Abstract
Retrons have been recently identified as bacterial defense systems that employ a tripartite of reverse transcriptase, non-coding RNA (ncRNA) and its derived multi-copy single stranded DNA (msDNA) to sequester effector activity. Phage invasion activates retrons, triggering effector activity and inducing abortive infection and cell growth arrest. Ec78 differs from other retrons by leveraging the Septu defense system, a stand-alone ATPase–nuclease pair (PtuAB), by reshaping the phage sensing and molecular assembly processes of PtuAB. To elucidate how Ec78 hijacks PtuAB, we determined electron cryomicroscopy structures of Ec78 as well as the retron-displaced PtuAB. We show that the Ec78-associated ATPase, PtuA, acquired unique elements that enable its interactions with the reverse transcriptase and the msDNA, and self-assembly when displaced by the retron. By biochemical and mutational analyses, we also show that the retron-displaced PtuAB forms a tetramer, unlike its stand-alone counterpart, that restricts the host. However, in the presence of the retron, the retron-displaced PtuAB confers a well-controlled immune response, eliciting ATP hydrolysis- and msDNA-regulated targeting to host factors. Our studies reveal an evolutionary principle for retrons to co-opt conserved enzyme modules for defense in response to different cellular needs.
Similar content being viewed by others
Main
Once considered selfish genetic elements1, the widely distributed bacterial genetic elements known as retrons have recently been shown to play a role in anti-phage defense2,3. Retrons protect the bacterial hosts from invading phages through a broad spectrum of effector proteins that include membrane proteins, DNA-binding proteins, metabolic enzymes, ATPases and nucleases3. Investigating how retrons and their effectors contribute to this collaborative process under different selection pressures would reveal their evolutionary capacity and better serve the retron-based biotechnology applications.
Retrons encode a reverse transcriptase (RT) and a non-coding RNA (ncRNA, also referred to as msrRNA) that serves both as the template and the primer for synthesizing multi-copy single-stranded DNA (msDNA, also referred to as RT-DNA) by the RT, leading to a tripartite system comprised of RT, ncRNA and msDNA4. The msDNA is covalently linked to the initiating guanine nucleotide downstream of the conserved UUA motif of the ncRNA via a 2′–5′ branched linkage. In some cases, the host XseA-XseB exonuclease (ExoVII) separates the msDNA from the ncRNA prior to the retron-mediated defense mechanism is activated4,5. Furthermore, the template region of the ncRNA is removed by the host RNase H, a process required for the retron-mediated defense6, leaving behind a short template–primer duplex with the msDNA. The mature RT–ncRNA–msDNA tripartite sequesters and neutralizes the cytotoxic activities of the effectors, which are often encoded as polycistrons along with retrons themselves3,4,7. Once activated by phage infections, effector activities lead to abortive infection and cell death. Like CRISPR–Cas, retron systems provide bacteria with another non-coding-RNA-mediated defense against invading phages8. The unique production of msDNA by retrons has been harnessed for genome engineering as well as prime-editing, in conjunction with CRISPR–Cas9, across diverse systems9,10.
Retron Ec78 (retron-Eco7) is a type I-A retron. Of the 11 known types, these retrons are unique in partnering with the stand-alone Septu defense system, which is an aggregate of PtuA and PtuB proteins (collectively PtuAB)2,3,11. The recruitment of PtuAB by Ec78 or other type I-A retrons suggests that Septu- and Ec78-mediated defenses are regulated differently. Indeed, unlike Septu, which is sensitized by the phage tail fiber12, Ec78 produces msDNA, consistent with its response to DNA-targeting proteins13. Despite this difference, the Septu- and the Ec78-associated PtuAB share a high degree of similarity. Both PtuA proteins belong to the family of ABC ATPases14, whereas both PtuB proteins share a conserved HNH domain characteristic of the ββα-Me (metal) superfamily of nucleases15. The ABC ATPase domain connects PtuA to the overcoming lysogenization defect (OLD) nucleases, which have a role in anti-phage defense, and the Mr11–Rad50 proteins, involved in DNA repair14. Like the recently characterized OLD nuclease complex GajA–GajB, whose nuclease function is inhibited by ATP16,17, stand-alone PtuAB also exhibits reduced DNA-nicking activity with increasing ATP levels18. However, the dependence of Ec78-associated PtuA on ATP remains unknown and its PtuAB targets host tRNATyr (ref. 19). The distinct host substrates might explain in part why the stand-alone PtuAB targets a broad spectrum of phages11,12, whereas the Ec78-associated PtuAB is specific for the T5 phage12. The molecular basis underlying the defense function of the Ec78 retron remains unclear. Importantly, the evolutionary process by which PtuAB was co-opted into the more complex retron system under the control of the RT, the ncRNA and the msDNA tripartite assembly is elusive.
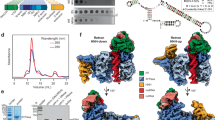
To elucidate the evolutionary basis for retrons to acquire stand-alone defense systems, we performed cryogenic electron cryomicroscopy (cryo-EM) and biochemical analysis on the recombinantly expressed Escherichia coli ECONIH5 retron Ec78 (Fig. 1a,b and Extended Data Fig. 1). Ec78 encodes an unprocessed ncRNA of 144 nucleotides (nt) in length, including 8-nt flanking inverted repeats (IRa1 and IRa2) and four RNA stem loops (stem loops I–IV) of varying sizes. The largest stem loop, stem loop IV, with a 22-base-paired stem, serves as the primary template for msDNA synthesis (Fig. 1a), after which stem loop IV is removed by RNase H. This leaves behind the ncRNA without stem loop IV, a short template–primer duplex, and the msDNA (Fig. 1a). We showed that the Ec78 ncRNA is further processed on retron, leaving behind 40 nt without stem loop I and the two inverted repeats. In addition, the msDNA is debranched, enabling PtuAB to bind. We also showed that, unlike the stand-alone PtuAB, the PtuAB heterocomplex is the cytotoxic unit of the Ec78 retron, requiring both the ATPase and HNH activities. We identified the structural elements acquired by PtuAB that enables its switch from the stand-alone to the retron-based defense, thereby illuminating the adaptive process for the Ec78 retron.
a, Key features of the Ec78 retron components. Top, amino acid residues of the motifs in the RT, PtuA and PtuB relevant to the study are listed. Mutated residues are shown in red. Bottom, secondary structures of the ncRNA before and after processing and msDNA. Key regions are colored and labeled. msr, multicopy single-stranded RNA; msd, multicopy single-stranded DNA. NET refers to the N-terminal extension of PtuA. CTE denotes the C-terminal extension of PtuA. b, Cophylogenetic analysis of retron RTs and their effectors. Retron types are color-coded and labeled. The effectors containing the HNH, TOPRIM or ATPase domains are color-coded and labeled. Some known retrons are marked. c, Plaque formation from the E. coli 11303 cells carrying an empty vector, the wild type (WT) or mutant Ec78 when challenged with serially diluted phage T5 phage. d, Results of cell growth analysis in DH10B cells transformed with plasmids encoding various Ec78 components and variants of PtuAB. ‘Induced’ indicates the plates containing IPTG; ‘Ec78’ denotes the transformation of the single plasmid encoding all retron-effector components; ‘PtuAB+RT–ncRNA’ denotes cotransformation of the plasmid encoding RT and ncRNA and that encoding PtuAB. The top five and bottom three rows cover two different starting cell densities, respectively, to reveal the range of toxic activity of PtuAB (Source Data Fig. 1).
Results
PtuAB complex is the retron-sequestered toxin unit
It has previously been shown that the PtuAB components of Ec78 induce cell growth arrest19. To understand how each PtuAB enzymatic function contributes to this activity, we conducted both phage challenge and cell growth assays on cells expressing different combinations of Ec78 components and their ATPase- and HNH-deficient variants (Fig. 1c,d). We constructed expression plasmids encoding the intact retron (WT Ec78), RT-ncRNA, PtuAB, PtuA, PtuB and their ATPase- or HNH-deficient variants. Individual plasmids, along with an empty vector, were transformed into E. coli strain 11303 or DH10B cells for the phage challenge or growth assay, respectively. Although intact Ec78 protected the host against T5 phage infection, neither the ATPase-deficient mutant, Ec78(PtuA-E396Q) nor the HNH-deficient mutant, Ec78(PtuB-H57A), offered such protection (Fig. 1c), indicating that both enzymatic activities of the PtuAB effector are required. Furthermore, expression of PtuAB in DH10B cells severely impaired growth, whereas the intact retron, PtuA or PtuB individually did not, indicating that PtuAB is the toxin unit (Fig. 1d). In accordance, the ATPase-deficient mutant, PtuA-E396Q–PtuB, or the HNH-deficient mutant, PtuA–PtuB-H57A, in PtuAB coexpression cells rescued the growth defects (Fig. 1d). These results confirm that Ec78 protects the host from phage infection and that PtuAB is the functional defense unit, in stark contrast to that of the stand-alone PtuAB system, whose activity is strictly controlled before phage invasion18,20. Therefore, in the Ec78 retron, the RT–ncRNA–msDNA entity exerts control over the adapted PtuAB for its defense function.
The PtuAB complex targets host tRNATyr and plasmid DNA
Previous efforts to identify phage determinants that evade Ec78 defense identified tRNATyr as a potential target19. To confirm that PtuAB targets host tRNATyr specifically, we transformed BL21(DE3) pLysS cells with plasmids expressing tRNATyr−GTA-1, tRNATyr−GTA-2, tRNAAla, tRNAHis, tRNAser, tRNAArg, tRNAMet or no tRNA (null), respectively, along with that encoding PtuAB (Fig. 2a). We showed that expression of both tRNATyr (Ec-tRNA-Tyr-GTA-1 and Ec-tRNA-Tyr-GTA-2) molecules, but not the null or other tRNAs, notably reduced the high levels of PtuAB toxicity, indicating that PtuAB specifically targets tRNATyr (Fig. 2a and Extended Data Fig. 2a), consistent with the previous finding that mutant phages overexpressing tRNATyr can evade retron Ec78 (ref. 19). The low-level toxicity of PtuAB in the non-expressing DH10B cells can also be rescued by the expression of tRNATyr and, to a lesser extent, tRNAAla (Extended Data Fig. 2b).
a, Results of cell growth analysis in BL21(DE3)pLysS cells cotransformed with the plasmid encoding PtuAB and that encoding various E. coli tRNAs. ‘Induced’ indicates the plates containing IPTG. b, PAGE gel analysis of in vitro cleavage reactions of the native tRNATyr by Ec78, effectors and their variants in the presence or absence of DNase I. The location of msDNA and ncRNA is labeled. ‘Ec78 WT released PtuAB’ denotes peak 3 of the gel filtration elution samples of the wild-type Ec78 (Extended Data Fig. 1a). ‘PtuAB (coexpressed with tRNA)’ denotes the wild-type PtuAB complex isolated from the expression cells coexpressed with tRNATyr. ‘PtuA’ and ‘PtuB’ denote the two subunits purified separately. ‘PtuA+PtuB’ denotes the mixture of the separately purified PtuA and PtuB. ‘M’ denotes molecular size markers. The faint msDNA band in the Ec78 treated with DNase I was due to incomplete digestion. c, Agarose gel analysis of in vitro cleavage reactions of plasmid DNA by Ec78, effectors and their variants. The labels are the same as described for b. ‘M’ denotes molecular size markers. The different amount of nicking might be due to slight differences in the PtuAB enzymes purified under different conditions. Experiments in b and c were repeated independently with similar results (Source Data Fig. 2).
We purified the intact Ec78 complex and various catalytic variants of Ec78 or PtuAB (Extended Data Fig. 1a,b). Given its cell toxicity, we reconstituted the wild-type PtuAB complex by either copurifying them from the cells overexpressing tRNATyr (PtuAB (coexpressed with tRNA)) or from the verified gel-filtration fractions during Ec78 purifications (released PtuAB) (Extended Data Fig. 1a,b). Among all purified retron and protein samples, wild-type PtuA exhibited the highest ATP-hydrolysis activity, with the ATPase-deficient mutants retaining some detectable activity (Extended Data Fig. 1c), unlike the ATPase-deficient PtuA of Septu18. We subsequently performed in vitro tRNA-cleavage assays with these samples. To our surprise, neither PtuAB, PtuB nor DNase-I-treated Ec78 could degrade in-vitro-transcribed tRNATyr−GTA-2 (Fig. 2b). Furthermore, DNase-I-treated Ec78 also did not degrade the native tRNATyr isolated from E. coli21 in the presence or absence of divalent ions or ATP (Extended Data Fig. 2c). In addition, the ATPase-deficient PtuA–PtuB-E396Q showed no direct binding activity to tRNATyr (Extended Data Fig. 2d).
We then tested the ability of Ec78 and its subcomplexes to cleave other types of nucleic acids (Fig. 2c and Extended Data Fig. 2e–g). We observed DNA-nicking activity by the released PtuAB and the tRNA-coexpressed PtuAB, but not other variants (Fig. 2c). Whereas the ATPase active site of PtuA is required, addition of 10 mM ATP did not impact the observed nicking activity (Fig. 2c). Notably, the same PtuAB complexes did not cut synthetic DNA (double or single-stranded) or RNA oligonucleotides (Extended Data Fig. 2e–g), suggesting a requirement for the supercoiled DNA topology in the PtuAB activity. Though the DNA nicking activity by the Ec78 PtuAB resembles that of the stand-alone Septu system18,20, its insensitivity to ATP inhibition is consistent with a distinct retron-mediated control mechanism. Given the amount of the released PtuAB fraction reduced in the Ec78_PtuA-E396Q variant (Extended Data Fig. 1a), ATPase hydrolysis is thus required for the release of the active PtuAB.
Molecular basis for Ec78 retron to sequester PtuAB
To uncover the molecular basis for Ec78’s ability to sequester PtuAB, we carried out cryo-EM structural analysis of the purified retron–effector complex. The sample-preparation process revealed that the assembly is sensitive to ATP hydrolysis. The wild-type retron displayed significant heterogeneity in assembly (Extended Data Fig. 1a), particularly with respect to PtuAB displacement. Therefore, PtuA-E396Q, the ATPase-deficient mutant with reduced heterogeneity, was used for structural determination (Table 1 and Extended Data Fig. 3).
The cryo-EM map revealed well-resolved density for both the ncRNA and the msDNA, enabling unambiguous assignment of their nucleotides (Fig. 3 and Extended Data Fig. 3). Surprisingly, only stem loop II (rU27 to rU48), a linker (rC49-rU58) and the template region (rG59–rU67) of the ncRNA were observed. The template region formed base pairs with msDNA (dA69–dC77). Furthermore, the msDNA is debranched at its 2′–5′ linkage to the initiation guanine nucleotide and lacks the first four nucleotides (dT1–dA4) (Fig. 3a), unlike that previously observed in the Ec86 retron22,23. We confirmed the processing of ncRNA and msDNA by analyzing the nucleic acids copurified with either the intact retron or RT alone on PAGE gels (Fig. 3b). We further showed that PtuAB is not required in the processing of both nucleic acids, because the processed products are also present in the RT-alone sample (Fig. 3b). To assess whether RT influences the processing activities, we replaced Lys146 and Arg143 by alanine in the RT (RT(RK)), which are found near the msDNA-cleavage site. We showed that the RT(RK) variant reduced processing (Fig. 3b), and its retron assembly no longer mitigates PtuAB toxicity (Extended Data Fig. 4), suggesting that, although host enzymes are likely responsible for the processing activities, RT exerts a notable influence.
a, The secondary and tertiary structures of the ncRNA and msDNA observed in the Ec78 structures. Only the RT is retained for simplicity with its characteristic domains labeled. b, Gel analysis results of the nucleic acids extracted from the purified Ec78, RT–ncRNA and RT-R143A K146A–ncRNA (RT(RK)) (right) or from the RNase-A-treated cell extract (left). RNA markers are the Low Range ssRNA Ladder (New England Biolab). c, Density maps (top) and cartoon representations (bottom) of the Ec78 retron complex. The 4:2:1 complex is shown to the left with one PtuB in trans with respect to the top RT, and the 4:2:2 complex is shown to the right with the corresponding PtuB in cis with respect to the top RT. Orange dots indicate the bound ADP molecules. Insets show the close-up views of the density around the two selected ADP molecules in the 4:2:1 complex. Experiments in b were repeated independently with similar results (Source Data Fig. 3).
The retron-effector particles have two major classes. The first is a 4:2:1 ratio of PtuA, PtuB and RT–ncRNA–msDNA, with four PtuA subunits, two PtuB subunits and one RT–ncRNA–msDNA complex (Fig. 3c). The second class exhibits a 4:2:2 stoichiometry, featuring an additional RT–ncRNA complex at the distal end of the msDNA hairpin, where only the terminal 12 msDNA nucleotides (dC65–dC77) are visible (Fig. 3c). This suggests that the complete Ec78 retron assembly accommodates a single msDNA molecule. There are other classes with either reduced PtuB occupancy or different combinations of PtuB orientations (cis versus trans to RT), without other structural differences. These classes led to reconstructions at lower resolutions and are thus not modeled (Table 1 and Extended Data Fig. 3). Interestingly, although we used PtuA-E396Q in structural determination, which showed significantly reduced ATPase activity (Extended Data Fig. 2c), ADP instead of ATP was trapped in three of the four subunits (Fig. 3c). This result contrasts with the stand-alone PtuAB structures in which ATP molecules are trapped, consistent with its lack of ATP hydrolysis18,20. Since there are no other differences in molecular interactions among these classes of structures, we focus the subsequent structural analysis primarily on the 4:2:1 assembly.
A single RT subunit forms the head of the Ec78 retron assembly, from which a long msDNA hairpin emerges to form the central spine of the assembly (Fig. 3c). This differs from the previously observed Ec86 retron structure, which is capped by RT dimers and bordered by two antiparallel msDNA strands22,23. In the 4:2:2 class, a second RT is positioned at the distal end of the msDNA hairpin but with its own msDNA largely disordered, leading to an architecture that is essentially the same (Fig. 3c). The lack of the dimerization α-helix at the amino terminus of Ec78 RT is responsible for the different oligomerization state than that of Ec86.
The 3′ overhang of the msDNA is anchored to the RT-bound ncRNA through a short RNA–DNA duplex (template–primer) that feeds into the catalytic Palm domain of RT, and its hairpin extends approximately 110 Å from the RT–ncRNA head (Fig. 3a). Four PtuA subunits, forming a dimer of dimers, construct a diamond-shaped platform with a central groove that adheres along the length of the msDNA hairpin, with two PtuB proteins capping the outer edges (Fig. 3c). If IRa1 and stem loop I of the ncRNA and the 2′–5′ linkage of the msDNA were retained, they would obstruct the assembly of PtuAB, suggesting that removal of these ncRNA elements, including msDNA debranching, is a required mechanism for Ec78 to achieve sequestration of the toxicity.
The PtuA platform forms an extensive interface with the msDNA, totaling ~2,000 Å2 buried solvent-accessible area (Fig. 4a). However, the contacts are non-specific and mostly occur between the loops in the helix-grip fold of PtuA and the minor groove of the msDNA (Fig. 4b,c). Most of these amino acids are positively charged and are not conserved in Septu PtuA (Extended Data Fig. 5a). In addition, a unique helix–loop–helix insertion into the RecA-like nucleotide-binding domain of PtuA forms a helical flap with the corresponding element of an opposing PtuA subunit encircling the msDNA (Figs. 3c and 4b). Thus, the assembly of PtuA with msDNA is based on shape complementarity, similar to other ABC ATPases, such as the Rad50 ATPase24, that act on DNA non-specifically. Notably, the loop forming a number of contacts with msDNA (m loop) is significantly longer in the stand-alone PtuA, which would obstruct its assembly with DNA (Fig. 4b and Extended Data Fig. 5a). In addition, the helical flap encircling the msDNA is deleted in the stand-alone PtuA (Fig. 4b and Extended Data Fig. 5a). The distinct elements in Ec78 PtuA that evolved for binding msDNA also enable it to partner with Ec78, reflecting a unique evolutionary path for retron acquisition.
a, Binding of msDNA to the central groove formed by PtuA. RT, PtuA and PtuB molecules are shown in surface representation. The other dimer of PtuAB is omitted for clarity. b, Superimposition of the Ec78 PtuA and the Septu PtuA (gray ribbons) structures reveals that unique protein elements in Ec78 PtuA evolved for contacting msDNA (m loop and helical flap) and the RT–ncRNA complex (N-terminal extension, NTE). The loop equivalent to the m loop in Septu PtuA would obstruct its ability to contact msDNA in the same manner. c, PtuA residues, mostly positively charged, making close contacts (within 3.5 Å) with the msDNA. Conservation of these residues is shown in Extended Data Fig. 5a. d, Close-up views illustrating structural features of PtuA interactions with the RT–ncRNA–msDNA tripartite. The characteristic domains of the RT are indicated and color coded. Nucleotides of the ncRNA and msDNA are labeled. NTE denotes the N-terminal extension of PtuA. Inset highlights the specific contacts of first α-helix of NTE with both the Finger and Thumb domains. e, The functional importance of msDNA-interacting residues. Left, phage challenge assay results with E. coli strain 11303 cells carrying an empty vector, the WT or mutant Ec78 in the msDNA-binding interface when challenged with serially diluted phage T5 phage (Source Data Fig. 4e). Right, cell growth arrest assay of the DH10 cells carrying an empty vector, the WT or mutant Ec78. f, Functional importance of the unique PtuA elements. Left, plaques challenge assay results with the E. coli strain 11303 cells carrying an empty vector, the wild-type (WT) or mutant Ec78 in PtuA when challenged with serially diluted phage T5 phage (Source Data Fig. 4f). Right, cell growth arrest assay of the DH10 cells carrying an empty vector, the wild-type (WT) or mutant Ec78. NTE denotes the N-terminal extension of PtuA; helical flap denodes the loop wrapping around the msDNA.
The association of PtuA with the Ec78 retron is further enhanced by its contacts with RT, which buries ~690 Å2 of solvent-accessible surface (Fig. 4c). This interaction is largely mediated by the N-terminal extension (NTE) of PtuA, an element also distinctly absent from the stand-alone PtuA (Extended Data Fig. 5a). The first α-helix of NTE is wedged between the characteristic Finger and Thumb domains of the RT, forming several specific protein–protein contacts (Fig. 4c). In addition, the first α-helix of the NTE also contacts the ncRNA (Fig. 4c), albeit non-specifically, further stabilizing the PtuA–retron association. The observed NTE–RT–ncRNA interaction is another significant feature acquired by the Ec78 PtuA.
To confirm the functional roles of the observed interfaces, we performed phage challenge and cell growth assays with Ec78 variants containing mutations in the msDNA-binding and PtuA–RT interfaces. Consistent with their interactions with the msDNA, substitutions of Arg126, Arg163 and Lys119 reduced immunity against T5 infection and, in parallel, caused notable arrest of cell growth (Fig. 4e). Similarly, removal of the helical flap residues (Δ261–274) markedly reduced immunity while increasing cell growth toxicity (Fig. 4f), underscoring its critical role in msDNA binding and sequestering cell toxicity. Interestingly, mutations in the N-terminal residues Asn4 and Tyr5, or deletion of residues 2–65 (Δ2–65), reduced immunity but had minimal impact on cell growth (Fig. 4f). The same result was also observed for the carboxy-terminus deletion mutation (Δ524–550), indicating that both the N and C termini of PtuA are required to elicit immunity, as well as cell toxicity.
Mechanism of PtuAB activation
To determine whether there are distinct sequence features in the PtuAB proteins adapted to function with type I-A retrons, we performed phylogenetic analysis of both proteins. The HNH nuclease PtuB revealed a somewhat scattered distribution between retron-associated and retron-independent sequences (Extended Data Fig. 6a), suggesting minimal evolutionary pressure for co-opting stand-alone PtuB to function within the type I-A retrons. In the retron structures, PtuB employs its helical C-terminal domain (CTD) to interact with the C-terminal extension of PtuA (Extended Data Fig. 6b). A pair of CTEs from each PtuA dimer sandwiches one PtuB subunit, forming a shared nine-helix bundle, in which three helices are from PtuB (Extended Data Fig. 6b). Both CTD of PtuB and CTE of PtuA seem to be well conserved between the Ec78 and Septu systems (Extended Data Fig. 5), consistent with the critical role of CTE of PtuA in cell toxicity (Fig. 4f). Furthermore, a comparison of the overall structure (Extended Data Fig. 6c) and the active site (Extended Data Fig. 6d) of Ec78 retron PtuB with stand-alone PtuB and the HNH domain of AceCas9 (ref. 25) indicates a high degree of structural similarity, consistent with the observed DNA-nicking activity in released PtuAB (Fig. 2c).
Unlike PtuB, the ATPase PtuA exhibits distinct clustering between the retron-associated and retron-independent sequences (Fig. 5a), suggesting that the evolution of the PtuA subunit from the stand-alone Septu system facilitated its integration into retron function. This is further supported by the observed structural features unique to the retron-associated PtuA for binding the RT–ncRNA–msDNA tripartite complex that are distinctively absent from the stand-alone PtuA (Fig. 4b and Extended Data Fig. 5a).
a, Phylogenetic analysis of both retron type I-A (black) and Septu (orange) PtuA sequences. b, Density maps and the corresponding cartoon representations of the Ec78 retron-displaced PtuAB complex in two orthogonal views. Orange dots indicate the bound ADP molecules. Inset shows the density map around one of the bound ADP molecules. c, Structural comparison between the retron-displaced (green) and the retron-bound PtuAB (gray), both in tetrameric forms, when a single PtuA subunit is aligned as indicated. The msDNA model is from the 4:2:1 Ec78 structure and shows clashes with the retron-displaced PtuA. d, Schematic oligomers of the retron-displaced (tetramer) and the Septu (hexamer) PtuAB. The hexameric structure of Septu PtuAB aligned with a PtuA subunit is shown to the right. The PtuB molecules are included in the assembly but not highlighted.
To further analyze the molecular basis for PtuAB activation, we obtained its structure in the absence of the retron. We introduced the Gln361 to leucine mutation within the signature motif (357-LSDGQR-362) of PtuA14 (Extended Data Fig. 5a) to abrogate the wild-type toxicity, which permitted protein purification and structure determination (Table 1 and Extended Data Fig. 7). Single-particle reconstruction led to a major class of PtuAB particles that was refined to 3.2 Å (Fig. 5b).
The PtuA-Q361L–PtuB complex forms a similar diamond-shaped platform, comprising four PtuA and two PtuB subunits (Fig. 5b), significantly different from the hexameric and inflammasome-like assembly of stand-alone PtuAB18. Although the free PtuA subunit seems to maintain the tetrameric form, free PtuB is monomeric (Extended Data Fig. 6e), suggesting that PtuA is the primary driver of PtuAB complex assembly. Retron-displaced PtuA-Q361L–PtuB assembles largely through dimerization of two helical flaps that otherwise encircle msDNA when in complex with retron (Fig. 5b). Compared with the retron-sequestered PtuAB-E396Q, the retron-displaced PtuA-Q361L–PtuB displays a substantially narrower central groove and a more compressed helical flap (Fig. 5c), both of which would exclude msDNA from binding. Notably, the retron-displaced PtuA-Q361L–PtuB, bearing Q361L in the ATPase domain, also traps ADP as opposed to ATP on all four sites (Fig. 5b), consistent with its detectable ATPase hydrolysis activity (Extended Data Fig. 1c). Notably, the stand-alone Septu PtuAB naturally carries a leucine in place of Gln361 and lacks ATPase activity, suggesting that this residue alone does not account for the enzymatic difference, and the retron-associated PtuA has evolved enhanced ATPase function.
The unique involvement of the helical flap in Ec78 PtuA oligomerization prevents it from forming the same inflammasome-like hexameric assembly observed for the stand-alone PtuAB complex18 (Fig. 5d). This difference might sufficiently explain the differences in both ATP hydrolysis and defense targets between the two PtuAB systems. It is remarkable that through addition of the elements to the well-conserved ATPase core, the PtuAB proteins are co-opted for sensing different phage determinants and targeting different host substrates.
The conformational closure of the retron-displaced PtuAB suggests a possible mechanism of its activation (Fig. 6). In the presence of the msDNA and the associated RT–ncRNA, PtuA is locked in a conformation that prevents rapid displacement of PtuB. In the absence of the msDNA, for instance as a result of the phage attack, and in conjunction with ATP hydrolysis, PtuAB disassembles, which activates PtuB for its nuclease activity, leading to growth arrest. This msDNA-mediated sequestration mechanism is consistent with the detected phage escape mutants in the D15 DNase13.
Two colors are used for PtuB to represent its cis or trans association with PtuA. Left, Ec78 activation model. The RT-ncRNA-msDNA sequestered PtuAB undergoes some release of the PtuAB molecules with the help of ATP hydrolysis without causing cell growth arrest. Upon phage infection, the tetrameric PtuAB is released from the tripartite and, in the presence of ATP hydrolysis, PtuAB disassembles to target genome DNA, leading to tRNATyr molecule degradation. ‘Pi’ denotes released inorganic phosphate. Right, Septu activation model. The hexameric PtuAB complex is activated by phage proteins and, upon the reduction of ATP levels, releases PtuAB or PtuB molecules to target genomic DNA.
Ec78 retron facilitates efficient genome editing in human cells
To assess the ability of the Ec78 retron to produce DNA to support genome editing in human cells, we transformed the Ec78 retron encoding a msDNA with flanking homology arms to a EMX1 site (Extended Data Fig. 8a) into HEK293T cells expressing an endogenous SpyCas9 targeting the same EMX1 site. Previous genome-editing studies using more than 100 retrons found that efficient production of msDNA led to replacement of the target site by the retron-synthesized msDNA. We thus included a previously identified, highly efficient type II-A retron, Mestre-1531, targeting the same site as the control. Next-generation sequencing showed that the genome-editing efficiency of Ec78 was at least comparable to that of Mestre-1531 (Extended Data Fig. 8b). These results highlight the Ec78 retron as a highly effective genome-editing enzyme supported with three-dimensional structure information.
Discussions
We report structural and functional characterization of the first retron example that co-opts a stand-alone anti-phage system for retron-mediated defense. In contrast to the protein-controlled Septu PtuAB, the Ec78 PtuAB possesses strong ATP hydrolysis activity and is neutralized by the RT–ncRNA–msDNA tripartite. Upon being displaced, the PtuAB indiscriminately restricts the host, leading to growth arrest. The Ec78 PtuAB acquired three protein elements: the N-terminal extension to interact with the RT and ncRNA, a helical flap and a number of positively charged loops to entrap msDNA. The incorporation of these tripartite interacting elements eliminates its ability to self-assemble into the inflammasome-like complex, typical of its stand-alone counterpart in Septu, that allows protein-mediated control, such as the sensing of phage tail fibers.
The observed DNA-nicking activity of Ec78 in vitro is potentially linked to its tRNA-targeting activity in vivo through unidentified factors or processes. One possibility is that activated PtuAB induces DNA damage upon phage infection, which triggers tRNA fragmentation, similar to that described in tRNA-repair studies26. Targeting tRNA as an immunity strategy has been observed in other defense systems such as the ATPase–TOPRIM nuclease pair, AriA–AriB, of the phage antirestriction induced system (PARIS) system, which is also a retron-independent abortive immunity system27,28. However, AriB contains the TOPRIM-like domain and functions when released from AriA27,28 whereas PtuB contains a HNH domain and requires PtuA for function. Interestingly, however, another type I-A retron, Ec83, that also partners with PtuAB, does not seem to target host tRNA19, raising the possibility that the PtuAB system offers a versatile platform for sensing additional phage triggers and targeting different nucleic acids in response to different environmental pressures.
The function of Ec78 retron RT resembles that of Ec86 retron22,23 and that of the diversity-generating retroelements (DGR)29, for which three-dimensional (3D) structures are now available, in using ncRNA to template DNA synthesis. Unlike the latter RTs, however, Ec78 RT has evolved specific features for its partner effector. It adopts the characteristic ‘right-hand’ architecture typical of group II intron family of RTs, comprising three distinct subdomains—Finger, Thumb and Palm—that are all preserved in the three RT enzymes. Structural comparison reveals a high similarity between Ec78 and Ec86, with a r.m.s. deviation of 1.155 over 121 Cα atoms (Extended Data Fig. 9). However, Ec78 RT exhibits two distinguishing features: the absence of an N-terminal helix preceding the Finger domain, and a particularly long, positively charged loop in the Palm domain (Lys141–Lys146). In Ec86 RT, the N-terminal helix facilitates RT dimerization, whereas its absence in Ec78 RT accounts for the monomeric state of RT in the Ec78 retron system (Extended Data Fig. 9). The positively charged loop plays a critical role in stabilizing the 5′ end of the msDNA, with the side chain of Arg143 forming a hydrogen bond with the phosphate group of terminal dG5, and Arg142, Arg143 and Lys146 engaging in electrostatic interactions with dG5 and dG6. We found that the R143A K146A substitutions impaired the msDNA biogenesis required for retron assembly, leading to cytotoxicity when the mutant retron is expressed in cells.
Consistent with the structural data, phylogenetic analyses also revealed co-evolved features that support the specific assembly. We observed distinct clusters of RTs based on types, with only a minor proportion scattered throughout the tree. For instance, the positively charged loop that impacts msDNA debranching is highly conserved among type I-A retrons, whereas the N-terminal helix required for dimerization is a common feature in type II retrons (such as Ec86). Notably, this clustering pattern is more pronounced in larger subtypes with a greater number of members, whereas smaller subtypes, with fewer members, exhibit greater dispersion. These findings suggest that coevolutionary selection plays a dominant role in retron evolution, while modular exchange occurs to a lesser extent, particularly in smaller subtypes. These findings suggest a potential coevolutionary relationship between RT and its associated effector in the retron system, highlighting structural adaptations that contribute to functional specificity in each retron type.
The unique production of msDNA by retrons has been harnessed for genome applications, in conjunction with CRISPR–Cas9, in bacteria, yeast and human cells9,10. Transfecting the plasmid carrying the RT–msrRNA cassette in targeted cells produces msDNA by the RT, providing an amplifiable single-stranded DNA that act as the homology-dependent repair (HDR) template to repair the double-stranded break generated by CRISPR–Cas9. Owing to the continuous template production, the retron-mediated HDR is more efficient than that mediated by synthetic DNA10. In addition, retron RTs have been repurposed for prime-editing, in which the fused nickase CRISPR–Cas9 nicks the DNA while the RT produces the desired edited DNA from the prime editing guide RNA9. Retron reverse transcriptases present a promising, compact alternative for prime editing, achieving efficiencies comparable to that of the larger M-MLV RT through protein engineering9. Our results indeed showed highly efficient genome editing with Ec78 retron. However, the implications of the distinctive biogenesis of Ec78 ncRNA and msDNA in genome editing remain unclear, highlighting a need for further research.
Methods
Cloning, protein expression and purification
For protein expression, the operon sequences of E. coli ECONIH5, including its native promoter and coding sequences of ncRNA, RT, PtuA and PtuB, were PCR amplified from the pLG008 plasmid2. The PCR product was cloned into a modified polycistronic expression plasmid, pST44 (ref. 30), in which a C-terminal 3C protease-cleavable His-tag was introduced in PtuB by Gibson assembly (Supplementary Data File 1). For the coexpression of PtuA and PtuB, the coding sequences of PtuA and PtuB were individually cloned into the first and second multiple cloning sites of a modified pRSF-Duet expression vector, in which an N-terminus of His-Sumo tag was introduced on PtuA. For the toxicity assay, the sequences encoding PtuA and PtuB were cloned either separately or together into expression vectors. When cloned separately, each gene was inserted into either the pBAD vector or the pRSF-Duet vector. For coexpression, the ptuA and ptuB genes were cloned together either in the pRSF-Duet vector or as part of a co-operonic construct in the pBAD vector. All the mutations were performed using the QuikChange mutagenesis method with Q5 DNA polymerases.
The expression plasmids were transformed into either the Nico21 (DE3) (NEB) or the BL21-CondonPlus (DE3)-RIPL (Agilent) cells. Transformed cells were plated onto LB agar plates containing appropriate antibiotics for selection. A single colony from the plate was used to inoculate 50 ml of LB medium supplemented with the same antibiotics, and the primary culture was grown overnight at 37 °C. Eight milliliters of the overnight culture were transferred into 1 L of LB medium, with a total volume of 6 L prepared for subsequent purification. The cells were grown at 37 °C until the optical density at 600 nm (OD600) reached 1.0. The temperature was lowered to 19 °C, and after 30 minutes, the culture was induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Induction was allowed to proceed overnight. After overnight induction, cells were pelleted by centrifugation, snap-frozen in liquid nitrogen and stored at –80 °C if not used for purification immediately.
For protein purification of the retron complex, the cell pellets were resuspended and lysed by sonication in 80 ml of lysis buffer (50 mM Tris-Cl, 500 mM NaCl, 5% glycerol, 10 mM Imidazole, 5 mM TCEP). The lysate was clarified by centrifugation, and the supernatant was loaded into a 5 ml HisTrap HP column (Cytiva) using a peristaltic pump. Subsequent washing and elution steps were also performed with the peristaltic pump. The column was sequentially washed with 200 ml of low-imidazole wash buffer (50 mM Tris-Cl, 500 mM NaCl, 5% glycerol, 25 mM imidazole) followed by 50 ml of high-imidazole wash buffer (50 mM Tris-Cl, 500 mM NaCl, 5% glycerol, 40 mM imidazole). Protein was then eluted with 20 ml of elution buffer (50 mM Tris-Cl, 500 mM NaCl, 5% glycerol, 400 mM imidazole, 5 mM TCEP). The eluted protein was then diluted with low-salt buffer (50 mM Tris-Cl, 100 mM NaCl, 5% glycerol, 5 mM TCEP) to achieve a final salt concentration of 300 mM NaCl. The diluted sample was then loaded onto a 5 ml HiTrap Q HP anion exchange column (Cytiva) using a peristaltic pump. The column, connected to an ӒKTA pure 25 chromatography system, was washed and eluted with a gradient of NaCl from 300 mM to 2 M in 25 mM Tris-Cl buffer (pH8.0) and 2 mM 1,4-dithiothreitol (DTT). The fractions of the elution peak were combined, concentrated and further purified by size-exclusion chromatography (SEC) using a Superose 6 10/300 column in a buffer containing 25 mM Tris-Cl (pH 8.0), 250 mM NaCl and 5 mM DTT. The intact complex was identified based on ultraviolet absorbance values at 260 nm and 280 nm, as well as SDS–PAGE analysis. The fraction with the highest protein concentration was used immediately for cryo-EM grid preparation. All remaining fractions were snap-frozen in liquid nitrogen and stored at –80 °C.
The protein purification of the PtuA and PtuB complex was carried out similarly as described above. To avoid the toxicity of co-expressing PtuA and PtuB, we expressed the two proteins separately and the cells were combined for co-purification. The wild-type PtuAB proteins could also be expressed when tRNATyr is co-expressed. For the non-toxic PtuAB mutants, cells co-expressing both proteins were collected as described. After HisTrap affinity purification, the eluted protein was loaded to a 5 ml HiTrap Heparin column instead of the HiTrap Q column. All other steps remained unchanged.
Cryo-EM sample preparation, data collection and 3D reconstruction
Cryo-EM grids were prepared using a Mark IV Vitrobot. Three microliters of the sample (3 mg ml−1) were applied to R 1.2/1.3 Carbon Quantfoil grids, which had been freshly glow-discharged in a Gatan Solarus 950. The grids were double-sided blotted for 4 s with a constant force of 0, in a chamber maintained at 100% relative humidity at 8 °C. They were then plunged into liquid ethane and stored in liquid nitrogen before screening.
Grids were initially screened on an Arctica Talos microscope equipped with a K3 camera operated at 200 kV. Good grids were subsequently transferred to a Titan Krios microscope, also equipped with a K3 camera and operated at 300 kV. Micrographs were automatically collected using SerialEM in low-dose mode at a nominal magnification of ×130,000 in a super-resolution mode, with an energy filter of 15 eV, leading to a corrected physical pixel size of 0.828 Å per pixel. A total dose of 50 e− per Å2 was applied over 50 frames, with a random defocus range of –1.0 to –1.8 µm.
The raw micrographs, initially in super-resolution, were frame-aligned using Patch Motion Correction with cropping to half the original resolution (0.828 Å per pixel), followed by Patch CTF Estimation. Particle picking was performed automatically using Blob Picker with a particle range size set to 100–180 Å. Subsequent particle inspection and extraction yielded a total of 639,730 particles. The extracted particles underwent two-rounds of 2D classification, resulting in 50,913 high-quality particles. These refined 2D templates were then used for particle picking through Template Picker, followed by another round of particle inspection, extraction and two rounds of 2D classification. Ultimately, 199,577 good particles were selected for 3D reconstruction using ab initio reconstruction, homogeneous refinement and non-uniform refinement. The resulting volume and selected particles were exported to Relion for further processing of the complete datasets, where they served as initial model and templates for topaz training.
The complete set of 20,372 raw videos of the Ec78 complex was imported to Relion (v5.0)31 for processing. The raw micrographs, initially in super-resolution, were frame-aligned using MotionCor2 (v1.6.4)32 with a binning factor of 2. CTF estimation was then performed using Gctf (v1.18)33 within Relion (v4.0). On the basis of a CTF resolution cutoff of 6.0 Å, a total of 19,745 micrographs were selected for further analysis. A subset of 999 randomly selected micrographs was used for topaz training. For topaz training, the cryoSPARC34 particle file was first converted to Relion-compatible star file using the PyEM package (csparc2star)35 before being imported to Relion. Subsequent 2D classification provided high-quality templates for automated particle picking from the selected 999 micrographs. The picked particles were inspected and refined through another round of 2D classification, and the resulting high-quality particles were used as input for Topaz training. The trained topaz model was then applied for automated particle picking across the entire dataset, yielding a total of 3,630,467 particles. The picked particles were extracted and downscaled by a factor of 2, resulting in a final pixel size of 1.656 Å per pixel. The extracted particles underwent three rounds of 2D classification, yielding a total of 1,235,478 high-quality particles for further processing. Three-dimensional classification was preformed using the initial volume from cryoSPARC, applying alignment and Blush regularization. Similar maps were combined, and the junk particles were discarded, resulting in two good classes: one consisting of 875,922 particles and the other 294,359 particles. For high-resolution reconstruction, those particles were separately re-extracted at the original physical pixel size of 0.828 Å per pixel. Subsequent 3D refinement and post-processing of these two classes generated two maps, leading to final resolution of 2.75 Å and 2.69 Å, respectively, following non-uniform refinement.
A data set of 17,040 raw micrographs was collected for the PtuAB complex. The same data processing and refinement steps were carried out, leading to a major class of particles with a reported resolution of 3.15 Å.
Both complexes were modeled based on the Septu PtuAB complex coordinates (8EE7) and the Ec86 coordinates (7V9X), manually adjusted in COOT36 and refined in PHENIX37.
Phylogenetic analysis
The phylogenetic analysis was performed with an in-house snakemake pipeline modified from https://github.com/vihoikka/Cas10_prober. In brief, 91,902 annotated genomes marked as ‘complete’ were downloaded on 22 August 2024. The genomes were analyzed through Defensefinder (ref) to identify RT and PtuAB sequences. For retron RT analysis, all organisms that annotated as retron systems were singled out, resulting in 8,143 sequences. These RT sequences were clustered with CD-hit (similarity cutoff 0.99) resulting in 1,935 representative sequences. To create a RT phylogenetic tree, proteins were aligned using Muscle 5.1 with the Super5 algorithm intended for large data sets. The RT tree was constructed from the alignment using FastTree 2.1.11 with the WAG + CA model and Gamma20-based likelihood and visualized in R4.1.1 using ggtree and ggplot.
PtuA and PtuB phylogenetic analyses were carried out similarly. A total of 6,295 PtuA and PtuB sequences from either Septu (5878) or Type I-A retrons (417) were extracted and clustered before the phylogenetic tree was constructed as described above.
ATPase assay
ATPase activity was measured using the ADP-Glo Kinase assay (Promega). The reactions were carried out with or without 500 μM ultrapure ATP in a reaction buffer containing 20 mM Tris pH8.0, 150 mM NaCl, 2 mM DTT, 10 mM MgCl2. Reactions containing 0.2 mg ml−1 of a protein complex were incubated at 37 °C for 45 minutes before adding ADP-Glo reagent to remove free ATP, followed by the detection reagent. Luminescence was measured on the Bio-tek multiplate reader. The reported luminescence reading for each sample was normalized against the readings from reactions without ATP. Mean values obtained from two technical replicates with bars indicating s.d.
Cell growth spot assay
An individual colony from previously transformed bacteria grown on agar plates with appropriate antibiotic(s) was inoculated in LB medium containing the appropriate antibiotic(s) and allowed to grow overnight at 37 °C with shaking. The next day, the overnight culture was used to inoculate a fresh culture that grew until the OD600 reached 0.25 or 0.5. Tenfold serial dilutions of these cultures were prepared in LB medium, and 7.5 μl of these dilutions was spotted onto LB agar plates containing the appropriate antibiotic(s) and inducer (IPTG). The plates were incubated overnight at 37 °C and were imaged the next day using a ChemiDoc Imaging System (Bio-Rad).
For tRNA rescue experiments, plasmids encoding E. coli tRNAs were constructed as described in Masuda et al.21 and cotransformed with that encoding PtuAB.
Non-coding RNA and msDNA analysis
The nucleic acids present in either RT or the intact retron-expressing cells were extracted using a Qiagen Plasmid Plus Mid kit and eluted. The nucleic acids co-purified with either RT or the intact retron were extracted by denaturing the purified proteins. The resulting nucleic acids were resolved on a 15% urea PAGE gel and visualized by SYBR Gold staining (ThermoFisher).
Plasmid nicking assay
Plasmid nicking assays were performed in a 10 μl reaction mixture containing 300 ng of pRSF-duet-1 plasmid and 200 nM protein in reaction buffer (10 mM HEPES (pH 7.5), 50 mM KCl, 10 mM MgCl2) in the presence or absence of 10 mM ATP. Reactions were incubated at 37 °C for 30 min. Products were analyzed by electrophoresis on 1% agarose gels and visualized by ethidium bromide staining.
Nuclease assay
For tRNA cleavage assay, purified PtuAB, PtuB and Ec78 at 1 mM concentration were incubated with either the native E. coli tRNATyr (ref. 21) to test their activity on tRNA cleavage. In-vitro-transcribed or native tRNATyr was heated to 95 °C for 5 min, and then immediately transferred to 65 °C. After adding MgCl2 to a final concentration of 10 mM, the tRNA was allowed to cool to room temperature. The cleavage assay was performed in a buffer containing 10 mM HEPES (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM DTT and 10 μM MnCl2 and at 37 °C for 1 hour. To test the effect of ATP on tRNA cleavage, 1 mM ATP was added. The reaction was stopped by the addition of RNA loading dye (Thermo Fisher), and was then heat-denatured at 95 °C for 3 min. The products were resolved on 15% urea–polyacrylamide gel electrophoresis gels, visualized by SYBR Gold staining and imaged using the Bio-Rad ChemiDoc system.
For DNA oligonucleotide nuclease assays, the same procedure was followed, except that proteins were incubated with 5′-FAM-labeled ssDNA or dsDNA (Supplementary Data File 1). Products were visualized on the Bio-Rad ChemiDoc system using multi-channel fluorescence detection. For ssRNA-cleavage assays, proteins were incubated with unlabeled ssRNA, and all subsequent steps were performed as described for the tRNA cleavage assay.
Plasmid construction for human cell transfection
Plasmids used for human cell transfection were adapted from the design described by Khan et al.10, in which the RT is expressed under a CAG promoter, whereas the non-coding RNA (ncRNA) and Cas9 guide RNA (gRNA) are driven by U6 and H1 promoters, respectively. The RT gene strand was codon-optimized and synthesized with the associated ncRNA through Twist Bioscience (Supplementary Data File 1). The a1/a2 region of the ncRNA was extended to 18 bp, and the msd donor stem was shortened to 9 bp, following modifications reported in the previous study10. The best-performing retron from Khan et al.10 (1531) was used as a performance benchmark. Both tested retrons carried ncRNA sequences designed to introduce a precise three-base edit in the EMX1 locus, converting GAAGGG to AAAGTT.
Phage plaque assay
T5 bacteriophage (ATCC 11303-B5) was propagated using the double agar overlay method. In brief, 3 ml of soft agar containing the host strain E. coli B (ATCC 11303) was overlaid onto Nutrient Agar plates (2.3% Nutrient Agar (BD 213000), 0.5% NaCl). Subsequently, 0.5 ml of rehydrated, freeze-dried T5 phage was added to each plate and incubated overnight at 37 °C. After incubation, the phage-containing top layer was scraped from the surface of the plates. The collected material was centrifuged at 1,000 r.p.m. for 25 min to pellet cellular debris and residual agar. The clarified supernatant was filtered through a 0.22-μm Millipore filter to remove remaining particulates. The resulting high-titer phage stock was stored at 4 °C for subsequent plaque assays.
For plaque assays, E. coli strains transformed with either an empty vector or plasmids expressing wild-type Ec78 or its variants were grown to early log phase. Cultures were mixed with soft agar supplemented with ampicillin (final concentration, 50 μg ml−1) and IPTG (final concentration 0.4 mM) and overlaid onto Nutrient Agar plates as described above. Then, 7 μl of tenfold serial dilutions of the T5 phage stock was spotted onto the surface of each plate. Plates were incubated overnight at 37 °C, and plaque formation was visualized and imaged using a ChemiDoc imaging system.
Human cell transfections
HEK293T cells constitutively expressing SpCas9 from a CBh promoter at the AAVS1 Safe Harbor locus (GeneCopoeia, SL502) were maintained in high-glucose DMEM supplemented with GlutaMax (Thermo Fisher Scientific, 10569010). For retron transfections, 200,000 cells per sample were nucleofected with 1,500 ng of plasmid using a Lonza 4D-Nucleofector X Unit (AAF-1003X) with the SF Cell Line Kit and pulse code DS150.
For DNA repair inhibition experiments, AZD7648 (1 μM; Selleck Chemicals, S8843) and PolQi2 (3 μM; MedChem Express, HY-150279) were added to the culture medium before seeding nucleofected cells into 24-well plates. After 48 h, genomic DNA (gDNA) was extracted using the Quick-DNA Microprep Kit (Zymo Research) and eluted in 30 μl of ultra-pure, nuclease-free water. Following PCR amplification of the EMX1 locus and subsequent gel purification, amplicons were subjected to nanopore sequencing through Plasmidsaurus, yielding approximately 5,000 reads per sample. Sequencing reads were analyzed using the CRISPResso platform to quantify the frequency of precise genome-editing events.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates of the cryo-EM structures of Ec78 and PtuAB have been deposited in the Protein Data Bank under the identifiers 9NNB, 9NNH and 9NNK and in the Electron Microscopy Data Bank under the entries EMD-49570, EMD-49575 and EMD-49576, respectively. Sequencing data associated with this study are available on National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (PRJNA1330876). Source data are provided with this paper.
References
Rice, S. A. & Lampson, B. C. Bacterial reverse transcriptase and msDNA. Virus Genes 11, 95–104 (1995).
Gao, L. et al. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369, 1077–1084 (2020).
Millman, A. et al. Bacterial retrons function in anti-phage defense. Cell 183, 1551–1561.e12 (2020).
Bobonis, J. et al. Bacterial retrons encode phage-defending tripartite toxin-antitoxin systems. Nature 609, 144–150 (2022).
Lima, T. M. & Lim, D. A novel retron that produces RNA-less msDNA in Escherichia coli using reverse transcriptase. Plasmid 38, 25–33 (1997).
Shimamoto, T., Shimada, M., Inouye, M. & Inouye, S. The role of ribonuclease H in multicopy single-stranded DNA synthesis in retron-Ec73 and retron-Ec107 of Escherichia coli. J. Bacteriol. 177, 264–267 (1995).
Mestre, M. R., Gonzalez-Delgado, A., Gutierrez-Rus, L. I., Martinez-Abarca, F. & Toro, N. Systematic prediction of genes functionally associated with bacterial retrons and classification of the encoded tripartite systems. Nucleic Acids Res. 48, 12632–12647 (2020).
Mayo-Munoz, D., Li, H., Mestre, M. R. & Pinilla-Redondo, R. The role of noncoding RNAs in bacterial immunity. Trends Microbiol. 33, 208–222 (2024).
Doman, J. L. et al. Phage-assisted evolution and protein engineering yield compact, efficient prime editors. Cell 186, 3983–4002.e26 (2023).
Khan, A. G. et al. An experimental census of retrons for DNA production and genome editing. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02384-z (2024).
Doron, S. et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120 (2018).
Stokar-Avihail, A. et al. Discovery of phage determinants that confer sensitivity to bacterial immune systems. Cell 186, 1863–1876.e16 (2023).
Azam, A. H. et al. Viruses encode tRNA and anti-retron to evade bacterial immunity. Preprint at bioRxiv https://doi.org/10.1101/2023.03.15.532788 (2023).
Dot, E. W., Thomason, L. C. & Chappie, J. S. Everything OLD is new again: how structural, functional, and bioinformatic advances have redefined a neglected nuclease family. Mol. Microbiol. 120, 122–140 (2023).
Yang, W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 44, 1–93 (2011).
Yang, X. Y. et al. Molecular basis of Gabija anti-phage supramolecular assemblies. Nat. Struct. Mol. Biol. 31, 1243–1250 (2024).
Li, J. et al. Structures and activation mechanism of the Gabija anti-phage system. Nature 629, 467–473 (2024).
Li, Y. et al. PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense. Nat. Struct. Mol. Biol. 31, 413–423 (2024).
Azam, A. H. et al. Evasion of antiviral bacterial immunity by phage tRNAs. Nat. Commun. 15, 9586 (2024).
Wang, S. et al. Landscape of new nuclease-containing antiphage systems in Escherichia coli and the ounterdefense roles of bacteriophage T4 genome modifications. J. Virol. 97, e0059923 (2023).
Masuda, I. et al. A genetically encoded fluorescent tRNA is active in live-cell protein synthesis. Nucleic Acids Res. 45, 4081–4093 (2016).
Wang, Y. et al. Cryo-EM structures of Escherichia coli Ec86 retron complexes reveal architecture and defence mechanism. Nat. Microbiol. 7, 1480–1489 (2022).
Carabias, A. et al. Retron-Eco1 assembles NAD+-hydrolyzing filaments that provide immunity against bacteriophages. Mol. Cell 84, 2185–2202.e12 (2024).
Gut, F. et al. Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11–Rad50. Mol. Cell 82, 3513–3522.e6 (2022).
Das, A. et al. Coupled catalytic states and the role of metal coordination in Cas9. Nat. Catal. 6, 969–977 (2023).
Hughes, K. J., Chen, X., Burroughs, A. M., Aravind, L. & Wolin, S. L. An RNA repair operon regulated by damaged tRNAs. Cell Rep. 33, 108527 (2020).
Burman, N. et al. A virally encoded tRNA neutralizes the PARIS antiviral defence system. Nature 634, 424–431 (2024).
Deep, A., Liang, Q., Enustun, E., Pogliano, J. & Corbett, K. D. Architecture and activation mechanism of the bacterial PARIS defence system. Nature 634, 432–439 (2024).
Handa, S. et al. RNA control of reverse transcription in a diversity-generating retroelement. Nature 638, 1122–1129 (2025).
Tan, S., Kern, R. C. & Selleck, W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40, 385–395 (2005).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Asarnow, D., Palovcak, E. & Cheng, Y. asarnow/pyem: UCSF pyem v0.5 (v0.5). Zenodo https://doi.org/10.5281/zenodo.3576630 (2019).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Acknowledgements
Cryo-EM data were collected at the David Van Andel Advanced Cryo-Electron Microscopy Suite (RRID:SCR_023210) at the Van Andel Institute (Grand Rapids, MI). We thank the Van Andel Institute Cryo-EM Core staff for their assistance with data collection. We thank all lab members for their support and discussions. This work was supported by NIH grant R35 GM152081 to H.L. and R35 GM134931 to Y.H.
Author information
Authors and Affiliations
Contributions
B.W., R.H. and H.L. designed the experiments; B.W. expressed and purified the proteins and performed all biochemical and cryo-EM studies; R.H. designed and performed human genome editing experiments; Y.H. provided the native tRNA and tRNA-expression plasmids. B.W., R.H. and H.L. wrote the manuscript. All authors edited the manuscript and provided insightful comments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Yue Feng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Purification of Ec78 and PtuAB and their ATP hydrolysis activities.
a. Gel filtration profiles (top) and SDS-PAGE analysis (bottom) of Ec78 and the Ec78_PtuA(E396Q). b. Gel filtration profiles and SDS-PAGE analysis of PtuAB(H57A), PtuA(E396Q)B, and PtuA(Q361L)B. c. ATP hydrolysis activities of Ec78 and PtuAB. Two independent measurements were performed for each sample. Mean and standard deviations are displayed. For panels a and b, protein markers are Color Prestained Protein Standard, Broad Range (10-250 kDa) (New England BioLab).
Extended Data Fig. 2 Ec78 targets tRNATyr in cells.
a. Effects of tRNA co-expression and toxic effectors on bacterial growth rescue. Each condition includes three replicates, represented by the same color in the plot. Each combination has a corresponding control without IPTG induction. ‘AB’ represents PtuA-PtuB, ‘Lys’ represents tRNA (Lys), and ‘TyrU’ represents tRNA (TyrU). ‘w/o IPTG’ indicates no IPTG induction, while ‘w/ IPTG’ indicates IPTG induction. b. Results of cell growth analysis in DH10B cells co-transformed with the plasmid encoding PtuAB and that encoding various E. coli tRNATyr-GTA-1 and tRNAAla. “induced” indicates the plates containing IPTG. Kan+ and Amp+ reference to the presence of kanamycin and ampicillin in the respectively used agar plates. c. PAGE gel analysis of in vitro cleavage reactions comparing Ec78 and DNase I-treated Ec78 on native tRNATyr in the presence and the absence of metal ions or ATP. The location of msDNA and nsRNA are labeled. d. Analysis of direct interaction between PtuA(E396Q)B and in vitro transcribed tRNATyr. Upper left, gel filtration profile of PtuA(E396Q)B incubated with tRNATyr. Lower left, native PAGE gel analysis of the eluted fractions stained by Coomassie Blue (left) and SYBR Gold (right), respectively. Right, SDS-PAGE (upper) and Urea-PAGE (lower) analysis of the eluted fractions. Protein markers are Color Prestained Protein Standard, Broad Range (10-250 kDa) (New England BioLab). RNA markers are Low Range ssRNA Ladder (New England BioLab). e. Gel analysis of cleavage products of single- (ssDNA) or double-stranded DNA (dsDNA) by Ec78 released wild-type PtuAB. f. Gel analysis of cleavage products of single- (ssDNA) or double-stranded DNA (dsDNA) by wild-type PtuAB co-expressed with tRNA. g. Gel analysis of cleavage products of single-stranded RNA (ssRNA) by Ec78 released wild-type PtuAB and that co-expressed with tRNA. RNA markers are Low Range ssRNA Ladder (New England BioLab).
Extended Data Fig. 3 Cryo-EM image analysis and reconstruction results of the Ec78_PtuA(E396Q) complex.
a. Image and particle selection, and reconstruction scheme. Selected micrograph and 2D class averages are shown with scale bar indicating 100 Å. b. The Fourier Shell Correlation (FSC) curve, direction distribution and local resolution maps of the class 1 reconstruction, 4:2:1 complex. 0.143 FSC cutoff was used for resolution estimation. c. The Fourier Shell Correlation (FSC) curve, direction distribution and local resolution maps of the class 2 reconstruction, 4:2:2 complex. 0.143 FSC cutoff was used for resolution estimation.
Extended Data Fig. 4 Functional defects of RT mutant, Arg143Ala and Lys146Ala (RK).
Liquid culture growth analysis of the Ec78, Ec78_PtuA(RK), and PtuAB in BL21(DE3) cells. Optical density at 600 nm was followed for 10 hours for each culture using a BioTek plate reader spectrophotometer. Measurements were from three independent experiments and are plotted with standard deviations.
Extended Data Fig. 5 Sequence alignment of PtuA and PtuB.
Sequence alignment of PtuA (a) and PtuB (b). Key residues involved in catalytic activities and binding are indicates and labeled. Proteins used in the alignment are listed in Supplementary Data File 1.
Extended Data Fig. 6 Phylogenetic and structural analysis of PtuB.
a. Phylogenetic analysis of both retron Type I-A (black) and Septu (orange) PtuB sequences. b. Close-up view of the C-terminal domain of PtuB (CTD) interacting with a pair C-terminal extension (CTE) of PtuA. c. Superimposed PtuB structures between the Ec78 (beige) and the Septu (gray, PDB code: 8EE7) systems. The catalytic site is marked by a red dot. The region displaying the largest difference is marked by Ec78 PtuB residues. d. Comparison of the HNH domain active site among Ec78 PtuB, Septu PtuB (PDB code: 8EE7) and the Cas9 from Acidothermus cellulolyticus (PDB code: 8D2K). e. Gel filtration profiles of free PtuA and PtuB in comparison with that of PtuA(Q361L)B complex and caliberated molecular weight standards. The estimated molecular weight markers are marked. The eluted fractions for each run were verified for the presence of the corresponding proteins by SDS-PAGE (lower panel). Protein markers are Color Prestained Protein Standard, Broad Range (10-250 kDa) (New England BioLab).
Extended Data Fig. 7 Cryo-EM image analysis and reconstruction results of the PtuA(Q361L)B complex.
a. Image and particle selection, and reconstruction scheme. Selected micrograph and 2D class averages are shown with scale bar indicating 100 Å. b. The Fourier Shell Correlation (FSC) curve, direction distribution, and local resolution maps of the major class.
Extended Data Fig. 8 Ec78 retron precise editing.
a. Schematic of Ec78 retron precise editing targeting an EMX1 site. b. Quantification of Ec78 retron precise editing in comparison with no reverse transcriptase control (No RT), in the absence (Ec78) or the presence (Ec78+inhib) of inhibitors, AZD7648 and PolQi2, and the same experiments with the Mestre-1531 retron (RT 1531) with the same inhibitors. Bars represent means ± standard deviations of two biological replicates.
Extended Data Fig. 9 Structure comparison between the Ec78 and Ec86.
Top, comparison of the reverse transcriptases (RT) in two orthogonal views. Ec78 RT is multi-colored and Ec86 is colored light blue. The active site is marked by Asp187 and Asp96. Bottom, comparison of the non-coding RNA (ncRNA) and msDNA associated with the two RTs in the same two views. The protein domains and nucleic acids elements are labeled.
Supplementary information
Supplementary Data 1
List of proteins and oligos used in this study.
Source data
Source Data Figs. 1–5 and Extended Data Figs. 1, 2, 4, 5 and 8
Zip file containing uncropped plate images, uncropped gels and statistical source data for Figs. 1–5 and Extended Data Figs. 1, 2, 4, 5 and 8.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, B., Hoffman, R.D., Hou, YM. et al. Structural basis for retron co-option of anti-phage ATPase-nuclease. Nat Struct Mol Biol 33, 53–62 (2026). https://doi.org/10.1038/s41594-025-01702-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-025-01702-6
This article is cited by
-
Architecture and mechanism of a dual-enzyme retron system in prokaryotic immunity
Nature Communications (2025)
-
Structural mechanism of the Retron-Eco7 anti-phage defense system
Nature Communications (2025)