Abstract
Centrosomes ensure accurate chromosome segregation during cell division. Although the regulation of centrosome number is well established, less is known about the suppression of noncentrosomal microtubule-organizing centers (ncMTOCs). The E3 ligase TRIM37, implicated in Mulibrey nanism and 17q23-amplified cancers, has emerged as a key regulator of both centrosomes and ncMTOCs. Yet, the mechanism by which TRIM37 achieves enzymatic activation to target these mesoscale structures had thus far remained unknown. Here we elucidate the activation process of TRIM37, unveiling a process that initiates with TRAF domain-directed substrate recognition followed by B-box domain-mediated oligomerization and culminates in RING domain dimerization. Using optogenetics, we demonstrate that the E3 activity of TRIM37 is directly coupled to the assembly state of its substrates, being activated only when centrosomal proteins cluster into higher-order assemblies resembling MTOCs. This regulatory framework provides a mechanistic basis for understanding TRIM37-driven pathologies and echoes the restriction of the human immunodeficiency virus capsid by TRIM5, thus unveiling a conserved activation blueprint among TRIM proteins to control turnover of complexes assembled at the mesoscale level.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The BioID MS proteomics data were deposited to the ProteomeXchange Consortium through the PRIDE partner repository under accession number PXD061083. Source data are provided with this paper.
References
Conduit, P. T., Wainman, A. & Raff, J. W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16, 611–624 (2015).
Gould, R. R. & Borisy, G. G. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J. Cell Biol. 73, 601–615 (1977).
Schnackenberg, B. J., Khodjakov, A., Rieder, C. L. & Palazzo, R. E. The disassembly and reassembly of functional centrosomes in vitro. Proc. Natl Acad. Sci. USA 95, 9295–9300 (1998).
Mennella, V., Agard, D. A., Huang, B. & Pelletier, L. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 24, 188–197 (2014).
Woodruff, J. B., Wueseke, O. & Hyman, A. A.Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130459 (2014).
Nigg, E. A. & Holland, A. J. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19, 297–312 (2018).
Godinho, S. A. & Pellman, D.Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130467 (2014).
Phan, T. P. & Holland, A. J. Time is of the essence: the molecular mechanisms of primary microcephaly. Genes Dev. 35, 1551–1578 (2021).
Sanchez, A. D. & Feldman, J. L. Microtubule-organizing centers: from the centrosome to non-centrosomal sites. Curr. Opin. Cell Biol. 44, 93–101 (2017).
Li, J. et al. Neurl4, a novel daughter centriole protein, prevents formation of ectopic microtubule organizing centres. EMBO Rep. 13, 547–553 (2012).
Shiratsuchi, G., Takaoka, K., Ashikawa, T., Hamada, H. & Kitagawa, D. RBM14 prevents assembly of centriolar protein complexes and maintains mitotic spindle integrity. EMBO J. 34, 97–114 (2015).
Ozato, K., Shin, D. M., Chang, T. H. & Morse, H. C. 3rdTRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8, 849–860 (2008).
Avela, K. et al. Gene encoding a new RING–B-box–coiled-coil protein is mutated in Mulibrey nanism. Nat. Genet. 25, 298–301 (2000).
Kallijarvi, J., Avela, K., Lipsanen-Nyman, M., Ulmanen, I. & Lehesjoki, A. E. The TRIM37 gene encodes a peroxisomal RING–B-box–coiled-coil protein: classification of Mulibrey nanism as a new peroxisomal disorder. Am. J. Hum. Genet 70, 1215–1228 (2002).
Kettunen, K. M. et al. Trim37-deficient mice recapitulate several features of the multi-organ disorder Mulibrey nanism. Biol. Open 5, 584–595 (2016).
Balestra, F. R., Strnad, P., Fluckiger, I. & Gonczy, P. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev. Cell 25, 555–571 (2013).
Dominguez-Calvo, A., Gonczy, P., Holland, A. J. & Balestra, F. R. TRIM37: a critical orchestrator of centrosome function. Cell Cycle 20, 2443–2451 (2021).
Meitinger, F. et al. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 214, 155–166 (2016).
Meitinger, F. et al. TRIM37 prevents formation of condensate-organized ectopic spindle poles to ensure mitotic fidelity. J. Cell Biol. 220, e202010180 (2021).
Balestra, F. R. et al. TRIM37 prevents formation of centriolar protein assemblies by regulating Centrobin. eLife 10, e62640 (2021).
Andersen, C. L. et al. High-throughput copy number analysis of 17q23 in 3520 tissue specimens by fluorescence in situ hybridization to tissue microarrays. Am. J. Pathol. 161, 73–79 (2002).
Lastowska, M. et al. Gain of chromosome arm 17q predicts unfavourable outcome in neuroblastoma patients. Eur. J. Cancer 33, 1627–1633 (1997).
Sinclair, C. S., Rowley, M., Naderi, A. & Couch, F. J. The 17q23 amplicon and breast cancer. Breast Cancer Res. Treat. 78, 313–322 (2003).
Yeow, Z. Y. et al. Targeting TRIM37-driven centrosome dysfunction in 17q23-amplified breast cancer. Nature 585, 447–452 (2020).
Meitinger, F. et al. TRIM37 controls cancer-specific vulnerability to PLK4 inhibition. Nature 585, 440–446 (2020).
Meroni, G. & Diez-Roux, G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27, 1147–1157 (2005).
Hamalainen, R. H. et al. Novel mutations in the TRIM37 gene in Mulibrey nanism. Hum. Mutat. 23, 522 (2004).
Hamalainen, R. H. et al. Wilms’ tumor and novel TRIM37 mutations in an Australian patient with Mulibrey nanism. Clin. Genet. 70, 473–479 (2006).
Ganser-Pornillos, B. K. & Pornillos, O. Restriction of HIV-1 and other retroviruses by TRIM5. Nat. Rev. Microbiol. 17, 546–556 (2019).
Sanchez, J. G. et al. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc. Natl Acad. Sci. USA 111, 2494–2499 (2014).
Goldstone, D. C. et al. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc. Natl Acad. Sci. USA 111, 9609–9614 (2014).
Li, Y. L. et al. Primate TRIM5 proteins form hexagonal nets on HIV-1 capsids. eLife 5, e16269 (2016).
Yudina, Z. et al. RING dimerization links higher-order assembly of TRIM5α to synthesis of K63-linked polyubiquitin. Cell Rep. 12, 788–797 (2015).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Gheiratmand, L. et al. Spatial and proteomic profiling reveals centrosome-independent features of centriolar satellites. EMBO J. 38, e101109 (2019).
Gupta, G. D. et al. A dynamic protein interaction landscape of the human centrosome–cilium interface. Cell 163, 1484–1499 (2015).
Diaz-Griffero, F. et al. A B-box 2 surface patch important for TRIM5α self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83, 10737–10751 (2009).
Wagner, J. M. et al. Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5α. eLife 5, e16309 (2016).
Massiah, M. A. Zinc-binding B-box domains with RING folds serve critical roles in the protein ubiquitination pathways in plants and animals. In Ubiquitin Proteasome System (ed. Summers, M.) (IntechOpen, 2019).
Reymond, A. et al. The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 (2001).
Li, X. & Sodroski, J. The TRIM5α B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 82, 11495–11502 (2008).
Fletcher, A. J. et al. Trivalent RING assembly on retroviral capsids activates TRIM5 ubiquitination and innate immune signaling. Cell Host Microbe 24, 761–775 (2018).
Zeng, J. et al. Target-induced clustering activates Trim-Away of pathogens and proteins. Nat. Struct. Mol. Biol. 28, 278–289 (2021).
Park, H. et al. Optogenetic protein clustering through fluorescent protein tagging and extension of CRY2. Nat. Commun. 8, 30 (2017).
Bellaart, A. et al. TRIM37 prevents ectopic spindle pole assembly by peptide motif recognition and substrate-dependent oligomerization. Nat. Struct. Mol. Biol. (in the press).
Kiss, L. et al. Trim-Away ubiquitinates and degrades lysine-less and N-terminally acetylated substrates. Nat. Commun. 14, 2160 (2023).
Fu, J. & Glover, D. M. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2, 120104 (2012).
Lawo, S., Hasegan, M., Gupta, G. D. & Pelletier, L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14, 1148–1158 (2012).
Mennella, V. et al. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14, 1159–1168 (2012).
Sonnen, K. F., Schermelleh, L., Leonhardt, H. & Nigg, E. A. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open 1, 965–976 (2012).
Foss, S. et al. TRIM21—from intracellular immunity to therapy. Front. Immunol. 10, 2049 (2019).
Zhang, Z. Y. et al. TRIM11 protects against tauopathies and is down-regulated in Alzheimer’s disease. Science 381, eadd6696 (2023).
Brinkman, E. K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Ghetti, S. et al. CRISPR/Cas9 ribonucleoprotein-mediated knockin generation in hTERT-RPE1 cells. STAR Protoc. 2, 100407 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52, D368–D375 (2023).
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.10.04.463034 (2022).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Firat-Karalar, E. N. & Stearns, T. Probing mammalian centrosome structure using BioID proximity-dependent biotinylation. Methods Cell. Biol. 129, 153–170 (2015).
Mellacheruvu, D. et al. The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat. Methods 10, 730–736 (2013).
Thomas, P. D. et al. PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22 (2022).
Mi, H. et al. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721 (2019).
Knight, J. D. R. et al. ProHits-viz: a suite of web tools for visualizing interaction proteomics data. Nat. Methods 14, 645–646 (2017).
Shukla, A., Kong, D., Sharma, M., Magidson, V. & Loncarek, J. Plk1 relieves centriole block to reduplication by promoting daughter centriole maturation. Nat. Commun. 6, 8077 (2015).
Acknowledgements
This work was supported by research grants R01GM114119, R01GM133897 and R01CA266199 (to A.J.H) from the National Institutes of Health. M.v.B. was supported by the Queen Mary University of London and by the Biotechnology and Biological Sciences Research Council (BB/X013030/1). We thank M. R. Leung and N. K. Sinha for providing valuable experimental insights.
Author information
Authors and Affiliations
Contributions
Z.Y.Y. designed, performed and analyzed the majority of the experiments and prepared the figures. S.S., F.-C.C. and L.Y.X. assisted with the cloning and immunofluorescence analyses. M.v.B. performed the experiments to identify TRIM37-binding regions within Centrobin. Z.Y.Y. and A.J.H. conceptualized the study. A.J.H. supervised the study. Z.Y.Y. and A.J.H. cowrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Effect of TRIM37 mutations on the regulation of the centrosome and Centrobin assemblies.
Related to Fig. 1. (a) Immunoblot showing TRIM37 protein levels in parental and CRISPR–Cas9 edited RPE-1 TRIM37−/− cells. Ponceau-stained blot indicates loading. Representative data; n = 3 biological replicates. (b) Left, Tracking of Indels by Decomposition (TIDE) analysis histogram reveals a one base pair insertion ( + 1 bp) adjacent to the predicted cut site in the RPE-1 TRIM37−/− cell line. Right, representative Sanger sequencing traces used for TIDE analysis, highlighting the +1 bp insertion. (c) Representative images of RPE-1 TRIM37−/− cells and those expressing the indicated HA-tagged TRIM37 variants. Inset #1 denotes the centrosome, marked by CEP192, and inset #2 denotes the Centrobin assembly, identified by intense Centrobin staining that is non-centrosome localized. Representative data; n = 3 biological replicates. Scale bars, 5 μm. (d) Schematic representation of TRIM37 HA-tagged domain-specific deletion constructs compared to full-length (FL) protein. (e) Immunoblot showing expression levels of FL TRIM37 and the respective deletion mutants in RPE-1 tet-on TRIM37 cells. Ponceau-stained blot indicates loading. Representative data; n = 3 biological replicates.
Extended Data Fig. 2 Characterization of higher molecular weight (HMW) TRIM37 species.
Related to Fig. 4. (a) Immunoblot showing expression of WT TRIM37 and indicated mutants in RPE-1 tet-on TRIM37 cells. HMW TRIM37 species are prominently formed in the C18R mutant and indicated with an arrow. β-Actin, loading control. Representative data; n = 3 biological replicates. (b) Same as in (a) but with MG132 (10 μM) treatment to achieve proteasomal inhibition and stabilization of WT TRIM37. β-Actin, loading control. Representative data; n = 3 biological replicates. (c) Top, immunoblot showing detection of HMW TRIM37 species with increasing concentrations of DSS crosslinker. Vinculin is the loading and oligomerization control. Dotted lines indicate separate cropped sections of the same immunoblot. Representative data; n = 3 biological replicates. Bottom, Densitometric analysis of normalized HMW TRIM37 intensity upon increasing DSS concentrations relative to DMSO control ( − DSS). Mean ± s.e.m. (d) Sanger sequencing traces of the TRIM37 locus in parental and CRISPR–Cas9 edited RPE-1 TRIM37C18R cells, highlighting the mutation (TGT > CGT) responsible for the biallelic C18R residue substitution, denoted by an asterisk. (e) Top, immunoblot showing endogenous TRIM37 protein levels across the indicated cellular fractions. Validation markers include CEP192, Centrobin, and SAS6 for centrosomal proteins, and Lamin A/C for the nuclear fraction. Ponceau-stained blot indicates loading. Representative data; n = 3 biological replicates. WCE, whole-cell extract; exp, exposure. Bottom, Densitometric analysis of endogenous TRIM37 enrichment in indicated fractions relative to WCE. P values, one-way ANOVA with post hoc Dunnett’s multiple comparisons test to evaluate enrichment of TRIM37 in each cellular fraction relative to WCE. Mean ± s.e.m. (f) Top, immunoblot showing detection of various HMW species of endogenous TRIM37 upon treatment with increasing concentrations of DSG crosslinker. Vinculin is the loading and oligomerization control. Representative data; n = 3 biological replicates. Bottom, Densitometric analysis of normalized HMW TRIM37 intensity upon increasing DSG concentrations relative to DMSO control ( − DSG). Mean ± s.e.m.
Extended Data Fig. 3 Defining the minimal TRIM37 domain architecture required for centrosome regulation.
a) Schematic of the miniTRIM37 (RBCC-TRAF) construct compared to full-length TRIM37. (b) Representative images of the localization and effect of indicated HA-tagged TRIM37 constructs on centrosomal CEP192 levels in RPE-1 tet-on TRIM37 cells. Arrows indicate the positions of centrosomes. Representative data; n = 3 biological replicates. Scale bars, 5 μm. (c) Quantification of centrosomal CEP192 signal upon doxycycline-induced expression of indicated HA-tagged TRIM37 constructs in RPE-1 tet-on TRIM37 cells from (b). n = 3 biological replicates, each with >90 cells. P values, one-way ANOVA with post hoc Tukey’s multiple comparisons test. Mean ± s.e.m. (d) Immunoblot showing total protein levels of indicated HA-tagged TRIM37 constructs in RPE-1 tet-on TRIM37 cells from (b-c). GAPDH, loading control. Representative data; n = 3 biological replicates. (e) Immunoblot showing detection of various higher molecular weight (HMW) species of miniTRIM37 upon treatment with increasing concentrations of DSG crosslinker. Vinculin is used as a loading and oligomerization control. Representative data; n = 3 biological replicates.
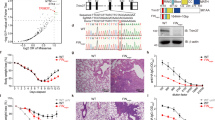
Extended Data Fig. 4 Impairment of TRIM37 oligomerization attenuates synthetic lethality with PLK4 inhibition in 17q23-amplified cells.
(a) Immunoblot showing TRIM37 protein levels in TP53−/− MCF-7 cells. TRIM37 wild-type (WT), TRIM37 knockdown (KD) via shRNA, and cells harboring the C109S mutation in approximately half of the TRIM37 alleles present (TRIM37C109S) were used. Vinculin, loading control. Representative data; n = 3 biological replicates. (b) Left, Representative data of a 10-d clonogenic survival of indicated MCF-7 cell lines from (a) treated with DMSO (control) or PLK4 inhibitor (PLK4i) (250 nM). Right, Quantification of relative growth in the presence PLK4i relative to DMSO. P values, one-way ANOVA with post hoc Dunnett’s multiple comparisons test to evaluate differences between each experimental condition (KD and C109S) and WT. Mean ± s.e.m. (c) Quantification of mitotic CEP192 foci in PLK4i-treated TP53−/− MCF-7 cells that lack centrosomes. n = 3, biological replicates, each comprising >30 cells. P values, one-way ANOVA with post hoc Dunnett’s multiple comparisons test to evaluate differences between each experimental condition (KD and C109S) and WT. Mean ± s.e.m. (d) Representative images for (c). Scale bars, 5 μm. (e) Representative Sanger sequencing traces for the TRIM37 locus in parental TP53−/− MCF-7 cells subjected to TRIM37 knockdown (KD) via shRNA, and CRISPR–Cas9 edited TRIM37C109S KI cells. The mutation (TGT > TCT) leading to the C109S residue substitution is denoted by an asterisk. Silent mutations introduced to prevent re-editing are highlighted.
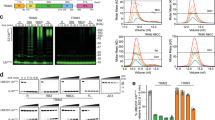
Extended Data Fig. 5 Substrate-independent clustering is sufficient to activate TRIM37.
Related to Fig. 7. (a) Top, schematic of the TRIM37G322V-mNeonGreen-CRY2 optogenetic fusion construct. The star denotes the TRAF domain mutation (G322V). Bottom, illustration of the blue light (BL)- activated optogenetic system enabling TRIM37 clustering independent of binding to a centrosome substrate. (b) Representative time-lapse images of RPE-1 cells expressing the optogenetic construct detailed in (a) incubated in the presence or absence of MG132. Timestamps indicate minutes post blue light exposure. Scale bar = 10 µm. (c) Quantification of mNeonGreen fluorescence intensity from (b), with each condition comprising >30 cells. Mean ± s.d. (d) RPE-1 cells expressing optogenetic constructs detailed in (a) were incubated with or without doxycycline (Dox) and MG132 (10 μM) in the absence or presence blue light for 3 h before immunoblotting for the indicated proteins. Higher molecular weight (HMW) TRIM37 species were prominently formed only in MG132 and BL-stimulated conditions and are indicated with an arrow. Ponceau-stained blot indicates loading. Representative data; n = 3 biological replicates. exp, exposure.
Supplementary information
Supplementary Video 1 (related to Extended Data Fig. 5)
Time-lapse of an RPE-1 cell expressing the optogenetic TRIM37 G322V–mNG–CRY2 construct, with blue-light pulses (470-nm filter) applied during imaging intervals to induce and sustain CRY2 clustering and to visualize TRIM37–mNG dynamics. Timestamps indicate minutes from the initial blue-light exposure.
Supplementary Video 2 (related to Extended Data Fig. 5)
Time-lapse of an RPE-1 cell expressing the optogenetic TRIM37 G322V–mNG–CRY2 construct, with blue-light pulses (470-nm filter) applied during imaging intervals to induce and sustain CRY2 clustering and to visualize TRIM37–mNG dynamics. Cells were incubated with MG132 1 h before blue-light exposure. Timestamps indicate minutes from the initial blue-light exposure.
Supplementary Video 3 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2 construct, incubated with doxycycline but in the absence of blue light. Timestamps indicate minutes from the first imaged frame. mCherry–CRY2 is displayed in grayscale.
Supplementary Video 4 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2 construct, subjected to blue-light pulses at each imaging interval, but incubated without doxycycline. Timestamps indicate minutes from the initial blue-light exposure. mCherry–CRY2 is displayed in grayscale.
Supplementary Video 5 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2 construct, incubated with doxycycline and subjected to blue-light pulses at each imaging interval. Timestamps indicate minutes from the initial blue-light exposure. mCherry–CRY2 is displayed in grayscale.
Supplementary Video 6 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2–Centrobin567–836 construct, incubated with doxycycline but in the absence blue light. Timestamps indicate minutes from the first imaged frame. mCherry–CRY2–Centrobin567–837 is displayed in grayscale.
Supplementary Video 7 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2–Centrobin567–836 construct, subjected to blue-light pulses at each imaging interval but incubated without doxycycline. Timestamps indicate minutes after initial blue-light exposure. mCherry–CRY2–Centrobin567–836 is displayed in grayscale.
Supplementary Video 8 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2–Centrobin567–836 construct, incubated with doxycycline and subjected to blue-light pulses at each imaging interval. Timestamps indicate minutes from the initial blue-light exposure. mCherry–CRY2–Centrobin567–836 is displayed in grayscale.
Supplementary Video 9 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2–Centrobin567–836 construct and TRIM37–mNG WT subjected to blue-light pulses at each imaging interval. Timestamps indicate minutes from the initial blue-light exposure. mCherry–CRY2–Centrobin567–836 is displayed in magenta and TRIM37–mNG is displayed in green.
Supplementary Video 10 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2–Centrobin567–836 construct and TRIM37–mNG WT subjected to blue-light pulses at each imaging interval. Cells were incubated with MG132 1 h before blue-light exposure. Timestamps indicate minutes from the initial blue-light exposure. mCherry–CRY2–Centrobin567–836 is displayed in magenta and TRIM37–mNG is displayed in green.
Supplementary Video 11 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2–Centrobin567–836 construct and TRIM37–mNG C18R subjected to blue-light pulses at each imaging interval. Timestamps indicate minutes from the initial blue-light exposure. mCherry–CRY2–Centrobin567–836 is displayed in magenta and TRIM37–mNG is displayed in green.
Supplementary Video 12 (related to Fig. 7)
Time-lapse of an RPE-1 TRIM37−/− cell expressing the optogenetic mCherry–CRY2–Centrobin567–836 construct and TRIM37–mNG G322V subjected to blue-light pulses at each imaging interval. Timestamps indicate minutes from the initial blue-light exposure. mCherry–CRY2–Centrobin567–836 is displayed in magenta and TRIM37–mNG is displayed in green.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots and/or gels.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots and/or gels.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yeow, Z.Y., Sarju, S., Chang, FC. et al. Mesoscale regulation of microtubule-organizing centers by the E3 ligase TRIM37. Nat Struct Mol Biol 32, 1787–1799 (2025). https://doi.org/10.1038/s41594-025-01540-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01540-6
This article is cited by
-
TRIMming centrosomal assemblies
Nature Structural & Molecular Biology (2025)