colors (dots). Horizontal and vertical lines, averages of each group in the first and second PCA axes. 133 G7, 189 G10, 104 G20, 109 G31, 113 G37, 129 G38, and 141 G45 ROIs, 11 retinae. 88 ON ChAT, 62 OFF ChAT, 4 retinae. (j) Top, example motion responses of a G20 ROI in control (gray) and AChR block (black; 100 nM methyllycaconitine and 100 μM hexamethonium for nicotinic receptors, 3 μM atropine for muscarinic receptors) conditions. Bottom, changes in direction-selectivity index during AChR block. 133 G7, 189 G10, 104 G20, 109 G31, 113 G37, 129 G38, and 141 G45 ROIs, 11 retinae. (k) Hierarchical clustering of NaV (Scn1a, Scn3a) expressions in amacrine cell molecular groups. Boxes represent 25 th–75th percentiles and median; whiskers represent 1.5 times the interquartile range; error bars, s.e.m.
Extended Data Fig. 5 ROIs involved in the same single cell share high intrinsic noise correlation.
(a) Schematic of intrinsic noise correlation. Suppose that there are four ROIs, with groups of two belonging to different cells (left). The response variances across trials are shared among ROIs from the same neurons (right; for example, ROI#1 and #3). (b) Left, iGABASnFR2 was expressed selectively in SACs (green). The dendritic processes of a single SAC were visualized by AlexaFluor 594 loaded through a patch pipette (magenta). Dotted gray circle, cell body of the patched cell. Right, example ROIs for GABA imaging from specified (magenta) and unspecified (green) dendritic processes. (c) A correlation matrix of the 16 ROIs in (b). ROIs on a single SAC (ROIs #1-#6) had similar noise correlation. (d) Left, comparisons of variance in response profiles between ROIs within the same groups and shuffled ROIs (49 groups, P = 1.11 \(\times\) 10-9, Wilcoxon signed-rank test). Right, response variances computed in ROIs on a single SAC (light gray; 82 pairs) and ROIs on unspecified SAC processes (dark gray; 28 pairs) in ChAT-IRES-Cre mice. Since the analysis of GABA signals was restricted to iGABASnFR2 expressed in the membrane of SACs, the difference in variance between groups was low. Two-sided Mann–Whitney-Wilcoxon test, Bonferroni correction for multiple comparisons. (e) Comparisons of noise correlation between ROIs on the same (“Same neurites”; 57 pairs, P = 0.030381, Two-sided Mann–Whitney-Wilcoxon test) and different neurites (“Different neurites”; 11 pairs) of a single SAC, ROIs on neurites of unspecified cells with unspecified preferred direction (Multiple SACs; 142 pairs, P = 1.38 \(\times\) 10-6, Two-sided Mann–Whitney-Wilcoxon test), ROIs on neurites of unspecified cells with same preferred direction (Multiple SACs, same pref. dir.; 20 pairs). 3 retinae. Different neurites of the same SAC showed lower correlation than ROIs from the same neurites, but still showed higher correlation than neurites from different SACs. (f) A map denoting heterogeneous correlations in G2, G5, G12, G19, G21, G35, G42, and G47. Gray, correlations with more than 0.25 noise correlation (noise corr.). Colored, significantly coincident connections. (g) Heatmaps indicating distribution of observed G2, G5 (delayed-ON), G12, G19, G21 (ON), and G35, G42, G47 (OFF) groups. Yellow squares, average distribution for delayed-ON, ON, and OFF groups. (h) A summary of characteristics of groups with heterogeneous connectivity. *, P < .05; *** P < .001; boxes represent 25 th–75th percentiles and median; whiskers represent 1.5 times the interquartile range.
Extended Data Fig. 6 Statistical characterization of visual encodings in GABA response groups.
(a-c) Schematics for characterization of visual encodings. The six visual features were used to represent visual encodings in each group (a). We computed measures for the visual features for each ROI and summarized those as barcodes. Then we decomposed the barcodes of 49 groups by principal component analysis (PCA) and obtained feature weights and Hotelling’s T2 score for each group (c). (d) A response matrix consists of responsibility for the six features. 4149 ROIs, 6 retinae. (e) Bars, explained variance ratio for the obtained PC dimensions. Circles, cumulative explained variance. (f) PC scores in each PC dimension. Circles, 49 groups. Colored circles, groups identified as significantly informative based on dataset shuffling. PC dimensions with the highest PC scores are marked.
Extended Data Fig. 7 Spatial mapping of release sites relative to receptive field location.
(a) Left, receptive fields (purples) of four ROIs (cyan). Middle, receptive field and each ROI location (cyan dots). Left, four ROI locations are mapped relative to individual receptive field centers. The analysis were replicated 11 times independently. (b) Protocols of projective field estimation for each response group. The pooled ROI locations in a response group were mapped relative to receptive field centers (left; 220 ROIs, 17 retinae). The ROI location map was converted into a density map (middle left). T, D, N, and V denote temporal, dorsal, nasal, and ventral directions, respectively. Projective field orientation was quantified based on a histogram of angles of ROI locations relative to receptive field centers (middle right). Convex hulls of receptive field and projective field were used to compute size change index and overlap index (right). (c) Relationship between directional bias and orientational bias. Each circle represents a response group. k-means clustering provided four clusters: group#1 (orange; directional#1), group#2 (blue; directional#2), group#3 (yellow; orientational), and nonbiased (not shown). The two directional groups were integrated. (d) Protocols to measure angular tunings along the cardinal axes. First, based on receptive field length, ROI locations in a response group were normalized relative to the receptive field center. Next, ROI locations were divided into four parts: temporal, dorsal, nasal, and ventral, and ROI locations for each part were converted into density maps. Angles and distances from the center provided angular tunings along the cardinal axes (red arrows in right polar plot). (e) Glutamatergic signals on direction-selective ganglion cell dendrites measure by two-photon imaging at the depth of ON ChAT. AAV encoding SF-iGluSnFR was injected into the eyes of Oxtr-T2A-Cre mice to target posterior-tuned direction-selective cells. Based on the response kinetics, we identified 12 glutamatergic signal groups: 5 ON, 5 OFF, and 2 glutamatergic amacrine cell groups. Colored circles, location of glutamatergic signals of the groups (39 ROIs in a retina). (f) Properties of projective field of example bipolar cell groups: orientation of release sites relative to receptive field, and convex hulls of receptive field (purple) and projective field (cyan). 178 bipolar cell G1, 238 bipolar cell G2 ROIs, 8 retinae. (g) Relationship between receptive field size and overlap index. Gray, TTX-insensitive; pink, inhibited by TTX; purple, disinhibited by TTX; cyan circles, direction-selective groups. (h) Comparison of overlap index between TTX-insensitive (35 groups) and TTX-sensitive (14 groups) groups. P = 0.016339. Two-sided Mann–Whitney-Wilcoxon test. (i) Comparisons of directional tunings (fraction of preferred direction) in motion responses (gray) and projective fields for direction-selective groups (Fig. 4b). (j) Schematic of lateral inhibition in bipolar cell (BC) mediated by a displaced projective field of amacrine cell (AC). In the global stimulus, AC receives excitatory inputs from BC#2 and inhibits BC#1 laterally. *, P < .05; boxes represent 25 th–75th percentiles and median; whiskers represent 1.5 times the interquartile range.
Extended Data Fig. 8 Diverse spatiotemporal filtering by GABA signal groups.
(a) Schematic of synaptic transmission from amacrine cell (dark gray) to post-synaptic cell (light gray) during motion stimuli. (b) Spatiotemporal integration model to simulate light-evoked responses in individual GABA signal groups. The spatiotemporal profile of each visual stimulus is convolved with the spatial and temporal receptive field in each response group. (c) G14, G6, and G32 amacrine cell (cyan) and bipolar cell (black) models during motion stimuli of different speeds. (d) Top, modulatory efficacy of bipolar cell response by amacrine cell. Bottom, relationship between motion speed and modulatory efficacy in three example groups (G14, G6, and G32). (e) Matrix of motion speed tuning and modulatory efficacy for 49 groups (left) and matrix sorted by optimal speed (right). (f) Top, clustering based on speed tuning (converted from µm/s to °/s). Bottom, plausible functional relevance for speed tuning. Numbers indicate reference number (in g). (g) References for the plausible range of speed tunings in (f). Wallace et al.61, Martinez-Conde and Macknik62, Michaiel et al.63, Payne and Raymond64, Sakatani and Isa40, Meyer et al.41, Harrod and Baker65, Migliaccio et al.66.
Extended Data Fig. 9 Parameters to determine delays in the synaptic transmission from amacrine cell to bipolar cells in motion stimulus.
The response timing of individual amacrine cell types is affected by the response kinetics and receptive field size of each cell type (a), displacement of release sites (b), and motion speeds (c). The net response time course was simulated by the kernel convolution (Extended Data Fig. 8b) in motion stimulus for each amacrine cell type. The bipolar cell model (b) was based on the spatiotemporal receptive fields estimated by the glutamate imaging from direction-selective ganglion cell dendrites (Methods). The bipolar cell receptive field (BC receptive field) was placed at the same location as amacrine cell release site.
Extended Data Fig. 10 Potential mechanisms to generate direction selectivity in amacrine cells.
(a) Left, an amacrine cell (DS amacrine cell; black) may receive inputs from SACs (dark gray) and other non-direction-selective (non-DS) amacrine cells (light gray). Right, this DS amacrine cell would receive stronger inhibition along the preferred direction (PD) of SAC processes, while non-DS amacrine cells would provide symmetric inhibition (light gray). (b) Left, schematic of potential spatiotemporal wiring between bipolar cells and an amacrine cell. At each dendritic segment, the amacrine cell may receive sustained (“sust.”) inputs from the proximal bipolar cell type (dark gray) and transient (“trans.”) inputs from the distal bipolar cell type (light gray). Right, time courses of excitatory inputs at the dendritic segment [1] (cyan) during centrifugal (left) and centripetal (right) motion. The excitatory inputs would be summated in centrifugal motion, but not in centripetal motion, resulting in direction-selective activity at the release site.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Tables 1 and 2.
Source data
Source Data
Source data for Figs. 1–6 and Extended Data Figs. 1–10.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Matsumoto, A., Morris, J., Looger, L.L. et al. Functionally distinct GABAergic amacrine cell types regulate spatiotemporal encoding in the mouse retina. Nat Neurosci 28, 1256–1267 (2025). https://doi.org/10.1038/s41593-025-01935-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-025-01935-0
This article is cited by
-
Functional diversity of amacrine cells
Nature Reviews Neuroscience (2025)