Abstract
Imaging across different scales is essential for understanding healthy organ morphology and pathophysiological changes. The macro- and microscale three-dimensional morphology of large samples, including intact human organs, is possible with X-ray microtomography (using laboratory or synchrotron sources). Preparation of large samples for high-resolution imaging, however, is challenging due to limitations such as sample shrinkage, insufficient contrast, movement of the sample and bubble formation during mounting or scanning. Here, we describe the preparation, stabilization, dehydration and mounting of large soft-tissue samples for X-ray microtomography. We detail the protocol applied to whole human organs and hierarchical phase-contrast tomography at the European Synchrotron Radiation Facility, yet it is applicable to a range of biological samples, including complete organisms. The protocol enhances the contrast when using X-ray imaging, while preventing sample motion during the scan, even with different sample orientations. Bubbles trapped during mounting and those formed during scanning (in the case of synchrotron X-ray imaging) are mitigated by multiple degassing steps. The sample preparation is also compatible with magnetic resonance imaging, computed tomography and histological observation. The sample preparation and mounting require 24–36 d for a large organ such as a whole human brain or heart. The preparation time varies depending on the composition, size and fragility of the tissue. Use of the protocol enables scanning of intact organs with a diameter of 150 mm with a local voxel size of 1 μm. The protocol requires users with expertise in handling human or animal organs, laboratory operation and X-ray imaging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Image data used to create the figures present in this protocol paper are publicly available from the ESRF data repository (https://human-organ-atlas.esrf.eu) or from the corresponding authors.
Code availability
The sCT data were reconstructed using a custom code written in MATLAB 2017 available on GitHub (https://github.com/HiPCTProject/Tomo_Recon) and the software package PyHST2 (https://software.pan-data.eu/software/74/pyhst2). VGSTUDIO MAX 3.5 (Volume Graphics) was used for volume rendering.
References
Alho, E. J. L. et al. High thickness histological sections as alternative to study the three-dimensional microscopic human sub-cortical neuroanatomy. Brain Struct. Funct. 223, 1121–1132 (2018).
Kofler, L. et al. Three‐dimensional histology vs. serial section histology in the treatment of primary basal cell carcinoma: a randomized, prospective, blinded study of 569 tumours. J. Eur. Acad. Dermatol. Venereol. 35, 1323–1330 (2021).
Pichat, J., Iglesias, J. E., Yousry, T., Ourselin, S. & Modat, M. A survey of methods for 3D histology reconstruction. Med. Image Anal. 46, 73–105 (2018).
Eberle, A. L. & Zeidler, D. Multi-beam scanning electron microscopy for high-throughput imaging in connectomics research. Front. Neuroanat. 12, 112 (2018).
Hildebrand, D. G. C. et al. Whole-brain serial-section electron microscopy in larval zebrafish. Nature 545, 345–349 (2017).
Miranda, K., Girard-Dias, W., Attias, M., de Souza, W. & Ramos, I. Three dimensional reconstruction by electron microscopy in the life sciences: an introduction for cell and tissue biologists. Mol. Reprod. Dev. 82, 530–547 (2015).
Tainaka, K. et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924 (2014).
Zhao, S. et al. Cellular and molecular probing of intact human organs. Cell 180, 796–812.e19 (2020).
Cai, R. et al. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections. Nat. Neurosci. 22, 317–327 (2019).
Pailhé, R. et al. Qualitative and quantitative assessment of cartilage degeneration using full-field optical coherence tomography ex vivo. Osteoarthr. Cartil. 26, 285–292 (2018).
Raghunathan, R., Singh, M., Dickinson, M. E. & Larin, K. V. Optical coherence tomography for embryonic imaging: a review. J. Biomed. Opt. 21, 1 (2016).
Lefort, C. A review of biomedical multiphoton microscopy and its laser sources. J. Phys. Appl. Phys. 50, 423001 (2017).
Disney, C. M., Lee, P. D., Hoyland, J. A., Sherratt, M. J. & Bay, B. K. A review of techniques for visualising soft tissue microstructure deformation and quantifying strain ex vivo: soft tissue microstructure deformation and quantifying strain. J. Microsc. 272, 165–179 (2018).
Schueder, F. et al. Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT. Nat. Commun. 8, 2090 (2017).
Goth, W., Lesicko, J., Sacks, M. S. & Tunnell, J. W. Optical-based analysis of soft tissue structures. Annu. Rev. Biomed. Eng. 18, 357–385 (2016).
Edlow, B. L. et al. 7 Tesla MRI of the ex vivo human brain at 100 micron resolution. Sci. Data 6, 244 (2019).
Bouazizi, K. et al. Differentiation and quantification of fibrosis, fat and fatty fibrosis in human left atrial myocardium using ex vivo MRI. PLoS ONE 13, e0205104 (2018).
Verleden, S. E. et al. Small airway loss in the physiologically ageing lung: a cross-sectional study in unused donor lungs. Lancet Respir. Med. 9, 167–174 (2021).
Withers, P. J. et al. X-ray computed tomography. Nat. Rev. Methods Primers 1, 18 (2021).
Mastrogiacomo, M., Campi, G., Cancedda, R. & Cedola, A. Synchrotron radiation techniques boost the research in bone tissue engineering. Acta Biomater. 89, 33–46 (2019).
Longo, E. et al. 3D spatial distribution of nanoparticles in mice brain metastases by X-ray phase-contrast tomography. Front. Oncol. 11, 554668 (2021).
Hwu, Y., Margaritondo, G. & Chiang, A.-S. Q&A: why use synchrotron x-ray tomography for multi-scale connectome mapping? BMC Biol. 15, 122 (2017).
Yokawa, K. et al. Synchrotron radiation-based X-ray phase-contrast imaging of the aortic walls in acute aortic dissection. JVS Vasc. Sci. 1, 81–91 (2020).
Liu, Y., Nelson, J., Holzner, C., Andrews, J. C. & Pianetta, P. Recent advances in synchrotron-based hard x-ray phase contrast imaging. J. Phys. Appl. Phys. 46, 494001 (2013).
Gonzalez-Tendero, A. et al. Whole heart detailed and quantitative anatomy, myofibre structure and vasculature from X-ray phase-contrast synchrotron radiation-based micro computed tomography. Eur. Heart J. Cardiovasc. Imaging 18, 732–741 (2017).
Planinc, I. et al. Comprehensive assessment of myocardial remodeling in ischemic heart disease by synchrotron propagation based X-ray phase contrast imaging. Sci. Rep. 11, 14020 (2021).
Chin, A.-L. et al. A synchrotron X-ray imaging strategy to map large animal brains. Chin. J. Phys. 65, 24–32 (2020).
Endrizzi, M. X-ray phase-contrast imaging. Nucl. Instrum. Methods Phys. Res. Sect. Accel. Spectrometers Detect. Assoc. Equip. 878, 88–98 (2018).
Schulz, G. et al. High-resolution tomographic imaging of a human cerebellum: comparison of absorption and grating-based phase contrast. J. R. Soc. Interface 7, 1665–1676 (2010).
Shinohara, G. et al. Three dimensional visualization of human cardiac conduction tissue in whole heart specimens by high-resolution phase-contrast CT imaging using synchrotron radiation. World J. Pediatr. Congenit. Heart Surg. 7, 700–705 (2016).
Westöö, C. et al. Distinct types of plexiform lesions identified by synchrotron-based phase-contrast micro-CT. Am. J. Physiol. Lung Cell. Mol. Physiol. 321, L17–L28 (2021).
Borisova, E. et al. Micrometer-resolution X-ray tomographic full-volume reconstruction of an intact post-mortem juvenile rat lung. Histochem. Cell Biol. 155, 215–226 (2021).
Broche, L. et al. Individual Airway Closure Characterized In Vivo by Phase-Contrast CT Imaging in Injured Rabbit Lung. Crit. Care Med. 47, e774–e781 (2019).
Dutel, H. et al. Neurocranial development of the coelacanth and the evolution of the sarcopterygian head. Nature 569, 556–559 (2019).
Mansuit, R. et al. Development and growth of the pectoral girdle and fin skeleton in the extant coelacanth Latimeria chalumnae. J. Anat. 236, 493–509 (2020).
Walsh, C. L. et al. Imaging intact human organs with local resolution of cellular structures using hierarchical phase-contrast tomography. Nat. Methods https://doi.org/10.1038/s41592-021-01317-x (2021).
Dias, C. S. B., Neto, D. P. A., Baraldi, G. L. & Fonseca, M. C. Comparative analysis of sample preparation protocols of soft biological tissues for morphometric studies using synchrotron-based X-ray microtomography. J. Synchrotron Radiat. 26, 2013–2023 (2019).
Li, T., Schreibmann, E., Yang, Y. & Xing, L. Motion correction for improved target localization with on-board cone-beam computed tomography. Phys. Med. Biol. 51, 253–267 (2006).
Sun, T., Kim, J.-H., Fulton, R. & Nuyts, J. An iterative projection-based motion estimation and compensation scheme for head x-ray CT. Med. Phys. 43, 5705–5716 (2016).
Topal, E. et al. Multi-scale X-ray tomography and machine learning algorithms to study MoNi4 electrocatalysts anchored on MoO2 cuboids aligned on Ni foam. BMC Mater. 2, 5 (2020).
Topal, E., Löffler, M. & Zschech, E. Deep learning-based inaccuracy compensation in reconstruction of high resolution XCT data. Sci. Rep. 10, 7682 (2020).
Burger, M. et al. A variational reconstruction method for undersampled dynamic x-ray tomography based on physical motion models. Inverse Probl. 33, 124008 (2017).
Patzelt, M. et al. Ethanol fixation method for heart and lung imaging in micro-CT. Jpn. J. Radiol. 37, 500–510 (2019).
Ackermann, M. et al. The bronchial circulation in COVID-19 pneumonia. Am. J. Respir. Crit. Care Med. 205, 121–125 (2022).
Ackermann, M. et al. The fatal trajectory of pulmonary COVID-19 is driven by lobular ischemia and fibrotic remodelling. eBioMedicine 85, 104296 (2022).
Rodgers, G. et al. Impact of fixation and paraffin embedding on mouse brain morphology: a synchrotron radiation-based tomography study. In Proc. SPIE Developments in X-Ray Tomography XIII (eds Müller, B. & Wang, G.) 27 (SPIE, 2021); https://doi.org/10.1117/12.2595144
Rodgers, G. et al. Virtual histology of an entire mouse brain from formalin fixation to paraffin embedding. Part 1: data acquisition, anatomical feature segmentation, tracking global volume and density changes. J. Neurosci. Methods 364, 109354 (2021).
Gusnard, D. & Kirschner, R. H. Cell and organelle shrinkage during preparation for scanning electron microscopy: effects of fixation, dehydration and critical point drying. J. Microsc. 110, 51–57 (1977).
Metscher, B. D. MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev. Dyn. 238, 632–640 (2009).
Shirai, R. et al. Enhanced renal image contrast by ethanol fixation in phase-contrast X-ray computed tomography. J. Synchrotron Radiat. 21, 795–800 (2014).
Wilke, H.-J., Krischak, S. & Claes, L. E. Formalin fixation strongly influences biomechanical properties of the spine. J. Biomech. 29, 1629–1631 (1996).
Rouleau, L., Tremblay, D., Cartier, R., Mongrain, R. & Leask, R. L. Regional variations in canine descending aortic tissue mechanical properties change with formalin fixation. Cardiovasc. Pathol. 21, 390–397 (2012).
Vesper, E. O., Hammond, M. A., Allen, M. R. & Wallace, J. M. Even with rehydration, preservation in ethanol influences the mechanical properties of bone and how bone responds to experimental manipulation. Bone 97, 49–53 (2017).
Madi, K. et al. In situ characterization of nanoscale strains in loaded whole joints via synchrotron X-ray tomography. Nat. Biomed. Eng. 4, 343–354 (2020).
Gignac, P. M. et al. Diffusible iodine‐based contrast‐enhanced computed tomography (diceCT): an emerging tool for rapid, high‐resolution, 3‐D imaging of metazoan soft tissues. J. Anat. 228, 889–909 (2016).
Koç, M. M., Aslan, N., Kao, A. P. & Barber, A. H. Evaluation of X‐ray tomography contrast agents: a review of production, protocols, and biological applications. Microsc. Res. Tech. 82, 812–848 (2019).
Metscher, B. D. MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol. 9, 11 (2009).
Mittone, A. et al. Multiscale pink-beam microCT imaging at the ESRF-ID17 biomedical beamline. J. Synchrotron Radiat. 27, 1347–1357 (2020).
Xian, R. P. et al. A multiscale X-ray phase-contrast tomography dataset of a whole human left lung. Sci. Data 9, 264 (2022).
Saccomano, M. et al. Synchrotron inline phase contrast µCT enables detailed virtual histology of embedded soft-tissue samples with and without staining. J. Synchrotron Radiat. 25, 1153–1161 (2018).
Strotton, M. C. et al. Optimising complementary soft tissue synchrotron X-ray microtomography for reversibly-stained central nervous system samples. Sci. Rep. 8, 12017 (2018).
Wereszczyńska, B. Alcohol-fixed specimens for high-contrast post-mortem MRI. Forensic Imaging 25, 200449 (2021).
Roebroeck, A., Miller, K. L. & Aggarwal, M. Ex vivo diffusion MRI of the human brain: technical challenges and recent advances. NMR Biomed. 32, e3941 (2019).
Shepherd, T. M., Thelwall, P. E., Stanisz, G. J. & Blackband, S. J. Aldehyde fixative solutions alter the water relaxation and diffusion properties of nervous tissue: aldehyde fixation alters tissue mri properties. Magn. Reson. Med. 62, 26–34 (2009).
Wilke, J. et al. Appraising the methodological quality of cadaveric studies: validation of the QUACS scale. J. Anat. 226, 440–446 (2015).
Paganin, D., Mayo, S. C., Gureyev, T. E., Miller, P. R. & Wilkins, S. W. Simultaneous phase and amplitude extraction from a single defocused image of a homogeneous object. J. Microsc. 206, 33–40 (2002).
Mirone, A., Brun, E., Gouillart, E., Tafforeau, P. & Kieffer, J. The PyHST2 hybrid distributed code for high speed tomographic reconstruction with iterative reconstruction and a priori knowledge capabilities. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. 324, 41–48 (2014).
Gürsoy, D., De Carlo, F., Xiao, X. & Jacobsen, C. TomoPy: a framework for the analysis of synchrotron tomographic data. J. Synchrotron Radiat. 21, 1188–1193 (2014).
Lyckegaard, A., Johnson, G. & Tafforeau, P. Correction of ring artifacts in X-ray tomographic images. Int J. Tomo Stat. 18, 1–9 (2011).
Oishi, H. et al. Ex vivo lung CT findings may predict the outcome of the early phase after lung transplantation. PLoS One 15, e0233804 (2020).
Verleden, S. E. et al. Radiological analysis of unused donor lungs: a tool to improve donor acceptance for transplantation? Am. J. Transplant. 17, 1912–1921 (2017).
Ross, M. H. & Pawlina, W. Histology: A Text and Atlas: With Correlated Cell and Molecular Biology (Wolters Kluwer, 2020).
Acknowledgements
We thank S. Bayat (INSERM) for help during the test phase, P. Masson (LADAF) for dissections of donors’ bodies, H. Reichert (ESRF) and R. Tori for general support of the project and E. Boller, C. Muzelle, R. Homs, C. Jarnias, F. Cianciosi, P. Vieux, P. Cook, L. Capasso and A. Mirone for their help in setup developments and improvements. We also thank R. Engelhardt, A. Muller Brechlin, C. Petzold, N. Kroenke and M. Kuhel for help with histology and autopsies. This project has been made possible in part by grant number 2020-225394 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation and grant number CZIF2021-006424 from the Chan Zuckerberg Initiative Foundation, The ESRF funding proposals md1252 and md1290, the Royal Academy of Engineering (CiET1819/10). P.D.L. and C.L.W. gratefully acknowledge funding from the MRC (MR/R025673/1). M.A. acknowledges grants from the National Institutes of Health (HL94567 and HL134229). This work was supported by the German Registry of COVID-19 Autopsies (DeRegCOVID, www.DeRegCOVID.ukaachen.de; supported by the Federal Ministry of Health: ZMVI1-2520COR201) and the Federal Ministry of Education and Research as part of the Network of University Medicine (DEFEAT PANDEMIcs, 01KX2021).
Author information
Authors and Affiliations
Contributions
P.T., P.D.L., D.D.J., M.A., C.L.W. and W.L.W. conceptualized the project and designed experiments. M.A., C.W., P.T., A.B., S.E.V., C.L.W. and J.B. performed and contributed to autopsies and sample preparation. P.T. designed and built instrumentation and performed HiP-CT imaging. S.M. designed sample holders. P.T. designed and implemented tomographic reconstruction methods. J.B., P.T., C.L.W. and P.D.L. wrote the paper. All authors assisted in reviewing and revising the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Ali Erturk, Stuart Stock and Hiroki Ueda for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Walsh, C. L. et al. Nat. Methods 18, 1532–1541 (2021): https://doi.org/10.1038/s41592-021-01317-x
Ackermann, M. et al. Am. J. Respir. Crit. Care Med. 205, 121–125 (2022): https://doi.org/10.1164/rccm.202103-0594IM
Ackermann, M. et al. eBioMedicine 85, 104296 (2022): https://doi.org/10.1016/j.ebiom.2022.104296
Extended data
Extended Data Fig. 1 Challenges faced during the development of the technique and their solutions.
This protocol was developed in an iterative manner overcoming all the different challenges related to soft tissue imaging, dose deposition and local tomography.
Extended Data Fig. 2 Basic information on human organs used in this protocol paper.
The heart, kidney, and brain data are present in the Human Organ Atlas (https://human-organ-atlas.esrf.eu). The liver data are not included in the Human Organ Atlas because of the large number of artifacts present in the images due to bubble formation during scanning. However, the images are available on request from the corresponding authors. The formation of these bubbles occurred because a crash in the beamline software caused the beam to remain in the same position for several hours, exceeding the organ’s dose threshold. Other livers have been scanned without artifacts.
Extended Data Fig. 3 List of human organs and biological samples compatible with this method and the preparation time for each step.
The process described in this protocol paper should work for all major organs, but only the organs listed here have been tested. The maximum degassing times listed here are specific to our vacuum degassing setup and are subject to change depending on the pump, the volume to be pumped, the pumping section, the quantity of gas to be evacuated, and the paths that the gas can take to leave the sample. As such, they should be adapted to each user vacuum setup by looking at the bubbling intensity as explained in Step 4A(ii) of the protocol.
Extended Data Fig. 4 The two types of large leak-proof container in PET in the custom-made container holders.
The left leak-proof container is a Medline Scientific container (cat. no. 129-0592) of 3 L. The right leak-proof container is a Lock & Lock container (cat. no. INL-403) of 2.1 L. Both are inside a custom-made container holder described in Extended Data Fig. 5.
Extended Data Fig. 5 Custom-made container holder drawings and 3D rendering.
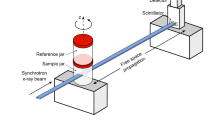
Two containers are represented inside the custom-made container holder. The bottom container contains the sample while the top container, referred to as the reference container, is used for the flat-field correction.
Extended Data Fig. 6 Beamline parameters of all the organ scans presented in this paper.
The optimal scanning settings are strongly dependent on the sample and on the beamline setup and equipment. The parameters presented here are given as examples. Quarter acquisition means one scan in half-acquisition plus one annular scan to increase the lateral field of view. Mo = molybdenum.
Supplementary information
Supplementary Video 1
Example of a sample mounted with insufficiently compacted agar, allowing rotation upon a slight movement.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Brunet, J., Walsh, C.L., Wagner, W.L. et al. Preparation of large biological samples for high-resolution, hierarchical, synchrotron phase-contrast tomography with multimodal imaging compatibility. Nat Protoc 18, 1441–1461 (2023). https://doi.org/10.1038/s41596-023-00804-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-023-00804-z
This article is cited by
-
Virtuelle Histopathologie des Pankreas: 3D-Einblicke mittels synchrotronbasierter Bildgebung
Die Pathologie (2026)
-
3D imaging of the human temporal bone by X-ray phase-contrast tomography
npj Imaging (2025)
-
Mapping the arterial vascular network in an intact human kidney using hierarchical phase-contrast tomography
npj Imaging (2025)
-
Revealing the unseen: 3D synchrotron X-Ray imaging of uterine vasculature in adenomyosis
Angiogenesis (2025)
-
Deep learning for 3D vascular segmentation in hierarchical phase contrast tomography: a case study on kidney
Scientific Reports (2024)