Abstract
Carvacrol (CAR) is a plant extract that has been reported to enhance antioxidant activity in animals. However, the effect of CAR on the intestinal health of rabbits is poorly understood. Here, we investigated whether CAR exerts protective effects on the intestinal health of rabbits following lipopolysaccharide (LPS) challenge and whether these effects were mediated via the reduction of intestinal inflammation and the regulation of the intestinal flora. Intestinal damage was assessed in LPS-challenged rabbits treated or not with CAR. The serum levels of inflammatory factors were assessed by enzyme-linked immunosorbent assay. Histopathological changes in the ileum and cecum were examined using hematoxylin and eosin staining. The relative gene expression levels of inflammatory factors and tight junction proteins in the rabbit cecum were determined by qRT-PCR. High-throughput sequencing analysis of the microbial 16S rRNA gene was performed using the Illumina NovaSeq Platform. The results showed that CAR can prevent intestinal inflammation and damage as well as mitigate gut dysbiosis in rabbits following LPS challenge. Our study provides a theoretical reference for the application of dietary CAR in rabbit production.
Similar content being viewed by others
Introduction
The intestinal tract is the main organ responsible for nutrient digestion and absorption as well as the first line of defense against pathogenic microorganisms and toxins1. This line of defense encompasses both mechanical and biological barriers. The mechanical barrier comprises intestinal epithelial cells, which separate the intestinal lumen from intestinal tissue, and tight junctions (TJs), consisting of proteins such as zonula occludens-1 (ZO-1), occludin, and claudin. In addition to organizing epithelial cells into apical and basal compartments and maintaining epithelial cell polarity, TJ proteins also regulate the permeability of cells to ions, water, and nutrients, thus preserving intestinal homeostasis2,3,4. The biological barrier, meanwhile, comprises a micro-ecosystem consisting of resident bacteria that are interdependent and exist in a symbiotic relationship with their host. Following pathological insult, however, this biological barrier can be disrupted, resulting in the translocation of bacteria and endotoxins from the intestinal lumen to intestinal tissue or the circulation, consequently leading to infection or intestinal inflammation5. During the breeding process, animals are subjected to a variety of stresses, such as uncomfortable living environments, differences in feed, and transportation, which affect the structure of their intestinal flora6,7,8. This dysbiosis is mainly manifested as an increase in the abundance of harmful microorganisms in the intestine and the production and release of large amounts of lipopolysaccharide (LPS), which is detrimental to health9.
LPS is a constituent of the outer wall of Gram-negative bacteria10 and comprises an O-antigen, a core oligosaccharide, and a lipid anchor (lipid A)11. Lipid A is the most conserved region of LPS. It is made up of six saturated acyl chains, which are double phosphorylated and are responsible for the proinflammatory effects of LPS. Furthermore, the abundant anion groups present in the core region of lipid A combine with divalent cations (Mg2+ and Ca2+), which increases lipid A molecular density, improves bacterial outer membrane stability, reduces permeability, and effectively resists the destructive effects of bacteriophages or bacteriocins12. In the body, LPS is recognized by LPS-binding protein (LBP), and is subsequently transported to the surface of immune cells, where it activates the Toll-like receptor 4 (TLR4) signaling pathway, leading to the production of cytokines and, consequently, an inflammatory response13. In vitro, 1 μg/mL LPS was shown to significantly downregulate the expression of occludin and claudin 1 in porcine intestinal epithelial cells (IPEC-J2)14, while in Caco-2 cells, treatment with 10 μg/mL LPS for 24 h significantly reduced the mRNA levels of ZO-1, claudin 1, and occludin, and activated the proinflammatory TLR4/NF-κB signaling pathway15. In vivo, intramuscular injection of LPS (500 μL/kg body weight) in broilers significantly increased the mRNA expression of interleukin 1 beta (IL-1β), IL-6, IL-8, and tumor necrosis factor alpha (TNF-α) in the small intestine, and activated the TLR/NF-κB pathway 2 h post-injection16.
Antibiotics act by killing bacteria or inhibiting their growth, thereby reducing the secretion of harmful substances produced by these bacteria, such as LPS, and mitigating their negative effects. Accordingly, antibiotics have long been used to inhibit bacterial proliferation and treat bacterial infections. However, antibiotics and their residues are associated with side effects, such as resistance and variability of effects and the destruction of animal intestinal microecology17,18. This highlights the need for the development of alternatives to antibiotics in the animal industry to alleviate the impact of harmful bacteria19. Among the many alternatives, plants and their constituents have become research hotspots owing to their antibacterial effects20, as well as their ability to alleviate inflammatory responses21 and improve intestinal health22.
The use of plant-based feed additives to alleviate or treat intestinal inflammation or intestinal injury caused by LPS has been widely studied in several species but not in rabbits. Carvacrol (CAR) is a monophenol widely present in various plants, fruits, and volatile oils, including oregano, thyme, and olive fruit23. Studies have shown that CAR has significant anticancer24 and antioxidative properties25; however, it is unclear whether CAR supplementation can protect the intestine of rabbits from LPS challenge. In this study, we hypothesized that dietary CAR can alleviate intestinal injury induced by LPS in rabbits by alleviating intestinal inflammation and regulating intestinal microflora.
Materials and methods
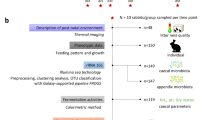
Experimental animals, diet, and design
All experiments were performed in accordance with the ARRIVE guidelines and were carried out following the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines; EU Directive 2010/63/EU for animal experiments; and the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised 1978) and were approved by the Ethics Committee of Animal Experimentation of Hebei Agricultural University (Protocol 2020006).
A total of 40-day-old Ira rabbits (20 male, 20 female; purchased from China Hebei Xingtai Weixian Kangming culture Co., LTD) of similar weight (1.25 kg) were placed in battery cages, one rabbit per cage. During the experiment, the temperature and photoperiod were maintained according to commercial conditions (20–23 °C and 12-h:12-h light/dark cycle, respectively). Each cage was equipped with one feeder and one nipple drinker. Each treatment group contained ten replicates (cages), with one sampling unit (rabbit) per cage. The basal diet was formulated to meet the nutrient and energy requirements of rabbits according to De Blas and Wiseman26. The CAR diet was formulated by adding 25 g/t CAR to the basal diet. CAR (purity ≥ 98%) was purchased from Fidi Feed Technology Co., Ltd (Beijing, China). All the diets were pelleted to a size of 4 × 10 mm (diameter × length). The composition and nutrient levels of the basal diet are shown in Table 1. The trial lasted for 28 days.
A 2 (with or without CAR) × 2 (with or without LPS) factorial arrangement of treatments in a completely randomized design was used in this study. Individually housed rabbits were randomly assigned to four treatment groups, as follows: A CON group (basal diet + saline), a CAR group (basal diet + carvacrol + saline), a LPS group (basal diet + LPS), and a CAR + LPS group (basal diet + CAR + LPS).
On day 28 of the trial (68 days of age), the rabbits assigned to the LPS groups were intramuscularly (thigh muscle) injected with 1 mL (200 μg/kg body weight) of Escherichia coli LPS (Escherichia coli O55: B5, L2880, Sigma-Aldrich). The remaining rabbits were injected with 1 mL of 0.9% saline (all injections were performed simultaneously at 08:00 h and all the animals were euthanized and sampled simultaneously at 12:00 h).
Sample collection
All the rabbits from each group were sampled 4 h post-injection. Blood samples were drawn from the ear vein using a sterile blood-sampling needle and centrifuged at 3500×g for 10 min at 4 °C to obtain serum. Serum samples were then immediately stored at − 80 °C until further analysis. Sodium pentobarbital was used as an anesthetic and was administered intravenously. The ileal and cecal segments were collected, washed, and immediately immersed in a 4% paraformaldehyde solution for histological examination. The cecum and cecal chyme were stored at − 80 °C for further analysis.
Measurement of inflammatory indicators in serum
Serum IL-1β, IL-6, IL-8, and TNF-α levels were detected using the respective enzyme-linked immunosorbent assay (ELISA) kits (Jiangsu Meimian Industrial Co., Ltd, Jiangsu, China).
Structural morphology of the intestine
The ileal and cecal segments were fixed in a 4% paraformaldehyde solution for 24 h, dehydrated, embedded in paraffin, cut into 5-μm-thick sections using a microtome (Kedee KD2508, Kedee BInstruments and Equipment Co., Ltd, Jinhua, China), fixed on slides, and finally stained with hematoxylin and eosin. Images of the ileum and cecum were acquired with a Nikon Eclipse E100 microscope (Nikon Inc., Tokyo, Japan) and analyzed with CaseViewer software (version 2.4, ServiceBio, Wuhan, China). Five sections of the slice were randomly selected from each sample for assessment of morphology. The average of five values from each rabbit was used for statistical analysis.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
The mRNA expression of ZO-1, occludin, claudin-1, IL-1β, IL-6, IL-8, TNF-α, Toll-like receptor 2 (TLR2), TLR4, myeloid differentiation primary response 88 (MYD88), nuclear factor kappa B (NF-κB), mitogen-activated protein kinase 1 (MAPK1), MAPK8, and MAPK14 was detected by qRT-PCR. The primers used in this study are listed in Table 2. The mRNA sequences were obtained from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/cgi-bin/bank). Total RNA was extracted from the cecum using Trizol as described by the manufacturer (TaKaRa, Dalian, China)27. Total RNA concentration and purity were determined by measuring the optical density at 260 and 280 nm. The extracted mRNA was reverse transcribed into cDNA according to the manufacturer's instructions (TaKaRa, Dalian, China). The reverse transcription conditions were 42 °C for 40 min and 65 °C for 10 min. Quantitative PCR involved a pre-denaturation step at 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. A melt curve analysis was applied at the end of the run to determine specific product amplification. Relative gene expression levels were evaluated using the 2−ΔΔCT method16 (CAR-treated samples versus the non-supplemented [control] samples) after normalization to the housekeeping gene.
Short-chain fatty acid (SCFA) analysis
Cecal digesta (0.5 g) was diluted (dilution factor of 3) and centrifuged at 12,000 rpm for 10 min at 4 °C. Then, 1 mL of supernatant was mixed with 0.2 mL of 25% ammonium metaphosphate solution containing the internal standard 2-ethylbutyric acid (2 EB), the mixture was soaked in ice for 40 min and then centrifuged at 12,000 rpm for 10 min at 4 °C to remove protein precipitates. The supernatant was used for SCFA analysis using a gas mass spectrometer (GC2014, Shimadzu Inc., Tokyo Japan).
16S rRNA gene sequencing and analysis
Total genomic DNA was extracted from cecal digesta using the OMEGA Soil DNA Kit (M5635-02; Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s instructions and was stored at − 20 °C before analysis. The quantity and quality of the extracted DNA were assessed using a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively. PCR amplification of the V3–V4 region of the bacterial 16S rRNA gene was performed using forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The reverse primer contained a sample-specific 7-nucleotide barcode. Amplification was performed using the following program: 98 °C for 5 min, followed by 25 cycles of 98 °C for 30 s, 53 °C for 30 s, and 72 °C for 45 s, with a final extension at 72 °C for 5 min. The amplicons were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After individual quantification, the amplicons were pooled in equal amounts, and paired-end sequenced (2 × 250 bp) at Shanghai Personal Biotechnology Co., Ltd (Shanghai, China) on the Illumina NovaSeq Platform using the NovaSeq 6000 SP Reagent Kit (500 cycles).
Analysis of sequencing data was mainly performed using QIIME2 and R software (v3.2.0). To estimate microbial diversity in individual samples, alpha diversity indices, such as the Chao1 richness estimator, Shannon diversity index, Simpson index, and Faith’s PD, at the amplicon sequence variant (ASV) level were calculated using the ASV table in QIIME2. Principal coordinate analysis (PCA) was performed based on Bray–Curtis and UniFrac distance metrics.
Data analysis
All data were analyzed using Statistical Package for the Social Sciences (SPSS) ver. 21 and were expressed as means ± SEM. Differences among groups were examined using one-way analysis of variance (ANOVA) followed by Duncan’s multiple-range test. Differences were considered significant at P < 0.05.
Results
Levels of inflammatory cytokines in serum
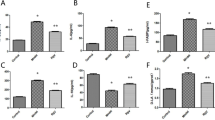
As shown in Fig. 1, LPS challenge increased the levels of IL-1β, IL-6, IL-8, and TNF-α in serum (P < 0.05), whereas the addition of CAR to the diet reduced the serum levels of IL-1β (P < 0.05). Additionally, dietary CAR alleviated the increase in IL-1β, IL-6, IL-8, and TNF-α levels resulting from LPS challenge (P < 0.05).
Expression of inflammatory genes in the cecum
To determine the genetic basis of the CAR-mediated improvement of the inflammatory reaction in LPS-challenged rabbits, the expression of proinflammatory genes and genes involved in the inflammatory signaling pathway was investigated by qPCR (Fig. 2). LPS treatment upregulated the expression of the IL-1β, IL-6, IL-8, TNF-α, TLR2, TLR4, MYD88, NF-κB, MAPK1, MAPK8, and MAPK14 genes (P < 0.05). In contrast, dietary CAR significantly reduced cecal MYD88 and NF-κB p65 expression (P < 0.05) and suppressed the LPS challenge-mediated increase in IL-1β, IL-6, IL-8, TNF-α, TLR2, TLR4, MYD88, NF-κB, MAPK1, MAPK8, and MAPK14 expression levels (P < 0.05). These results indicated that dietary CAR ameliorated cecal inflammation in LPS-challenged rabbits.
Intestinal morphology
We next assessed the effect of dietary CAR on intestinal morphology in response to LPS stimulation. As shown in Fig. 3, in the normal condition, ileal villi are closely connected. However, LPS stimulation significantly altered the morphology of the ileum. Compared with the controls, the villi were fractured and narrower, and the intestinal mucosa was significantly thinner. Under the influence of dietary CAR, LPS treatment did not result in any obvious injury to the rabbit ileum. Under normal conditions, the cecal mucus layer is closely connected to the sarcolemma. After LPS stimulation, the cecal sarcolemma was significantly damaged, and the space between the mucosal layer and the sarcolemma was enlarged, indicative of mucosa shedding. However, under the influence of dietary CAR, LPS did not cause obvious damage to the cecum of rabbits.
The effects of the different treatments on intestinal morphology are presented in Fig. 4. LPS challenge led to a decrease in ileal villus height (VH), villus width (VW), and mucosal thickness (MT) (P < 0.05). Meanwhile, dietary CAR significantly increase ileal VH (P < 0.05) and mitigated the negative effects of LPS treatment on ileum VH, VW, and MT and cecal DIG and MT (P < 0.05). These results suggested that CAR supplementation promoted the recovery of intestinal morphology following LPS challenge.
Expression of TJ genes in the cecum
The effects of the different treatments on the expression of TJ genes in the cecum of rabbits are shown in Fig. 5. LPS challenge significantly reduced the cecal expression of the ZO-1, claudin-1, and occludin genes (P < 0.05), whereas the opposite was observed with dietary CAR (P < 0.05). Furthermore, the addition of CAR to the diet of rabbits blocked the negative effects of LPS challenge on the expression of the three genes (P < 0.05). These results suggested that CAR supplementation fostered the recovery of intestinal barrier function in rabbits following LPS insult.
SCFAs in cecal digesta
The effects of treatment on SCFAs contents are presented in Fig. 6. LPS challenge led to a decrease in acetate, propionate, and butyrate contents (P < 0.05). However, dietary CAR significantly increased butyrate levels (P < 0.05) and alleviated the LPS challenge-mediated reduction in the contents of acetate, propionate, and butyrate (P < 0.05). These results suggested that CAR supplementation reversed the negative effects of LPS on cecal SCFAs contents.
Quantification of cecal bacteria
High-throughput sequencing of the 16S rRNA gene was performed to reveal the role of the cecal microbiota in the CAR-mediated recovery of intestinal injury induced by LPS challenge. As seen in the Venn diagram in Fig. 7 11,675, 12,828, 11,504, and 13,231 operational taxonomic units (OTUs) were identified in the CON, CAR, LPS, and CAR + LPS groups, respectively; 1212 OTUs were found to be shared among the four groups (Fig. 7). The microbial diversity within an individual sample was assessed by the Chao1, Faith’s PD, Shannon, and Simpson indices, but no alteration among all the groups was observed (Table 3). Next, PCA was performed on the OTUs to assess the similarities and differences between samples and groups (Fig. 8). The results revealed that the cecal microbiota in the LPS group was separated from that in the CON, CAR, and CAR + LPS groups. Taxonomic profiling indicated that Firmicutes accounted for most of the intestinal bacteria of rabbits (Fig. 9). No differences in relative bacterial abundance were observed among all the groups at the phylum level (Table 4). The relative abundance of the 20 dominant genera in each group at the genus level was also analyzed. Ruminococcus and Oscillospira were found to be the main genera in the four treatment groups (Fig. 10). As shown in Table 5, LPS challenge significantly reduced Ruminococcus abundance in the intestine of rabbits (P < 0.05). In contrast, CAR provision in the diet increased Ruminococcus abundance (P < 0.05) and alleviated the LPS stimulation-induced reduction in the relative abundance of members of this genus (P < 0.05). Combined, these results suggested that CAR supplementation mitigates the LPS stimulation-induced dysbiosis in the cecum of rabbits.
The correlations among inflammatory factors, TJ proteins, and gut microbiota
To further understand the role of the gut microbiota in the regulation of inflammation and TJ function, we performed a Spearman’s correlation analysis of the relationship among inflammatory factors, TJ proteins, and intestinal bacteria (Fig. 11). Eight microbial genera were found to be significantly correlated with inflammatory gene levels or the expression levels of genes coding TJ proteins. Among the genera upregulated by CAR, Ruminococcus was negatively correlated with the mRNA levels of IL-1β, IL-6, IL-8, TNF-α, TLR2, TLR4, MYD88, NF-κB, MAPK1, MAPK8, and MAPK14 (P < 0.05) and positively correlated (P < 0.05) with those of ZO-1, claudin 1, and occludin.
Discussion
The TLRs/MYD88/NF-κB pathway is the main pathway by which LPS induces inflammation in the intestine. TLRs are the major components of the innate immune system and their primary function is the recognition of intrinsic pathogen-associated molecules. LPS binds to and is recognized by TLRs, which is followed by the recruitment of the adaptor protein MYD8828. This leads to the activation of the NF-κB transcription factor, which stimulates the production and secretion of inflammatory factors such as TNF-α, IL-1β, IL-6, and IL-829, and, in turn, leads to intestinal damage30. In the present study, the expression of the TLR2 and TLR4 genes was significantly increased in the rabbit intestine upon LPS stimulation; however, the addition of CAR to the diet mitigated this upregulation. Studies have found that TLR2 recognizes LPS through its lipid A part31. When the mouse Tlr4 gene is mutated, it can no longer recognize LPS32, suggesting that CAR may reduce the ability of the intestine to recognize LPS by inhibiting the expression of TLR2 and/or TLR4, thereby reducing the inflammatory response. MYD88 acts as a downstream factor of TLRs. After recognizing LPS, TLRs bind to the TIR domain of MYD88, leading to its activation and, subsequently, to that of NF-κB29,33. Activated NF-κB migrates to the nucleus, where it stimulates the production of inflammatory factors via its function as a transcription factor34. In our study, CAR suppressed the LPS-induced upregulation of MYD88, NF-κB, TNF-α, IL-1β, IL-6, and IL-8, indicating that CAR inhibited the LPS-induced activation of MYD88 and the NF-κB pathway. Dietary CAR also reduced the secretion of TNF-α, IL-1β, IL-6, and IL-8, thereby alleviating the intestinal inflammatory response. TNF-α can activate the MAPK genes and thus promote the further release of inflammatory factors, leading to an inflammatory cascade35,36,37. Several studies have shown that CAR can also inhibit the activation of MAPK genes by directly inhibiting the phosphorylation of MAPK1, MAPK8, and MAPK1438,39,40, which blocks the release of inflammatory factors. In the present study, we found that the MAPK-encoding genes were downregulated by CAR supplementation, likely due to the reduced expression of TNFA, which mitigated the amplification of the inflammatory response. In summary, the mechanism by which CAR alleviates LPS-induced intestinal inflammation in rabbits may involve the inhibition of the activation of TLRs/NF-κB/TNF-α/MAPK pathway-related genes, leading to a decrease in the production and secretion of TNF-α, IL-1β, IL-6, and IL-8.
Intestinal integrity is crucial for the development of animals and intestinal damage often leads to feeding intolerance and weakened nutrient absorption capacity41. VH, VW, crypt depth, and MT are important indicators of intestinal integrity. Intestinal villi are formed by intestinal stem cells that continuously self-renew and migrate upward. The intestinal mucosa, composed of intestinal epithelial cells42, is made up of ring-shaped folds comprising numerous villi. In the present study, we found that CAR significantly suppressed the reduction in VH, VW, and ileal and cecal mucosa thickness in rabbits after LPS stimulation. These results indicated that CAR could reverse the intestinal injury induced by LPS. The intestinal mucosal barrier prevents the translocation of bacteria and macromolecules, such as LPS, from the intestinal lumen to intestinal tissue43. The mechanical barrier function of the intestinal mucosa is mediated by proteins such as ZO-1, occludin, and claudin that form TJs between epithelial cells44. Occludin acts directly on the ZO-1 protein, which serves as a scaffold between the former (and claudin) and the cytoskeleton45; claudin, meanwhile, plays a key role in intestinal wall charge selectivity and the resistance of TJs46. Cell culture studies have shown that TNF-α can induce the reorganization of ZO-1, occludin, and claudin in cells. During this process, some genes occur protein loss and mislocalization, which suggests that TNF-α can disrupt TJs, thereby negatively affecting intestinal barrier function47. Studies have reported that ZO-1 mislocalization disrupts the stability of TJ complexes in both rats and piglets48,49. In the present study, adding CAR to the diet alleviated the marked increase in TNF-α expression and the significant reduction in ZO-1, occludin, and claudin 1 genes expression in the rabbit intestine after LPS stimulation. This result suggested that CAR mitigates the inhibitory effects of LPS on the expression of ZO-1, occludin, and claudin 1 by inhibiting that of TNF-α, thus reducing intestinal permeability. As the first line of intestinal defense, when the mechanical barrier is disrupted, LPS can enter the circulation through the mucosal barrier, leading to the release of inflammatory factors50. In addition, the upregulation of TLRs51, TNF-α52, IL-853, NF-κB54, and MAPKs55 can promote apoptosis. Fukuda et al.56 showed that an increase in apoptosis reduced intestinal VH. In the present study, we found that CAR downregulated the LPS-induced increase in the expression of TLRs, TNF-α, IL-8, NF-κB, and MAPKs, suggesting that CAR can alleviate the intestinal injury induced by LPS by inhibiting the expression of these genes. The mechanism by which CAR alleviates LPS-induced intestinal injury in rabbits may involve the inhibition of the activation of TLRs/NF-κB/TNF-α/MAPK pathway-related genes. Increasing the expression of ZO-1, occludin, and claudin 1 and reducing the production of TLRs, TNF-α, IL-8, NF-κB, and MAPKs can alleviate the apoptosis of intestinal epithelial cells and intestinal stem cells, thereby reducing the destruction of the intestinal mucosa and villi and alleviating the LPS-induced intestinal damage. In this study, we demonstrated that CAR exerted a significant preventive effect on LPS-induced intestinal injury in rabbits. However, additional research is needed to further clarify this therapeutic effect of CAR.
A multitude of microorganisms colonize the gastrointestinal tract of animals, and these microorganisms interact with the intestinal epithelium, helping to maintain its integrity57. Intestinal microbes can benefit the host by regulating intestinal morphology58. In the present study, we found that LPS treatment modulated the structure of the cecal microbiota community in rabbits, which was similar to that previously reported21; however, this effect of LPS was alleviated by dietary CAR supplementation. Species-rich communities have been reported to enhance the stability of the intestinal microecology and reduce sensitivity to bacterial invasion and intestinal inflammation59. These results suggest that CAR may alleviate intestinal inflammation by regulating intestinal microbial community structure. In our study, CAR alleviated the LPS-induced reduction in the relative abundance of the genus Ruminococcus in the cecum of rabbits. Members of this genus are probiotics that promote gut health and play a crucial role in maintaining the metabolism of nutrients and microbial homeostasis60. In addition, Ruminococcus bacteria have been shown to produce SCFAs61, which are the main energy source of intestinal epithelial cells. SCFAs can promote the differentiation of intestinal epithelial cells as well as the development of intestinal villi, thereby maintaining intestinal morphology62. In addition, SCFAs provide energy for the differentiation of epithelial cells and consume 70% of intestinal oxygen63,64, thus generating a favorable environment for the growth and proliferation of Ruminococcus bacteria. The genus Ruminococcus was reported to be negatively correlated with inflammatory factors61. Sam et al.65 found that SCFAs can inhibit the secretion of inflammatory factors through the TLR pathway, which is consistent with the results of this study. Additionally, we found that Ruminococcus abundance and the relative mRNA expression levels of TJ proteins were positively correlated. Wan et al.66 demonstrated that SCFAs could significantly increase the relative expression of TJ protein-related genes by activating the AMPK signaling pathway, as well as inhibit the secretion of inflammatory factors, which also improved the relative expression levels of genes encoding TJ proteins. These results indicate that CAR can enhance the proliferation of Ruminococcus bacteria, which would be expected to lead to an increase in acetate, propionate, and butyrate contents in the cecum of rabbits. An increase in the levels of these SCFAs would subsequently result in a reduction of intestinal inflammation and the enhancement of the expression of intestinal TJ proteins in the cecum of rabbits, thereby alleviating the LPS-induced damage to intestinal morphology. These results provide a theoretical basis for understanding the relationship between CAR-mediated regulation of LPS-induced intestinal injury and the gut microbiota; however, further microbiome-based studies are needed to elucidate the mechanism underlying the anti-inflammatory effects of Ruminococcus bacteria.
Conclusion
In summary, we found that dietary CAR supplementation can alleviate and reverse LPS-induced intestinal injury in rabbits by reducing the inflammatory response and increasing the relative abundance of Ruminococcus bacteria. In addition, our results confirmed that CAR exerted an anti-inflammatory effect in LPS-treated rabbits, which may be related to the inhibition of the expression of genes in the TLRs/NF-κB/TNF-α/MAPK pathway (Supplementary Information).
Data availability
The datasets generated and/or analysed during the current study are available in the CNCB repository, GSA ID: CRA010835.
References
Yamada, T. et al. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat. Immunol. 17(6), 687–694 (2016).
Peterson, L. W. & Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 14(3), 141–153 (2014).
Liu, Z. et al. Aryl hydrocarbon receptor activation maintained the intestinal epithelial barrier function through Notch1 dependent signaling pathway. Int. J. Mol. Med. 41(3), 1560–1572 (2018).
Wilms, E. et al. Intestinal barrier function is maintained with aging—A comprehensive study in healthy subjects and irritable bowel syndrome patients. Sci. Rep. 10(1), 475 (2020).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464(7285), 59–65 (2010).
Xiong, Y., Yi, H., Wu, Q., Jiang, Z. & Wang, L. Effects of acute heat stress on intestinal microbiota in grow-finishing pigs, and associations with feed intake and serum profile. J. Appl. Microbiol. 128(3), 840–852 (2020).
Castillo, M. et al. Application of 16S rRNA gene-targetted fluorescence in situ hybridization and restriction fragment length polymorphism to study porcine microbiota along the gastrointestinal tract in response to different sources of dietary fibre. FEMS. Microbiol. Ecol. 59(1), 138–146 (2007).
Perry, E. et al. Effect of road transport on the equine cecal microbiota. J. Equine. Vet. Sci. 68, 12–20 (2018).
Torres, P. J. et al. Gut microbial diversity in women with polycystic ovary syndrome correlates with hyperandrogenism. J. Clin. Endocrinol. Metab. 103(4), 1502–1511 (2018).
Tang, N. et al. Dihydromyricetin suppresses TNF-α-induced NF-κB activation and target gene expression. Mol. Cell. Biochem. 422(1–2), 11–20 (2016).
Mazgaeen, L. & Gurung, P. Recent advances in lipopolysaccharide recognition systems. Int. J. Mol. Sci. 21(2), 379 (2020).
Molinaro, A. et al. Chemistry of lipid A: At the heart of innate immunity. Chemistry 21(2), 500–519 (2015).
Gao, J. & Guo, Z. Progress in the synthesis and biological evaluation of lipid A and its derivatives. Med. Res. Rev. 38(2), 556–601 (2018).
Wu, J. et al. Betaine attenuates LPS-induced downregulation of Occludin and Claudin-1 and restores intestinal barrier function. BMC. Vet. Res. 16(1), 75 (2020).
Lan, H. et al. Sinapic acid alleviated inflammation-induced intestinal epithelial barrier dysfunction in lipopolysaccharide-(LPS-) treated Caco-2 cells. Mediators. Inflamm. 8, 5514075 (2021).
Liu, S. D. et al. Effect of carvacrol essential oils on immune response and inflammation-related genes expression in broilers challenged by lipopolysaccharide. Poult. Sci. 98(5), 2026–2033 (2019).
Clark, S. et al. Extension Education Symposium: The future of biosecurity and antimicrobial use in livestock production in the United States and the role of extension. J. Anim. Sci. 90(8), 2861–2872 (2012).
Booth, A., Aga, D. S. & Wester, A. L. Retrospective analysis of the global antibiotic residues that exceed the predicted no effect concentration for antimicrobial resistance in various environmental matrices. Environ. Int. 141, 105796 (2020).
Kogut, M. H. Issues and consequences of using nutrition to modulate the avian immune response. J. Appl Poultry. Res. 26, 605–612 (2017).
Windisch, W., Schedle, K., Plitzner, C. & Kroismayr, A. Use of phytogenic products as feed additives for swine and poultry. J. Anim. Sci. 86(14), 140–148 (2008).
Kong, L., Wang, Z., Xiao, C., Zhu, Q. & Song, Z. Glycerol monolaurate attenuated immunological stress and intestinal mucosal injury by regulating the gut microbiota and activating AMPK/Nrf2 signaling pathway in lipopolysaccharide-challenged broilers. Anim. Nutr. 10, 347–359 (2022).
Ding, H., Cao, A., Li, H., Zhao, Y. & Feng, J. Effects of Eucommia ulmoides leaf extracts on growth performance, antioxidant capacity and intestinal function in weaned piglets. J. Anim. Physiol. Anim. Nutr. 104(4), 1169–1177 (2020).
Bruix, J. & Llovet, J. M. Major achievements in hepatocellular carcinoma. Lancet 373(9664), 614–616 (2009).
Jung, C. Y., Kim, S. Y. & Lee, C. Carvacrol targets AXL to inhibit cell proliferation and migration in non-small cell lung cancer cells. Anticancer. Res. 38(1), 279–286 (2018).
Zou, Y., Wang, J., Peng, J. & Wei, H. Oregano essential oil induces SOD1 and GSH expression through Nrf2 activation and alleviates hydrogen peroxide-induced oxidative damage in IPEC-J2 cells. Oxid. Med. Cell. Longev. 2016, 5987183 (2016).
De Blas, C. & Wiseman, J. Nutrition of the rabbit[M] 3rd edn. (CABI Publishing, 2020).
Cui, J. et al. Effects of ammonia on hypothalamic-pituitary-ovarian axis in female rabbits. Ecotoxicol. Environ. Saf. 227, 112922 (2021).
Francisco, S. et al. Induction of TLR4/TLR2 interaction and heterodimer formation by low endotoxic atypical LPS. Front. Immunol. 12, 748303 (2022).
Kamm, K., Schierwater, B. & DeSalle, R. Innate immunity in the simplest animals-placozoans. BMC Genomics 20(1), 5 (2019).
He, Y. et al. Polystyrene nanoplastics deteriorate LPS-modulated duodenal permeability and inflammation in mice via ROS drived-NF-κB/NLRP3 pathway. Chemosphere 307(1), 135662 (2022).
Yang, R. B. et al. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature 395(6699), 284–288 (1998).
Poltorak, A. et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282(5396), 2085–2088 (1998).
Zhou, J. et al. Icariside II attenuates lipopolysaccharide-induced neuroinflammation through inhibiting TLR4/MyD88/NF-κB pathway in rats. Biomed. Pharmacother. 111, 315–324 (2019).
Onomoto, K., Onoguchi, K. & Yoneyama, M. Regulation of RIG-I-like receptor-mediated signaling: Interaction between host and viral factors. Cell. Mol. Immunol. 18(3), 539–555 (2021).
Christofferson, D. E. et al. A novel role for RIP1 kinase in mediating TNFα production. Cell. Death. Dis. 3(6), e320 (2012).
Almagro, M. C., Goncharov, T., Newton, K. & Vucic, D. Cellular IAP proteins and LUBAC differentially regulate necrosome-associated RIP1 ubiquitination. Cell. Death. Dis. 6(6), e1800 (2015).
Kim, S. W. et al. Protein kinase C-associated kinase regulates NF-κB activation through inducing IKK activation. Int. J. Oncol. 45(4), 1707–1714 (2014).
Somensi, N. et al. Carvacrol suppresses LPS-induced pro-inflammatory activation in RAW 264.7 macrophages through ERK1/2 and NF-kB pathway. Int. Immunopharmacol. 75, 105743 (2019).
Lee, K. P. et al. Carvacrol inhibits atherosclerotic neointima formation by downregulating reactive oxygen species production in vascular smooth muscle cells. Atherosclerosis 240(2), 367–373 (2015).
Zou, Y., Xiang, Q., Wang, J., Peng, J. & Wei, H. Oregano essential oil improves intestinal morphology and expression of tight junction proteins associated with modulation of selected intestinal bacteria and immune status in a pig model. Biomed. Res. Int. 2016, 5436738 (2016).
Wang, T., Huo, Y. J., Shi, F., Xu, R. J. & Hutz, R. J. Effects of intrauterine growth retardation on development of the gastrointestinal tract in neonatal pigs. Biol. Neonate. 88(1), 66–72 (2005).
Itoh, H. et al. Regeneration of small intestinal mucosa after acute ischemia-reperfusion injury. Dig. Dis. Sci. 47(12), 2704–2710 (2002).
Edelblum, K. L. & Turner, J. R. The tight junction in inflammatory disease: Communication breakdown. Curr. Opin. Pharmacol. 9(6), 715–720 (2009).
Ren, W. et al. Serum amino acids profile and the beneficial effects of l-arginine or l-glutamine supplementation in dextran sulfate sodium colitis. PLoS ONE 9(2), e88335 (2014).
Shin, K., Fogg, V. C. & Margolis, B. Tight junctions and cell polarity. Annu. Rev. Cell. Dev. Biol. 22, 207–235 (2006).
Popova, O. P., Kuznetsova, A. V., Bogomazova, S. Y. & Ivanov, A. A. Claudins as biomarkers of differential diagnosis and prognosis of tumors. J. Cancer Res. Clin. Oncol. 147(10), 2803–2817 (2021).
Xiao, K. et al. TGF-β1 protects intestinal integrity and influences Smads and MAPK signal pathways in IPEC-J2 after TNF-α challenge. Innate. Immun. 23(3), 276–284 (2017).
Jurkiewicz, D., Michalec, K., Skowronek, K. & Nałęcz, K. A. Tight junction protein ZO-1 controls organic cation/carnitine transporter OCTN2 (SLC22A5) in a protein kinase C-dependent way. Biochim. Biophys. Acta. Mol. Cell. Res. 1864(5), 797–805 (2017).
Zhang, J. et al. Protective effects of baicalin on peritoneal tight junctions in piglets challenged with Glaesserella parasuis. Molecules 26(5), 1268 (2021).
Ye, G. et al. Challenges in diagnosing community-acquired carbapenem-susceptible Acinetobacter baumannii enterogenic sepsis: A case report. Medicine 98(26), e16248 (2019).
Hsu, L. C. et al. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature 428(6980), 341–345 (2004).
Cnop, M. et al. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: Many differences, few similarities. Diabetes 542, S97–S107 (2005).
Hu, L. et al. Postnatal nutritional restriction affects growth and immune function of piglets with intra-uterine growth restriction. Br. J. Nutr. 114(1), 53–62 (2015).
Jiang, J., Qi, L., Wei, Q. & Shi, F. Maternal stevioside supplementation ameliorates intestinal mucosal damage and modulates gut microbiota in chicken offspring challenged with lipopolysaccharide. Food. Funct. 12(13), 6014–6028 (2021).
Yue, J. & López, J. M. Understanding MAPK signaling pathways in apoptosis. Int. J. Mol. Sci. 21(7), 2346 (2020).
Fukuda, K. et al. A diarylpentanoid curcumin analog exhibits improved radioprotective potential in the intestinal mucosa. Int. J. Radiat. Biol. 92(7), 388–394 (2016).
Macdonald, T. T. & Monteleone, G. Immunity, inflammation, and allergy in the gut. Science 307(5717), 1920–1925 (2005).
Pan, D. & Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes. 5(1), 108–119 (2014).
Ott, S. J. et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53(5), 685–693 (2004).
Ahn, J. R. et al. Ruminococcus gnavus ameliorates atopic dermatitis by enhancing Treg cell and metabolites in BALB/c mice. Pediatr. Allergy Immunol. 33(1), e13678 (2022).
Guo, Y. et al. Inulin supplementation ameliorates hyperuricemia and modulates gut microbiota in Uox-knockout mice. Eur. J. Nutr. 60(4), 2217–2230 (2021).
Marzocco, S. et al. Supplementation of short-chain fatty acid, sodium propionate, in patients on maintenance hemodialysis: Beneficial effects on inflammatory parameters and gut-derived uremic toxins, a pilot study (PLAN Study). J. Clin. Med. 7(10), 315 (2018).
Kelly, C. J. & Colgan, S. P. Breathless in the gut: Implications of luminal O2 for microbial pathogenicity. Cell. Host. Microbe. 19(4), 427–428 (2016).
Roediger, W. E. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut 21(9), 793–798 (1980).
Sam, Q. H. et al. The divergent immunomodulatory effects of short chain fatty acids and medium chain fatty acids. Int. J. Mol. Sci. 22(12), 6453 (2021).
Wan, J. et al. Alterations in intestinal microbiota by alginate oligosaccharide improve intestinal barrier integrity in weaned pigs. Food Funct. 71, 104040 (2020).
Acknowledgements
The authors would like to thank the College of Animal Science and Technology of Agricultural University of Hebei for their laboratory work during this study.
Funding
This study was supported by a Natural Science Foundation of Hebei Province, China (C2020204186), Agricultural Science Park (Base) Project of Hebei Province, China (20536603D) and Modern Agriculture Industry Technology System of Rabbit, China (CARS-43-B-2).
Author information
Authors and Affiliations
Contributions
Conceptualization, D.W. and S.L.; methodology, D.W., M.X., S.L., and B.C.; formal analysis and data curation, D.W., A.Y., J.F., S.Z., X.W.; writing—original draft preparation, D.W.; writing—review and editing, H.J., S.L., and B.C.; visualization and supervision S.L. and B.C.; project administration and funding acquisition, S.L. and B.C. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, D., Xia, M., Yan, A. et al. Carvacrol attenuated lipopolysaccharide-induced intestinal injury by down-regulating TLRs gene expression and regulating the gut microbiota in rabbit. Sci Rep 13, 11447 (2023). https://doi.org/10.1038/s41598-023-38577-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-023-38577-w