Abstract
The kinesin-9 family comprises two subfamilies specific to ciliated eukaryotic cells, and has recently attracted considerable attention because of its importance in ciliary bending and formation. However, only scattered data are available on the motor properties of kinesin-9 family members; these properties have not been compared under identical experimental conditions using kinesin-9 motors from the same species. Here, we report the comprehensive motor properties of two kinesin-9 molecules of Tetrahymena thermophila, TtK9A (Kif9/Klp1 ortholog) and TtK9B1 (Kif6 ortholog), using microtubule-based in vitro assays, including single-motor and multi-motor assays and microtubule-stimulated ATPase assays. Both subfamilies exhibit microtubule plus-end-directed, extremely slow motor activity, both in single and multiple molecules. TtK9A shows lower processivity than TtK9B1. Our findings indicate that the considerable slow movement of kinesin-9 that corresponds to low ATP hydrolysis rates is a common feature of the ciliary kinesin-9 family.
Similar content being viewed by others
Introduction
Cilia and flagella are highly conserved organelles found across eukaryotes and are characterized by hair-like structures composed of several hundred proteins1. Cilia play an essential role in cellular dynamics via performing dual functions. First, they function as motile apparatuses that facilitate fluid flow in diverse biological processes. For instance, various single-cell organisms, such as algae, ciliates, and flagellates, move via beating cilia and flagella, whereas airway cilia contribute to mucus clearance2,3. Second, cilia act as sensory antennae, detect extracellular stimuli, and transduce them into cellular responses. For example, primary cilia in the kidney epithelium translate mechanical signals into intracellular calcium fluxes4. Additionally, immotile nodal cilia, located at the node periphery, exhibit mechano- or chemo-sensitive properties to sense the nodal flow direction, which is crucial for determining left–right asymmetry5,6,7. Hence, cilia are indispensable for various biological processes, including locomotion and development2,8,9. Ciliary dysfunction or defects, also known as ciliopathies, further highlight the critical importance of cilia10,11.
Most motile cilia feature a “9 + 2” configuration with well-organized microtubules, wherein the motor proteins, axonemal dyneins, provide power to bend the cilia10,12. In addition, several kinesins act as motor proteins within cilia13. Of these, kinesin-9 family members are expressed exclusively in ciliated organisms such as protists, and in tissues with motile cilia, such as the testis, trachea, and brain. The kinesin-9 family consists of two subfamilies, kinesin-9A (Klp1 in Chlamydomonas and Kif9 in vertebrates) and kinesin-9B (Kif6 in vertebrates), each with distinct functions, both of which have been implicated in hydrocephalus14,15,16. Kinesin-9A is an axonemal component protein localized along one of the central pair of microtubules (C2)17,18,19. The loss of kinesin-9A function causes defective ciliary motility in multiple species20,21,22,23. In an in vitro single-molecule motility assay, Xenopus truncated kinesin-9A (Kif9) showed slow processive movement along microtubules, although its directionality has not been examined20. Structural studies have revealed that kinesin-9A motors are aligned along the C2 microtubule protofilament and adopt two states, positioned 8 nm apart17, suggesting that kinesin-9A movement may regulate dynein-induced ciliary bending17,18,24. However, it has not been examined whether kinesin-9A can work in an array to generate ensemble forces. Vertebrate Kif9 does not only function in the axoneme, but localizes to the mitotic spindle and is required for maintaining proper spindle length in Hela cells25. Loss of Kif9 results in misaligned chromosomes. By contrast, kinesin-9B (mouse Kif6) localizes to both the axoneme and the apical cell cortex in ependymal cells14. Loss of kinesin-9B function disrupts the directionality of multiciliated ciliary motility in ependymal cells of the mouse brain, causing severe hydrocephalus14,15,16. Using in vitro single-molecule assays, mouse and zebrafish Kif6 have also been found to be plus-end-directed, slow processive motors14. However, a limited number of studies have reported on comparable in vitro motor properties of two kinesin-9 subfamilies in the same species; directionality, multiple motor motility, and ATP hydrolysis rates are yet to be characterized.
In this study, we characterized the motor properties of two kinesin-9 subfamilies in the ciliate Tetrahymena using in vitro microtubule gliding assays, single-molecule motility assays, and ATPase rate measurements. Our findings show that both kinesin-9A and kinesin-9B motors display remarkably slow velocities, consistent with the quantified low ATPase rates, and are plus-end-directed motors in both single and multiple molecules.
Results
Tetrahymena kinesin-9 exhibits extremely slow plus-end-directed motility
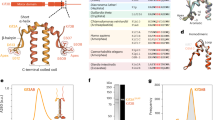
The ciliate Tetrahymena thermophila possesses four kinesin-9 genes, one kinesin-9A and three kinesin-9B genes22,26. We termed these kinesins TtK9A (NCBI Protein Database accession no. XP_001022313.1), TtK9B1 (XP_001024804.2), TtK9B2 (XP_001025897.4), and TtK9B3 (XP_001020926.2). In this study, the motor activities of TtK9A and TtK9B1, which were phylogenetically close to human KIF9 and KIF6, respectively (Fig. 1a), were investigated. Recent studies have shown that full-length kinesin-9 motor proteins of Xenopus20 and mice14 cannot bind to microtubules in vitro, possibly because of auto-inhibition involving the tail domain. In particular, in Chlamydomonas Klp1, the tail domain forms a complex in axonemes17. To avoid the effects of possible regulation, we created C-terminal tail domain-deleted TtK9A and TtK9B1 constructs, which contained the N-terminal 473 and 506 residues, respectively; they were fused to the Avi-tag and His-tag. These constructs, termed TtK9A_473 and TtK9B1_506, respectively, were assumed to be dimeric because of the presence of sufficient coiled-coil regions, as predicted by the Marcoil software27 (Fig. 1b, c). The truncated constructs expressed in Escherichia coli were purified using immobilized metal affinity chromatography and affinity binding to microtubules (Fig. 1d).
Tetrahymena kinesin-9 constructs used in this study. (a) Phylogenetic tree constructed with full-length kinesin-9 family amino acid sequences using maximum likelihood phylogenetic analyses, and 500 bootstrap resampling values (on nodes). Kinesin-9 can be divided into two subfamilies: kinesin-9A and kinesin-9B. The scale shows the estimated branch length, which represents the number of changes per site. (b) Coiled-coil (CC) probability plots for TtK9A (red) and TtK9B1 (blue) generated by Marcoil. (c) Schematic diagram of wild-type Tetrahymena kinesin-9 and the truncated kinesin-9 constructs used in this study. All constructs have a biotinylated peptide and affinity tag (AviHis). Values in brackets are theoretical molecular weights. (d) SDS-PAGE analysis of purified proteins (asterisks): 9A, 9A-GFP, 9A-SG, 9B1, and 9B1-GFP represent TtK9A_473, TtK9A_473-mEGFP, TtK9A_473-mStayGold, TtK9B1_506, and TtK9B1_506-mEGFP, respectively.
To test the motor properties of multiple kinesin-9 molecules, in vitro microtubule gliding assays were conducted using TtK9s fused to a biotinylated peptide (Avi-tag) at the C-terminus, which allowed their C-terminal stalk domains to be anchored to the streptavidin-coated glass surface (Fig. 2a). We found that lawns of either TtK9A_473 or TtK9B1_506 molecules drove microtubule gliding (Fig. 2b, c) with extremely slow velocities: 1.3 ± 0.1 and 13 ± 1 nm s−1 (mean ± SD), respectively (Fig. 2d, e and Supplementary Movie 1). In a control experiment using the well-studied rat kinesin-1 motor protein RK430 (1–430 aa)-Avi, the microtubule gliding velocity driven by dimeric rat kinesin-1 was 703 ± 36 nm s−1 (mean ± SD), which is consistent with previous results28,29 (Supplementary Fig. 1a). To determine the directionality driven by multiple kinesin-9 molecules, motility assays were performed using polarity-marked microtubules with a brighter-labeled fluorescent segment at the minus end and a more dimly labeled segment at the plus end30. Similar to other N-kinesins, such as kinesin-131, both TtK9 constructs directed microtubule gliding, which was led by the brighter minus-end, indicating plus-end-directed motility (Supplementary Fig. 2 and Supplementary Movie 2). These unidirectional gliding motions of microtubules have not been reported for the kinesin-9 family. This motion demonstrates that TtK9s can work in multi-motor ensembles.
Microtubule gliding assays driven by Tetrahymena kinesin-9 molecules. (a) Schematic diagram of the in vitro microtubule gliding assay. Biotinylated kinesins are anchored to the streptavidin-coated glass surface. (b, c) Kymographs of microtubule gliding driven by TtK9A_473 and TtK9B1_506. (d, e) Histograms of microtubule gliding velocities for TtK9A_473 (n = 114) and TtK9B1_506 (n = 160). Each histogram is fitted to a Gaussian distribution; mean ± SD. Only gliding microtubules that were > 2 μm in length, moved > 2 µm, and did not cross each other were analyzed.
Tetrahymena kinesin-9 motors display plus-end-directed processivity at low levels
Next, we examined motilities of single TtK9 molecules using a total internal reflection fluorescence (TIRF) microscope. Investigating the extremely slow motion of single-molecule kinesin-9A requires a more photostable fluorescent label than mEGFP. Thus, for the TtK9A fluorescent tag, mStayGold, a monomeric, highly photostable, and bright green fluorescent protein derived from StayGold32 with the E138D mutation33, was used; whereas for TtK9B1, mEGFP was used (Fig. 1c). To demonstrate the high photostability and brightness of mStayGold, single-molecule photobleaching assays were performed using a high-intensity laser (12% output power of the 15 mW 488-nm laser), with TtK9A_473-mStayGold bound to an anti-His-tag antibody-coated glass surface and TtK9A_473-mEGFP as a control (Supplementary Fig. 3a). Using our equipment, mStayGold (also known as StayGold-E138D) was found to be almost twice as bright as mEGFP (Supplementary Fig. 3b). Fitting results of the bleaching distributions revealed that mStayGold was approximately twice as photostable as mEGFP with \({k}_{\text{bleach}}\) values of 0.02 and 0.05 s−1, respectively (Supplementary Fig. 3e, f). Moreover, both mStayGold- and mEGFP-fused TtK9A molecules predominantly exhibited one or two photobleaching steps (Supplementary Fig. 3c, d) but not more than three steps, similar to other dimeric kinesins, such as kinesin-1 and -1434,35.
In single-molecule motility assays, polarity-marked microtubules were attached to the glass surface via anti-β-tubulin antibodies to measure single-molecule motile properties of truncated Ttk9 motors (Fig. 3a). At concentrations of 3–10 pM truncated TtK9A and TtK9B1, truncated Ttk9 motors exhibited microtubule plus-end-directed activities (Fig. 3b–e), although TtK9B1 often displayed pausing on the microtubule (Fig. 3e asterisks). In contrast to truncated TtK9 motors, full-length TtK9A-mStayGold and full length TtK9B1-mEGFP, even at a 100-fold higher concentration, could not be analyzed as they hardly bound to microtubules under our assay conditions (Supplementary Fig. 4). The plus-end-directed activity of a single TtK9B1_506 molecule was consistent with that of mouse Kif614. However, the directionality of a single kinesin-9A molecule has not been previously reported. The single-molecule motility parameters of TtK9 motor proteins were analyzed; the average velocity, dwell time, and run length of TtK9A_473-mStayGold were 1.3 ± 0.8 nm s−1, 94 ± 5 s, and 77 ± 4 nm, whereas those of TtK9B1_506-mEGFP were 9 ± 5 nm s−1, 70 ± 22 s, and 336 ± 24 nm, respectively (Fig. 3f–h, j–l). The fluorescence intensity volume data support Ttk9A_473 and TtK9B1_506 to be dimers and unaggregated because the sum of two Gaussians fit each intensity volume distribution well; the Gaussian center values were almost doubly proportional (Fig. 3i, m). The TtK9 single-molecule velocities were comparable to those of kinesin-9A or kinesin-9B that have been previously reported: 7.2 ± 0.3 nm s−1 (Xenopus Kif9)20, 12.2 ± 2 nm s−1 (mouse Kif6), and 15.5 ± 4.4 nm s−1 (zebrafish Kif6)14. TtK9A_473-mStayGold displayed a much shorter run length than Xenopus Kif9(1–461)_mNeonGreen (790.6 ± 33.1 nm)20. Truncated kinesin-1 dimer fused to mEGFP (RK430-mEGFP) was used as a control (Supplementary Fig. 1b–e). The velocity and run length of dimeric rat kinesin-1 was comparable to previous results28,36. Overall, we demonstrated that both TtK9s possessed plus-end-directed processivity, albeit at low (especially kinesin-9A) levels.
Single-molecule assays of Tetrahymena kinesin-9 dimers. (a) Schematic diagram of in vitro single-molecule assays. Fluorescently labeled kinesins move on polarity-marked microtubules fixed to the anti-β-tubulin-antibody-coated glass surface, as observed by a total internal reflection fluorescence microscope. (b, c) Sequential merged images of TtK9A_473-mStayGold or TtK9B1_506-mEGFP (green) moving on polarity-marked microtubules (magenta). Both kinesins move towards the dim plus-end of the microtubule (arrowheads). The plus ( +) and minus ( −) signs refer to the plus- and minus-ends of the microtubule. (d) Trajectories of TtK9A_473-mStayGold with a run length > 250 nm (n = 15). (e) Trajectories of TtK9B1_506-mEGFP with a run length > 600 nm (n = 21). Asterisks indicate typical temporal pauses of TtK9B1_506. (f–i) Histograms of the motility parameters for TtK9A_473-mStayGold (n = 101). Only tracking data for motors that moved > 100 nm were analyzed. (f) Velocity fitted to a Gaussian distribution; mean ± SD, (g) dwell time and (h) run length fitted to exponential distributions; mean ± SEM and (i) intensity volume for all tracking data fitted to the sum (black) of two Gaussians (gray); center 1 and center 2 refer to the centers of their respective Gaussians. (j–m) Histograms for TtK9B1_506-mEGFP (n = 99). Only tracking data for motors that moved > 100 nm were analyzed.
ATPase activities of Tetrahymena kinesin-9 dimers are low as well
Finally, the microtubule-stimulated ATPase activities of TtK9 dimers were determined. The maximum ATPase rate (\({k}_{\text{cat}}\)) was 0.22 ± 0.01 s−1 per dimer for TtK9A_473 and 2.2 ± 0.1 s−1 per dimer for TtK9B1_506; \({K}_{0.5,\text{ MT}}\) was 0.7 ± 0.1 μM for TtK9A_473 and 0.4 ± 0.1 μM for TtK9B1_506 (Fig. 4a, b). As expected from the low velocities measured by microtubule gliding (Fig. 2) and single-molecule movement (Fig. 3), the \({k}_{\text{cat}}\) values were low. Assuming that TtK9 motors take 8-nm steps per ATP turnover, their respective maximum ATPase rate values are comparable to the corresponding velocities of single or multiple molecules; that is, the number of steps/ATP hydrolyzed is approximately 137.
Microtubule-stimulated ATPase activities of Tetrahymena kinesin-9 dimers. (a, b) Data (circles with error bars: SD [n = 4]) for TtK9A and TtK9B1 are fitted to Michaelis–Menten model (lines) with \({k}_{\text{cat}}\) values of 0.22 ± 0.01 s−1 and 2.2 ± 0.1 s−1 per dimer and \({K}_{0.5,\text{ MT}}\) of 0.7 ± 0.1 and 0.4 ± 0.1 μM, respectively (mean ± SEM).
Discussion
Recent studies have shown that kinesin-9 family members play crucial roles in the cilia; however, information regarding their motility is limited. To address this gap, we performed a comprehensive investigation employing in vitro microtubule gliding assays (Fig. 2), single-molecule motility assays (Fig. 3), and ATPase rate measurements (Fig. 4) of T. thermophila kinesin-9 molecules (TtK9s). The experiments were performed under equivalent conditions such that the two kinesin-9 molecules could be compared. Our results revealed that both TtK9A and TtK9B1 motors exhibited exceedingly slow velocities, consistent with the low ATPase rates measured, and demonstrated plus-end directionality in both single-molecule and multi-motor microtubule gliding assays. Additionally, mStayGold was used as a fluorescent tag, which was brighter and more photostable than conventional mEGFP (Supplementary Fig. 3); it is useful not only for extremely slow motors, but also for all single-molecule motility assays. Our findings suggest that the observed slow velocities may be a common characteristic in the kinesin-9 family, and may be attributed to the kinesin-9 specific extended loop (Supplementary Fig. 5a). Kinesin-9 has an extended loop 2 compared to other kinesin family members, except for the kinesin-8 family. A previous study has revealed the kinesin-8 (Candida albicans Kip3) specific extended loop 2 contacts with α-tubulin38. According to the predicted structures generated by AlphaFold339, the extended loop 2 of kinesin-9 is very close to helix-12 of α-tubulin (Supplementary Fig. 5b). We speculate that this loop may interact with the microtubule surface, increasing the dwell time of the microtubule-bound state and limiting the ATPase cycle similar to that of kinesin-838.
Previous studies have revealed that kinesin-9A motors are localized along the C2 microtubule17,18,19 and may be active in the axoneme17,20, implying that ciliary beating is regulated via changing the central apparatus conformation17,18,20,24,40. Consistent with previous studies, we demonstrated that TtK9A motors exhibited motility in vitro. TtK9A showed a low velocity compared to that in kinesin-9A reported in Xenopus (Figs. 2d and 3f), while TtK9A demonstrated lower processivity than Xenopus Kif9 (Fig. 3h)20. This difference in processivity may be because they are from different species, such as the differences in yeast and human cytoplasmic dynein41,42, or potentially due to different buffer conditions. This may be owing to the collisions of TtK9A with the surface caused by helical tracking along the microtubule surface, as reported for other kinesin family members43,44,45. TtK9A that is anchored to the central apparatus moves only a limited longitudinal distance over the C2 microtubule and may not require high processivity.
The ATPase activity rate of TtK9A is considerably lower than the ciliary beating frequency of Tetrahymena46,47. This apparent inconsistency with the hypothesis that kinesin-9A regulates ciliary beating can be explained by considering the spatial arrangement of kinesin-9A on the C2 microtubule. Kinesin-9A motors are tightly aligned and bind to each other through associated proteins, forming a well-organized array17. This structured configuration suggests that kinesin-9A may move in a metachronal motion such as a “stadium wave.” These structural constraints suggest that kinesin-9A may exhibit an ATP hydrolysis rate synchronized with the ciliary beating frequency, somehow signaling via radial spokes to control axonemal dyneins spatiotemporally and axoneme bending24. Further investigation is needed to determine the synchronization between the kinesin-9A array and ciliary movement.
Cellular biological and biochemical assays with kinesin-9B motors have provided strong evidence that the motors localize to basal bodies and axonemes in punctate patterns14,16, align with the ciliary rotational polarity14,22, and are plus-end-directed processive motors14. Our experimental results agree with the motor in vitro activities reported for zebrafish Kif6 and mouse Kif6 (Fig. 3c, j)14. Kinesin-9B localization resembles those of intraflagellar transport (IFT) proteins16; a recent study reported that mouse Kif6 moved bidirectionally with or without IFT-B particles along axonemes in ependymal cells at a velocity comparable to that of IFT15. However, a previous study14 and our current study demonstrated that kinesin-9B moved at much slower velocities in vitro than IFT, indicating that kinesin-9B may not be a motor like kinesin-2 that drives IFT but may be a component of IFT or a cargo carried by IFT. Kinesin-9B knockdown in Trypanosoma disrupts the asymmetrical arrangement of the paraflagellar rod (PFR, a trilaminar lattice-like structure linked to the axonemal microtubule) along the axonemes, suggesting that the trypanosome kinesin-9B serves as a linker protein for PFR in Trypanosoma22. Mammalian cell cilia do not have a PFR, but a kinesin-9B deficiency or mutation disrupts the basal foot (protein complex protruding from the basal body) alignment of mouse multiciliate brain ependymal cells14,15, suggesting that mammalian kinesin-9B may also enable the asymmetrical arrangement of the basal foot. Tetrahymena do not have a PFR, but possess several basal body accessory structures: post-ciliary microtubules, transverse microtubules, and kinetodesmal fibers48. The three types of kinesin-9B in Tetrahymena most likely have distinct functions and achieve the asymmetrical arrangement of these structures around basal bodies. Further cell biological studies are necessary to elucidate the function of kinesin-9B in Tetrahymena.
Methods
Multiple sequence alignment and phylogenetic tree of the kinesin-9 family
Multiple sequence alignments and a phylogenetic tree of the full-length kinesin-9 family amino acid sequences were generated using SeaView version 5.0.549. Multiple sequence alignment was performed using Clustal Omega50 with the default parameters. Maximum likelihood phylogenetic trees were constructed using PhyML version 3.151 with the LG model52. The tree was internally validated using bootstrapping resampling (n = 500). The following protein sequences of kinesin-9 family members were used: Homo sapiens (NCBI Protein Database accession numbers NP_001400904.1 and NP_659464.3), Mus musculus (NP_001157041.1 and NP_796026.2), Xenopus laevis (XP_018124453.1 and XP_018118191.1), Danio rerio (XP_001922460.1 and NP_001410600.1), Strongylocentrotus purpuratus (XP_030830023.1 and XP_030854411.1), Tetrahymena thermophila (XP_001022313.1, XP_001024804.2, XP_001025897.4, and XP_001020926.2), Chlamydomonas reinhardtii (P46870.1 and XP_042928646.1), and Trypanosoma brucei (XP_846252.1 and XP_846346.1).
Plasmid construction
To construct C-terminally truncated Tetrahymena kinesin-9A and kinesin-9B dimers, coiled-coil predictions were calculated using Marcoil27, and the genes from 1–473 amino acids of TtK9A (XP_001022313.1) and 1–506 amino acids of TtK9B1 (XP_001024804.2) were cloned into different pColdIII (Takara Bio Inc., Shiga, Japan) vectors: pColdIII-AviHis, pColdIII-mEGFP-AviHis, and pColdIII-mStayGold-AviHis (only for TtK9A_473). To construct full length Tetrahymena kinesin-9A and kinesin-9B, the genes from 1–717 amino acids of TtK9A and 1–847 amino acids of TtK9B1 were cloned into different pColdIII vectors: pColdIII-mEGFP-AviHis (for TtK9B1) and pColdIII-mStayGold-AviHis (for TtK9A). AviHis refers to Avi-tag (GLNDIFEAQKIEWHE; Avidity LLC, Colorado, USA), which is biotinylated by BirA in E. coli53 and 6 × His-tag. The sequence of mStayGold (also known as StayGold-E138D), a monomeric green fluorescent protein derived from StayGold32 with the E138D mutation33, was used. Truncated constitutively active kinesin-1 (Rattus norvegicus KIF5C; residues 1–430, RK430)36 was used as a control in all experiments. All constructs were verified using DNA sequencing.
Kinesin expression and purification
All expression plasmids were transformed into E. coli strain BL21 Star (DE3) cells (Invitrogen, Thermo Fisher Scientific, MA, USA) using the pBirAcm plasmid, which encoded BirA. Kinesin expression was induced via adding 0.1 mM isopropyl β- d-1-thiogalactopyranoside and > 50 µM biotin at 15 °C, and cells were harvested after 24 h54. The cells were pelleted, resuspended in lysis buffer (500 mM NaCl, 80 mM PIPES-KOH, 1 mM MgCl2, 1 mM EGTA, 5 mM ATP, 1 mM DTT, 0.1% CHAPS, 0.1% Tween20, 10% glycerol, and protease inhibitors; pH 6.8), and sonicated on ice. After centrifuging the lysate, Ni–NTA affinity chromatography was performed on a HisTrap HP column (Cytiva, MA, USA) using an AKTA system (Cytiva), followed by buffer exchange into the desalting buffer (80 mM NaCl, 20 mM potassium phosphate, 1 mM MgCl2, 1 mM DTT, 20 μM ATP; pH 7.4) using a HiTrap Desalting column (Cytiva). Paclitaxel-stabilized microtubules were mixed with purified kinesins supplemented with 1 mM AMP-PNP and incubated for 15 min at 25 °C. After removing the unbound kinesins via centrifugation (20 min, 305,000 × g, 23 °C), the microtubule-bound kinesins were eluted with an ATP-containing buffer (10 mM ATP, 13 mM MgCl2, 200 mM potassium acetate, 20 μM paclitaxel, 16 mM PIPES-KOH, 1 mM EGTA; pH 7.4) via incubating for 10 min at 25 °C. Microtubules were removed using centrifugation (20 min, 245,000 × g, 23 °C)55. RK430 was purified using the same method as that used for TtK9A and TtK9B1. Supernatants were snap-frozen and stored in liquid nitrogen. The concentration of each kinesin was estimated using sodium dodecyl-sulfate polyacrylamide gel electrophoresis on 10% acrylamide gels using bovine serum albumin (BSA) as standards (Thermo Fisher Scientific) loaded on the same gel. Gels were stained with Quick-CBB PLUS (FUJIFILM Wako Pure Chemical Corporation, Tokyo, Japan) and imaged using a CCD camera (CSFX36BC3, Toshiba-Teli Corporation, Tokyo, Japan). Bands containing kinesins and BSA standards were quantified using the ImageJ software (NIH)56.
Tubulin purification
Tubulin was purified from porcine brains via four cycles of temperature-controlled polymerization and depolymerization in a high-molar PIPES buffer to remove the contaminating microtubule-associated proteins57. Purified tubulin in BRB80 buffer (80 mM PIPES-KOH, 1 mM MgCl2, and 1 mM EGTA; pH 6.8) was snap-frozen and stored in liquid nitrogen.
Cy5-labeled microtubule polymerization
The purified tubulin was polymerized and labeled with Cy5 mono-reactive NHS ester (Cytiva). Only polymerizable tubulin was collected after depolymerization, polymerization, and depolymerization cycles. The collected Cy5-labeled tubulin was snap-frozen and stored in liquid nitrogen. To polymerize the Cy5-labeled microtubules, 70% unlabeled tubulin and 30% Cy5-labeled tubulin were incubated in BRB80 buffer containing 7.5 mM GTP and 15 mM MgCl2 at 37 °C for 30 min. Paclitaxel was added at a molar ratio of at least ten times the tubulin quantity and incubated for another 10 min. Cy5-labeled microtubules were diluted to 10 µM in BRB80 buffer containing 20 µM paclitaxel, snap-frozen, and stored in liquid nitrogen. Cy5-labeled polarity-marked microtubules were prepared as described previously58. First, bright short microtubules (labeled tubulin: unlabeled tubulin = 1:2) were polymerized with 0.5 mM GMP-CPP (a non-hydrolyzable GTP analog, Jena Bioscience, Jena, Germany). The dim long segments were elongated on the plus end (labeled tubulin: unlabeled tubulin = 1:9) via including N-ethyl maleimide-treated tubulin, which inhibited minus-end polymerization.
Microtubule gliding assays
Microtubule gliding assays were performed in flow chambers assembled from plasma-cleaned coverslips (24 × 36 mm and 18 × 18 mm, Matsunami Glass, Osaka, Japan) attached to each other using double-sided tape (Scotch W-12, 3M)54. For gliding assays, the flow chamber comprised one flow chamber volume (5 μL) of diluted (5,000-fold) 0.1-µm diameter microbeads (carboxylate-modified, red fluorescent [580/605], Thermo Fisher Scientific), which were incubated for 30 s for drift correction. A concentration of one flow chamber volume of 5 mg mL−1 biotinylated-BSA (Sigma-Aldrich, MO, USA) was non-specifically absorbed onto the glass surface, incubated for 5 min, and then rinsed with four volumes of BRB80 buffer. The chamber surface was sequentially coated with one volume of 1 mg mL−1 streptavidin (FUJIFILM Wako Pure Chemical Corporation) for 5 min, four volumes of 1 mg mL−1 casein (Nacalai Tesque Inc., Kyoto, Japan) for 3 min, one volume of either 0.05 μM TtK9A_473 or 0.1 μM TtK9B1_506 for 5 min, four volumes of 1 mg mL−1 casein for 3 min, and then four volumes of 50 nM Cy5-labeled or polarity-marked microtubules in BRB80 buffer containing 0.4 mg mL−1 casein and 20 μM paclitaxel for 3 min. Finally, four volumes of motility buffer (BRB80 buffer containing 3 mM ATP, 3 mM MgCl2, 20 μM paclitaxel, ATP regeneration system, oxygen scavenger system, 10 mM DTT, and 0.4 mg mL−1 casein) were applied to the chamber. All chambers were sealed with grease (Apiezon M Grease; M&I Materials Ltd., Manchester, UK). Assuming that all of the 0.05 μM TtK9A_473 or 0.1 μM TtK9B1_506 applied was absorbed onto the glass surfaces of the flow chamber, the corresponding density would be 1.4–2.8 × 103 motors μm-2. This high motor density indicated that microtubule gliding in this condition was driven by multiple motors.
Assays were performed at 25 °C with temperature-control equipment59. Microtubule gliding was observed using a fluorescence microscope (Eclipse Ti-E equipped with a Perfect Focus System, Nikon Corporation, Tokyo, Japan) with a stable stage (KS-N, Chukousya-Seisakujo, Tokyo, Japan) and a stage controller (QT-CM2-35, Chuo Precision Industrial Co., Ltd., Tokyo, Japan) using illumination from an LED light source (D-LEDI, Nikon Corporation), 100 × /1.49 NA, Plan-Apochromat objective lenses (Nikon Corporation), and a Cy5 filter set (Semrock, NY, USA). The images were acquired using an EM-CCD camera (iXon X3 DU897E; Andor Technologies, Belfast, UK). Images of TtK9A and TtK9B1 were acquired every 100 s for 80 min and every 10 s for 6 min 40 s, respectively, with an exposure time of 0.5 s.
Non-polarity-marked microtubules were tracked using Fluorescence Image Evaluation Software for Tracking and Analysis (FIESTA)60. Tracking data were corrected for drift using red beads as references. Only gliding microtubules that were > 2 μm in length, moved > 2 µm, and did not cross each other were analyzed. Microtubule gliding velocities were calculated via linearly fitting the travel distance versus time plots. Three independent measurements were conducted for each twice-purified construct. Polarity-marked microtubules were analyzed to determine directionality. For TtK9A, out of 154 polarity-marked microtubules, 149 showed plus-end and 5 showed minus-end directionality. For TtK9B1, out of 135 polarity-marked microtubules, 128 showed plus-end and 7 showed minus-end directionality. Two independent measurements were conducted for each construct.
Single-molecule motility assays
The chamber of the single-molecule motility assay was prepared in the same method as for the gliding assay. One flow chamber volume of 1 mg mL−1 Protein G (Sigma-Aldrich) was non-specifically absorbed onto the glass surface, incubated for 5 min, and then rinsed with four volumes of P12 buffer (12 mM PIPES-KOH, 4 mM MgCl2, 1 mM EGTA; pH 6.8). The chamber surface was sequentially coated with two volumes of 5 µg mL−1 anti-β-tubulin antibody solution (monoclonal, Santa Cruz Biotechnology, TX, USA) for 5 min, four volumes of 15 mg mL−1 BSA (Sigma-Aldrich) blocking solution for 5 min, four volumes of diluted Cy5-labeled polarity-marked microtubules in BRB80 buffer containing 0.4 mg mL−1 casein and 20 μM paclitaxel for 1 min, and then four volumes of blocking solution containing 20 µM paclitaxel for 5 min. Finally, four volumes of the kinesin solution (3–10 pM TtK9A_473-mStayGold or TtK9B1_506-mEGFP in P12 buffer containing 3 mM ATP, 20 μM paclitaxel, ATP regeneration system, oxygen scavenger system, 10 mM DTT, 6 mg mL−1 BSA, and 0.4 mg mL−1 casein) were applied to the chamber. All chambers were sealed with grease. The assays were performed at room temperature (23–24 °C) with a lens heater (25 °C; Tokai Hit, Shizuoka, Japan).
Fluorescent images of kinesins and Cy5-labeled polarity-marked microtubules were captured using a TIRF microscope (IX71 with a TIRF illumination module IX2-RFAEVA-2, Olympus Corporation, Tokyo, Japan) equipped with 100 × /1.45 NA, Plan-Apochromat objective lenses (Olympus) and a lens heater (Tokai Hit, Japan). Kinesins were illuminated using a 488-nm laser (OBIS 488-15 LS FP 15 mW, Coherent, PA, USA), which was further attenuated using neutral density filters with a total internal reflection angle. Microtubules were illuminated with epi-illumination using a 632-nm laser (632 05-LHP-991 30 mW, Melles Griot Inc., NY, USA). Kinesin and microtubule images were acquired simultaneously using an EM-CCD camera (iXon DV887DCS-BV, Andor Technologies) with a custom-designed optical path splitter61. Images were acquired at 1 frame s−1 for 600–900 s, with an exposure time of 1 s. For TtK9B1_506-mEGFP (with excitation [Ex] and emission [Em] maxima of 488 and 507 nm, respectively)62, the 488-nm laser intensity was twice that of TtK9A_473-mStayGold (Ex, 497 nm and Em, 504 nm)33 because of its lower intensity (0.36 mW for TtK9B1_506-mEGFP and 0.18 mW for TtK9A_473-mStayGold).
Single molecules were tracked using FIESTA60. Tracking data were corrected for drift using fixed microtubules as references. Only the tracking data for motors that bound to the microtubule for 5 s, moved > 100 nm towards the microtubule plus end, did not reach the plus end, and did not cross each other were analyzed. For TtK9A_473-mStayGold, out of 957 molecules, 101 moved > 100 nm towards the plus end, 11 molecules moved > 100 nm towards the minus end, and the others did not move > 100 nm (845 molecules). For TtK9B1_506-mEGFP, out of 155 molecules, 99 moved > 100 nm towards the plus end, 1 molecule moved > 100 nm towards the minus end, and the others did not move > 100 nm (55 molecules). Single-molecule velocities of the kinesin-9 motors were calculated via linearly fitting the distance to the origin versus time plot. The means of the velocities were calculated via fitting a Gaussian distribution to each histogram. The means of the dwell times and run lengths were calculated via fitting an exponential distribution to each histogram. Assays for each twice-purified construct were performed in at least six independent measurements.
Measuring microtubule-stimulated ATPase rates
ATPase measurements of kinesin-9 molecules were performed using a pyruvate kinase/lactate dehydrogenase-linked assay as previously described58. The decrease in NADH (Sigma-Aldrich) absorbance at 340 nm via the catalytic reaction of pyruvate kinase/lactic dehydrogenase enzymes (Sigma-Aldrich) with phosphoenol-pyruvate (Sigma-Aldrich) and ADP produced by ATPase activity of kinesins in the presence of microtubules (0.18–18.5 μM) was measured. Assays were performed in buffer BRB80 containing 3 mM ATP, 3 mM MgCl2, and 97 or 83 nM TtK9A_473 or 31 or 7 nM TtK9B1_506. Assays were performed at 25 °C with a temperature-control equipment (Shimadzu, Kyoto, Japan). Four independent measurements were conducted for the assays in each twice-purified construct.
Statistics and reproducibility
Tracking data for microtubules were obtained from three independent experiments, tracking data for single molecules were obtained from at least six independent experiments, microtubule stimulated ATPase rates were obtained from four independent experiments, and sample sizes are indicated in detail in the text or the Methods section.
Data availability
All samples used in this study are available from the corresponding author upon reasonable request. The source data for graphs in the main figures are provided as Supplementary Data.
Code availability
Tracking data were analyzed using Python. These scripts are available upon reasonable request.
References
Pazour, G. J., Agrin, N., Leszyk, J. & Witman, G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 (2005).
Ginger, M. L., Portman, N. & McKean, P. G. Swimming with protists: Perception, motility and flagellum assembly. Nat. Rev. Microbiol. 6, 838–850 (2008).
Bustamante-Marin, X. M. & Ostrowski, L. E. Cilia and mucociliary clearance. Cold Spring Harb. Perspect. Biol. 9, a028241. https://doi.org/10.1101/cshperspect.a028241 (2017).
Praetorius, H. A. & Spring, K. R. Bending the MDCK cell primary cilium increases intracellular calcium. J. Membr. Biol. 184, 71–79 (2001).
Katoh, T. A. et al. Immotile cilia mechanically sense the direction of fluid flow for left-right determination. Science 379, 66–71 (2023).
Djenoune, L. et al. Cilia function as calcium-mediated mechanosensors that instruct left-right asymmetry. Science 379, 71–78 (2023).
Tanaka, Y., Morozumi, A. & Hirokawa, N. Nodal flow transfers polycystin to determine mouse left-right asymmetry. Dev. Cell 58, 1447-1461.e6. https://doi.org/10.1016/j.devcel.2023.06.002 (2023).
Lyons, R. A., Saridogan, E. & Djahanbakhch, O. The reproductive significance of human Fallopian tube cilia. Hum. Reprod. Update 12, 363–372 (2006).
Hirokawa, N., Tanaka, Y., Okada, Y. & Takeda, S. Nodal flow and the generation of left-right asymmetry. Cell 125, 33–45 (2006).
Horani, A. & Ferkol, T. W. Understanding primary ciliary dyskinesia and other ciliopathies. J. Pediatr. 230, 15-22.e1. https://doi.org/10.1016/j.jpeds.2020.11.040 (2021).
Fliegauf, M., Benzing, T. & Omran, H. When cilia go bad: Cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893 (2007).
Samsel, Z., Sekretarska, J., Osinka, A., Wloga, D. & Joachimiak, E. Central apparatus, the molecular kickstarter of ciliary and flagellar nanomachines. Int. J. Mol. Sci. 22, 3013. https://doi.org/10.3390/ijms22063013 (2021).
Li, L. & Ran, J. Regulation of ciliary homeostasis by intraflagellar transport-independent kinesins. Cell Death Dis. 15, 47 (2024).
Takagishi, M., Yue, Y., Gray, R. S., Verhey, K. J. & Wallingford, J. B. Kif6 regulates cilia motility and polarity in brain ependymal cells. Dis. Model. Mech. https://doi.org/10.1242/dmm.050137 (2024).
Fang, C. et al. Distinct roles of Kif6 and Kif9 in mammalian ciliary trafficking and motility. bioRxiv 2023.11.09.564704 (2023).
Konjikusic, M. J. et al. Mutations in Kinesin family member 6 reveal specific role in ependymal cell ciliogenesis and human neurological development. PLoS Genet. 14, e1007817. https://doi.org/10.1371/journal.pgen.1007817 (2018).
Han, L. et al. Cryo-EM structure of an active central apparatus. Nat. Struct. Mol. Biol. 29, 472–482 (2022).
Yokoyama, R., O’toole, E., Ghosh, S. & Mitchell, D. R. Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. U. S. A. 101, 17398–17403 (2004).
Bernstein, M., Beech, P. L., Katz, S. G. & Rosenbaum, J. L. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 125, 1313–1326 (1994).
Konjikusic, M. J. et al. Kif9 is an active kinesin motor required for ciliary beating and proximodistal patterning of motile axonemes. J. Cell Sci. 136, jcs259535. https://doi.org/10.1242/jcs.259535 (2023).
Meng, Z. et al. Identification of bi-allelic KIF9 loss-of-function variants contributing to asthenospermia and male infertility in two Chinese families. Front. Endocrinol. 13, 1091107. https://doi.org/10.3389/fendo.2022.1091107 (2022).
Demonchy, R. et al. Kinesin 9 family members perform separate functions in the trypanosome flagellum. J. Cell Biol. 187, 615–622 (2009).
Miyata, H., Oyama, Y., Kaneda, Y. & Ikawa, M. The motor domain of testis-enriched kinesin KIF9 is essential for its localization in the mouse flagellum. Exp. Anim. 71, 46–52 (2022).
Grossman-Haham, I. Towards an atomic model of a beating ciliary axoneme. Curr. Opin. Struct. Biol. 78, 102516. https://doi.org/10.1016/j.sbi.2022.102516 (2023).
Andrieu, G., Quaranta, M., Leprince, C. & Hatzoglou, A. The GTPase gem and its partner Kif9 are required for chromosome alignment, spindle length control, and mitotic progression. FASEB J. 26, 5025–5034 (2012).
Wickstead, B. & Gull, K. A “Holistic” Kinesin phylogeny reveals new kinesin families and predicts protein functions. MBoC 17, 1734–1743 (2006).
Delorenzi, M. & Speed, T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625 (2002).
Poulos, A., Budaitis, B. G. & Verhey, K. J. Single-motor and multi-motor motility properties of kinesin-6 family members. Biol. Open 11, bio059533. https://doi.org/10.1242/bio.059533 (2022).
Yajima, J. & Cross, R. A. A torque component in the kinesin-1 power stroke. Nat. Chem. Biol. 1, 338–341 (2005).
Hyman, A. A. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J. Cell Sci. Suppl. 14, 125–127 (1991).
Kim, A. J. & Endow, S. A. A kinesin family tree. J. Cell Sci. 113, 3681–3682 (2000).
Hirano, M. et al. A highly photostable and bright green fluorescent protein. Nat. Biotechnol. 40, 1132–1142 (2022).
Ivorra-Molla, E. et al. A monomeric StayGold fluorescent protein. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-02018-w (2023).
Pierce, D. W., Hom-Booher, N. & Vale, R. D. Imaging individual green fluorescent proteins. Nature 388, 338 (1997).
Popchock, A. R. et al. The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nat. Commun. 8, 13999. https://doi.org/10.1038/ncomms13999 (2017).
Yajima, J., Alonso, M. C., Cross, R. A. & Toyoshima, Y. Y. Direct long-term observation of kinesin processivity at low load. Curr. Biol. 12, 301–306 (2002).
Coy, D. L., Wagenbach, M. & Howard, J. Kinesin takes one 8-nm step for each ATP that it hydrolyzes. J. Biol. Chem. 274, 3667–3671 (1999).
Hunter, B. et al. Kinesin-8-specific loop-2 controls the dual activities of the motor domain according to tubulin protofilament shape. Nat. Commun. 13, 4198. https://doi.org/10.1038/s41467-022-31794-3 (2022).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Miyata, H. et al. Testis-enriched kinesin KIF9 is important for progressive motility in mouse spermatozoa. FASEB J. 34, 5389–5400 (2020).
Reck-Peterson, S. L. et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell 126, 335–348 (2006).
Trokter, M., Mücke, N. & Surrey, T. Reconstitution of the human cytoplasmic dynein complex. Proc. Natl. Acad. Sci. U. S. A. 109, 20895–20900 (2012).
Brunnbauer, M. et al. Torque generation of kinesin motors is governed by the stability of the neck domain. Mol. Cell 46, 147–158 (2012).
Mitra, A., Ruhnow, F., Girardo, S. & Diez, S. Directionally biased sidestepping of Kip3/kinesin-8 is regulated by ATP waiting time and motor-microtubule interaction strength. Proc. Natl. Acad. Sci. U. S. A. 115, E7950–E7959. https://doi.org/10.1073/pnas.1801820115 (2018).
Maruyama, Y. et al. CYK4 relaxes the bias in the off-axis motion by MKLP1 kinesin-6. Commun. Biol. 4, 180. https://doi.org/10.1038/s42003-021-01704-2 (2021).
Wood, C. R., Hard, R. & Hennessey, T. M. Targeted gene disruption of dynein heavy chain 7 of Tetrahymena thermophila results in altered ciliary waveform and reduced swim speed. J. Cell Sci. 120, 3075–3085 (2007).
Stoddard, D. et al. Tetrahymena RIB72A and RIB72B are microtubule inner proteins in the ciliary doublet microtubules. Mol. Biol. Cell 29, 2566–2577 (2018).
Bayless, B. A., Galati, D. F. & Pearson, C. G. Tetrahymena basal bodies. Cilia 5, 1. https://doi.org/10.1186/s13630-016-0022-8 (2015).
Gouy, M., Guindon, S. & Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. https://doi.org/10.1038/msb.2011.75 (2011).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008).
Cull, M. G. & Schatz, P. J. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 326, 430–440 (2000).
Yamagishi, M., Sumiyoshi, R., Drummond, D. R. & Yajima, J. Anchoring geometry is a significant factor in determining the direction of kinesin-14 motility on microtubules. Sci. Rep. 12, 15417. https://doi.org/10.1038/s41598-022-19589-4 (2022).
Yamagishi, M., Maruyama, Y., Sugawa, M. & Yajima, J. Characterization of the motility of monomeric kinesin-5/Cin8. Biochem. Biophys. Res. Commun. 555, 115–120 (2021).
Yamaguchi, S. et al. Torque generation by axonemal outer-arm dynein. Biophys. J. 108, 872–879 (2015).
Castoldi, M. & Popov, A. V. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32, 83–88 (2003).
Yamagishi, M. et al. Structural basis of backwards motion in kinesin-1-kinesin-14 chimera: Implication for kinesin-14 motility. Structure 24, 1322–1334 (2016).
Sato, Y., Yamagishi, M. & Yajima, J. Effect of temperature on actin filament corkscrewing driven by nonprocessive myosin IC. Biochem. Biophys. Res. Commun. 703, 149597. https://doi.org/10.1016/j.bbrc.2024.149597 (2024).
Ruhnow, F., Zwicker, D. & Diez, S. Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 100, 2820–2828 (2011).
Sugawa, M., Maruyama, Y., Yamagishi, M., Cross, R. A. & Yajima, J. Motor generated torque drives coupled yawing and orbital rotations of kinesin coated gold nanorods. Commun. Biol. 5, 1368. https://doi.org/10.1038/s42003-022-04304-w (2022).
Zacharias, D. A., Violin, J. D., Newton, A. C. & Tsien, R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002).
Acknowledgements
The authors thank Dr. M. Sugawa for the guidance related to the experimental technique. This work was supported by JSPS KAKENHI (Grant number JP23K18135 to J.Y. and Grant number JP23K14177 to M.Y.), MEXT KAKENHI Grant-in-Aid for Transformative Research Areas (Grant number JP23H04401 to J.Y.) and the Precise Measurement Technology Promotion Foundation (PMTP-F to J.Y.)
Author information
Authors and Affiliations
Contributions
J.Y. designed the project; H.I. performed the experiments and data analysis; H.I. and M.Y. prepared biological samples; J.Y. and M.Y. supervised the project; H.I. and J.Y. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Movie 1.
Supplementary Movie 2.
Supplementary Movie 3.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ishii, H., Yamagishi, M. & Yajima, J. Two Tetrahymena kinesin-9 family members exhibit slow plus-end-directed motility in vitro. Sci Rep 14, 20993 (2024). https://doi.org/10.1038/s41598-024-71280-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-71280-y