Abstract
Retinitis pigmentosa (RP) is a progressive and degenerative retinal disease resulting in severe vision loss. RP have been extensively studied for pathogenetic mechanisms and treatments. Yet there is little information about alterations of RP associated proteins in phosphodiesterase 6 beta (Pde6b) mutated model. To explore the roles of RP causing proteins, we performed a label free quantitative mass spectrometry based proteomic analysis in rd10 mouse retinas. 3737 proteins were identified at the degenerative time points in rd10 mice. 222 and 289 differentially expressed proteins (DEPs) (fold change, FC > 2, p < 0.05) were detected at 5 and 8 weeks. Based on Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, visual perception and phototransduction were severely affected. The downregulated DEPs were significantly enriched in cilium assembly and protein localization. 25 decreased DEPs causing autosomal recessive/dominant retinitis pigmentosa were visualized by heatmaps. Protein-protein interaction network represented 13 DEPs interacted directly with Pde6b protein. 25 DEPs causing RP were involved in phototransduction, visual perception, response to stimulus, protein localization and cilium assembly pathways. The significantly reduced expressions of DEPs were further validated by quantitative reverse transcription polymerase chain reaction (qPCR), Western blots (WB) and immunohistochemistry (IHC). This study revealed the molecular mechanisms underlying early and late stage of RP, as well as changes of RP-causing proteins.
Similar content being viewed by others
Introduction
RP is a common inherited retinal disease in clinic, affecting millions of individuals, and the prevalence rate of RP is about 1/4,000 worldwide1,2. Night blindness is the hallmark of RP, which often starts in adolescence and progresses to peripheral vision loss and eventually blindness3. Patients with RP may retain a few degrees of central vision by midlife, but in many cases the disease ends with complete blindness3. RP may be an autosomal dominant (ad), autosomal recessive (ar), X-linked, or simplex/multiplex disease4.
RP is an inherited retinopathy characterized by clinical and genetic heterogeneity that results in retinal dysfunction and eventual photoreceptor cell death5. More than 110 genes have been identified as the pathogenic genes of RP, in which 31 genes lead ad-RP, 66 genes caused ar-RP, and 3 genes caused X-linked RP (RetNet Database: https://sph.uth.edu). Mutations of over 50 genes are known related with non-syndromic RP. 12 genes mutations may cause Usher syndrome and 17 genes can lead Bardet-Biedl syndrome3. In most cases, mutations occur in rod and/or cone photoreceptors, which account for more than 80% of retinal cells6. It is estimated that up to 5% of ar-RP cases are caused by mutations in PDE6B gene which encodes the beta subunit of rod cGMP phosphodiesterase5.
Retinitis pigmentosa results in rigorous vision loss caused by photoreceptor degeneration under ill-defined mechanisms. According to previous studies, phosphodiesterase (PDE) plays a regulatory role in signal transduction. The PDE defects induced by Pde6b gene mutation could induce the accumulation of cGMP, leading to the final photoreceptor cell apoptosis7. The previous study in RP mouse model suggested the central role of STAT signaling during retinal degeneration by studying the up-regulated proteins in rd10 mice8. Although previous studies adopted molecular biological techniques, sequencing and transcriptomics, there was rare research centering on the roles and alterations of RP causative genes in the Pde6b mutated RP mouse model. In previous studies, rd1 and rd10 are two mainly used hereditary mouse models for RP research2,9,10. Rd10 is caused by a missense point mutation in exon 13 of Pde6b gene10. Rd1 mice are completely blind at 3 weeks of age4. However, rd10 mouse presents later onset and milder retinal degeneration, which makes it an applicable model for investigating the pathogenesis of RP. The aims of this study are: (1) to explore the roles of RP pathogenic proteins in the retinal degeneration mouse model by using proteomic analysis; (2) to discover the pathways influenced by RP pathogenic proteins; (3) to verify the critical pathogenic proteins by qPCR, WB and IHC. We believe the results of this extensive proteomic analysis in rd10 retinas at key time points can help unravelling the defects of molecular pathways in RP. Moreover, these data can help shed light on mechanisms of retinal degeneration processes, thus potentially providing new hints for prospective RP therapeutic strategies.
Materials and methods
Animal care and use
Rd10 mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Sanger sequencing11,12 was performed by Beijing Genomics Institute to confirm Pde6b mutation. 5/8 weeks C57BL/6 mice as wild type (WT) were purchased from Vital River Laboratory Animal Technologies Co. Ltd. The animals were kept under 12-h light-dark cycle and given a standard chow diet. ARVO (Association for Research in Vision and Ophthalmology) guidelines were followed for animal care. Experiments and procedures involving animals were permitted by the Ethics Committee of Henan Eye Hospital (Permit No: HNEECA-2021-10).
Real-time quantitative PCR analysis
Retinas of rd10 and WT mice were dissected and homogenized for total RNA extraction. The total RNA was extracted using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) as directed by the manufacturer. By using the PrimeScript® RT reagent kit (Takara Biotechnology, Dalian, China), complementary DNA (cDNA) was generated. qPCR was performed with ABI Prism 7500 system (Applied Biosystems, Foster City, CA, USA) based on what was previously described13. Supplementary Table S1 shows the primer sequences used for the qPCR assay.
Western blot analysis
In the experiment, retinas were dissected from WT and rd10 mice eyeballs, and then homogenized in RIPA lysis buffer (EpiZyme, Shanghai, China) with 1% protease inhibitor cocktail (EpiZyme)14. A BCA protein kit (Beyotime) was used to detect the protein concentration. Protein was loaded in a 10% SDS-polyacrylamide gel and transferred to polyvinylidenedifluoride (PVDF) membranes (Millipore Corporation, Burlington, MA, USA). A 1.5 h soak in 5% non-fat milk blocked PVDF membranes was followed by an overnight incubation with antibodies against Pde6b (PA1-722, Thermo Fisher Scientific), Pde6a (PA1-720, Thermo Fisher Scientific), Tulp1 (18971-1-AP, Proteintech, Chicago, USA), Prph2 (18109-1-AP, Proteintech), Rbp3 (14352-1-AP, Proteintech), Bbs2 (11188-2-AP, Proteintech), Bbs7 (18961-1-AP, Proteintech), Bbs4 (ER1904-15, Huabio), Nr2e3 (14246-1-AP, Proteintech), Rho (ET1704-12, Huabio), Rom1 (CSB-PA344843ESR2HU, Cusabio, Wuhan, China), Arl6 (12676-1-AP, Proteintech) and β-actin (ab8227, Abcam) at 4 °C. β-actin was used as the endogenous reference. The PVDF membranes were cut for incubating with above-mentioned antibodies. Western Blot Fast Stripping Buffer (EpiZyme, PS107) was used to strip the previous bands.
Electroretinography (ERG) assessment
Retinal functions were measured with electroretinography (ERG), according to the previous description15. Following dark adaptation overnight, mice were anesthetized with injection of 1.25% avertin (0.4 ml/20 g body weight) (Aibei Biotechnology Co., Ltd, Nanjing, China) intraperitoneally. Tropiamide eye drops (Santen Pharmaceutical, Osaka, Japan) were used to dilate the pupils 5 min prior to recording. In all procedures, dim red light was utilized to obtain full-field ERGs using RetiMINER-C, a visual electrophysiology system (AiErXi Medical Equipment Co., Ltd., Chongqing, China). The stimulus intensity ranged from − 3 to 1 log cd-s/m2 in dark-adapted ERGs. Light-adapted ERGs were recorded to stimuli of 0 to 1 log cd-s/m2 after 5 min of light adaptation. An analysis of the amplitudes of a- and b-waves was conducted to evaluate retinal function.
Optical coherence tomography (OCT) examination
The retinal structure was assessed with OCT according to a previously described method16,17. Briefly, age-matched rd10 and WT mice were anesthetized with 1.25% avertin solution of 0.4 mL/20 g body weight. Pupils were dilated by applying compound tropicamide eye drops at 5 min before measurement. Retinal structure was measured by swept-source optical coherence tomography (SS-OCT, VG200D SVision Imaging, China). The set up parameters of OCT are 8 mm length, 6 mm super depth and Star 18 line R32.
Immunofluorescent and immunohistochemical staining
The immunofluorescence was conducted according to the previously described methodology15. Eyes were enucleated and fixed with FAS eyeballs fixative solution (Servicebio Technology Co., Ltd., Wuhan, China) overnight. Eyeballs were embedded and flash-frozen in Optimum Cutting Temperature compound (Sakura Finetek., Tokyo, Japan). Serial 8 μm cryosections were cut through cornea-optic nerve axis. The sections were then overnight incubated with anti-Pde6b antibody at 4 °C. The slides were then incubated in dark with DAPI and anti-rabbit secondary antibody. Paraffin embedding and Hematoxylin-Eosin staining were conducted for immunohistochemistry. The paraffin sections were incubated with anti-Pde6a antibody, anti-Prph2 antibody, anti-Nr2e3 antibody, anti-Rho antibody, anti-Rom1 antibody, anti-Rbp3 antibody. Staining results were observed under a fluorescence microscope (Olympus, Tokyo, Japan).
Proteomic samples preparation
Currently, mass spectrometry is gaining prominence as a powerful tool for identifying and quantifying proteins and peptides18. This technique is increasingly being used in ophthalmic research. In this study, retinas were obtained from rd10 and WT mice at 5 weeks and 8 weeks. Three retinas from each group were collected and processed as previously19,20,21. Briefly, lysis of retinas was performed in the mixture of 50 mM NH4HCO3, 8 M Urea and 1% protease inhibitor cocktail on ice, followed by ultrasonic sonication for 2 min. 100 µg proteins were reduced with 10 mM DTT (Sigma-Aldrich, MO, USA) for 1 h and alkylated with 40 mM iodoacetamide (Sigma-Aldrich) in dark for 1 h. The samples were transferred to equilibrated ultrafiltration tubes (10 kDa) and centrifuged at 14,000 g for 15 min at room temperature. 3 µg trypsin (Promega, Madison, WI, USA) was added to samples and incubated overnight at 37 °C. After drying under vacuum, the peptides were dissolved in 0.1% FA (formic acid) and analyzed by LC-MS/MS. Proteomics could elucidate the molecular foundations of biology and disease, with liquid chromatography-tandem mass spectrometry (LC-MS/MS) serving as a widely employed platform for bottom-up proteomic analysis22.
LC-MS/MS-based label-free proteomic analysis
LC-MS/MS was performed using an Ekspert nanoLC 415 (AB Sciex, Framingham, MA, USA) coupled with a Triple TOF 6600 mass spectrometer (AB Sciex). The peptides were desalted using a C18 trap column (10 × 0.3 mm, particle size: 5 μm) and then loaded onto C18 analytical column (150 × 0.3 mm, particle size: 3 μm) following the gradient elution of 6-35% buffer B (0.1% FA, 2% water and 97.9% acetonitrile). The MS was operated in positive mode using data-dependent acquisition (DDA). In data-independent acquisition (DIA) analysis, SWATH variable window assay calculator (AB Sciex, version 1.1) was used to calculate the 100 scanning windows. TOF MS accumulation time was set to 0.25 s with a mass range of 100–1500 Da. Ion spray voltage floating was set at 2300 V.
Processing of LC-MS/MS data
Data from DDA were processed using ProteinPilot software (version 5.0.1). With trypsin, up to two cleavage sites were allowed to be missed during specific digestion mode. Based on the Uniprot Swiss mouse database (17,050 entries, released 2020.03), peptides and proteins were identified. Data processing was performed by using SWATH 2.0 plug-in for PeakView software (Version 2.2) to quantify the DIA data. The peptide length was limited from six amino acids to 5000 Da. The confidence threshold was set at 99%, and the protein FDR (false discovery rate) was set at 1%. The allowed maximum fragment mass deviation was 50 ppm. The retention time was adjusted by 1 ~ 2 endogenous peptides with high spectral quality per 10 min. The peak area was selected as a relative quantitative value.
Bioinformatic and statistical analysis
Normalized and log2 transformed protein intensities were calculated. T-tests were used to calculate p values for protein expression. In this study, differentially expressed proteins (DEPs) were defined as proteins with p < 0.05 and FC > 2. Principal component analysis (PCA) was performed using R ggplot packages. Volcano plots were generated by “ggplot2” in R studio. Heatmaps were created by “pheatmap” package. GO and KEGG pathways enrichment analyses were performed with DAVID (Database for Annotation, Visualization and Integrated Discovery) 6.8 (https://david.ncifcrf.gov) based on DEPs (p < 0.05 and FC > 2). Chord diagrams were created by “GOplot” in R studio. Protein-Protein Interaction (PPI) analysis was carried out and visualized with String (http://string-db.org/, version 11.5).
Experimental data were expressed as mean ± standard error of mean (SEM). An analysis of the statistical data was conducted with GraphPad Prism 8 software (GraphPad Software, San Diego, CA, USA). Unpaired t-tests and two-way ANOVA followed by Bonferroni correction were applied to experimental data. Statistical significance was defined as p value less than 0.05.
Results
Morphological assessment showed the Outer Nuclear Layer thickness decreased in RP progression
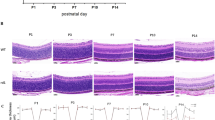
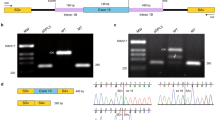
The qPCR and Western blot results revealed the almost undetectable Pde6b mRNA level and protein level at 8 weeks (Fig. 1A and B). The work flowchart of this study was shown as Supplementary Figure S1. Sanger sequencing was performed to identify the pathogenic homozygous mutation in Pde6b gene. Forward and reverse sequencing confirmed the c.1678 C > T mutation in rd10 mice (Supplementary Figure S2A and S2B). We used Pymol (version 2.4.0) to predict the 3D models of wild type and mutated Pde6b protein structures. In rd10 mice, the 560th amino acid Arginine (R) in yellow (Supplementary Figure S2C) mutated to Cysteine (C) in red (Supplementary Figure S2D). The results indicated the R560C mutation caused the absence of Pde6b protein in rd10 mice. To determine the retinal morphology changes in rd10 mice, retinal cryosections and immunofluorescence with DAPI and Pde6b antibody were conducted on wild type mice and rd10 mice at 5 weeks and 8 weeks. Most of the photoreceptors disappeared at 5 weeks and 8 weeks, although the thickness of the inner nuclear layer (INL) and the ganglion cell layer (GCL) were not yet affected. In Fig. 1C, 5 weeks rd10 mice revealed significantly decline in photoreceptor nuclei (blue) and Pde6b protein (red). The ONL of rd10 is reduced to barely detectable single layer by 8 weeks. Besides, SS-OCT results showed that the retinal ONL thickness and ONL/INL ratio measured in retinas of 5 weeks rd10 mice were remarkably decreased (Fig. 1D and E). Moreover, the retinal ONL thickness and ONL/INL ratio of 8 weeks rd10 showed a greater reduction (Fig. 1F). The results suggested a correlation between advancing age and the exacerbation of retinal degeneration in rd10 mice. These findings are consistent with the previous studies4,5.
The phenotype of rd10 mice containing Pde6b mutation. (A) The mRNA level of Pde6b decreased significantly in rd10 mice at 8 weeks (n = 3 retinas). (B) Pde6b protein expression was almost undetectable in rd10 mice at 8 weeks (n = 3 retinas). (C) The retinal histomorphology of rd10 mice (n = 3 eyes). Retinal cryosections with staining and Pde6b immunofluorescence in WT and rd10 mice (blue: DAPI, red: Pde6b). (D) The retinal ONL thickness of WT and rd10 mice at 5w and 8w (n = 4 eyes). (E) The ONL/INL ratio in the retinas of age-matched WT and rd10 mice (n = 4 eyes). (F) The OCT images of age-matched WT and rd10 mice (n = 6–8 mice per group). The results were mean ± SEM. Significance of difference (****p < 0.0001) was determined by unpaired t-test and two-way ANOVA with Bonferroni correction. WT: wild type; 5w: 5 weeks; 8w: 8 weeks; GCL: ganglion cell layer; INL: inner nuclear layer; ONL: outer nuclear layer.
Electrophysiological examination indicated retinal function impairment in rd10 mice
Flash ERG was performed to evaluate the retinal functions of age-matched WT and rd10 mice. In Fig. 2A, the scotopic ERG responses occurred a delay in the onset of a-wave and b-wave in rd10 mice at 5 weeks. The dark-adapted amplitudes of a-wave and b-wave were significantly lower in 5 weeks rd10, and almost undetectable in rd10 mice at 8 weeks (Fig. 2A, C and D). As for the photopic ERG, the average amplitudes of b-wave in 5 weeks rd10 mice remarkably declined, and the b-wave onset time also delayed obviously (Fig. 2B and E). The light-adapted b-wave of 8weeks rd10 mice was barely detectable. Overall, retinal functions of rd10 mice were incompetent at 5 weeks and completely disabled at 8 weeks. The above results indicated the Pde6b mutation impaired the visual functions severely.
Retinal function of age-matched WT and rd10 were assessed by dark- and light-adapted ERG (n = 6–8 mice per group). (A) (B) Representative dark- and light-adapted ERG responses in 5w and 8w of WT and rd10 mices were shown. (C) (D) (E) The ERG amplitudes vs. flash intensity profiles for the dark-adapted a-wave, dark-adapted b-wave, and light-adapted b-wave were summarized. The results were mean ± SEM. significance of difference(**p < 0.01, ***p < 0.001, ****p < 0.0001) was determined by using two-way ANOVA with Bonferroni correction. ERG: electroretinogram; 5w: 5 weeks; 8w: 8 weeks.
Proteomic profiling of wild type and rd10 mice retinas at 5 & 8 weeks
A proteomic analysis was conducted to unveil the molecular mechanisms behind alterations in the structures and functions of rd10 retinas with progression of retinitis pigmentosa. A diagram of the experimental strategy can be found in Fig. 3A. Based on the genotype (rd10 or WT) and age (5w or 8w) of the retinas, four groups were formed. According to Fig. 3B, 573 proteins significantly changed at 5w (p < 0.05), with 222 proteins changed by two-fold. Additionally, 722 proteins were significantly changed (p < 0.05) at 8w, and 289 proteins of them changed at least 2-fold. Venn diagram showed 111 overlapped DEPs with p < 0.05 and FC > 2 in retinas at 5w (pink) and 8w (blue) (Fig. 3B). Besides, there were 178 exclusive DEPs in 8w while 111 exclusive DEPs in 5w, illustrating the dramatic changes at the late stage of retinal degeneration in rd10 mice. The results showed a good agreement with previous research8. Based on the normalized log2 intensity values for individual proteins, principal component analysis (PCA) was performed to visualize the difference in gross protein differences between samples. Generally, the samples were segregated well by genotype WT vs. rd10 (PC1, 23.6% variance) and the age 5w vs. 8w (PC2, 15.6% variance) (Fig. 3C). Additionally, PC1 of rd10-5w/WT-5w is 23.6% variance and PC1 of rd10-8w/WT-8w is 32.3% variance (Fig. 3D and E), indicating that WT and rd10 retinas had significant differences at both 5 weeks and 8 weeks. Volcano plot indicated 84 up-regulated (red) and 138 down-regulated (green) proteins in rd10 retinas at 5 weeks (FC > 2, and p < 0.05) (Fig. 3F). Moreover, Fig. 3G showed 120 up-regulated (red) and 169 down-regulated (green) proteins in rd10 retinas at 8 weeks (FC > 2, and p < 0.05). It illustrated that more proteins changed at 8 weeks compared to those at 5 weeks. Supplementary Figure S3 showed the numbers of down-regulated proteins and up-regulated proteins in rd10 mouse retinas at 5w and 8w under the condition of p < 0.05 or the condition of p < 0.05 and FC > 2.
Proteome overview of WT and rd10 mice (n = 3 retinas per group). (A) The experimental strategy. Four groups of retinas were separated into two experiments based on age (5w or 8w) and genotype (WT or rd10). (B) Venn diagram showing the overlap of DEPs with p < 0.05 and FC > 2 in retinas at 5w (pink) and 8w (blue). (C) (D) (E) PCA analysis of gross protein differences, and between samples of WT and rd10 at 5w and 8w. (F) Volcano plot indicating 84 upregulated (red) and 138 downregulated (green) proteins in rd10 retinas at 5w (FC > 2, and p < 0.05). (G) Volcano plot showing 120 upregulated (red) and 169 downregulated (green) proteins in rd10 retinas at 8w (FC > 2, and p < 0.05). DEP: differentially expressed protein; PCA: principal component analysis; 5w: 5 weeks; 8w: 8 weeks.
Functional enrichment analysis of DEPs in rd10 mice
DEPs (p < 0.05 and FC > 2) were enriched with DAVID for GO and KEGG pathway analysis. According to KEGG pathway analysis, phototransduction played the most significant role in retinal changes. In addition, Purine metabolism, Metabolic pathways, Nucleotide metabolism, Cysteine and methionine metabolism, and PPAR signaling pathway were affected at 5 weeks (Fig. 4A). RP progression disrupted multiple pathways, including GABAergic synapse, metabolic pathways, purine metabolism, fructose and mannose metabolism (Fig. 4B).
Functional analysis of DEPs in rd10 retinas (n = 3 retinas per group). (A) (B) KEGG pathways enrichment with DEPs (FC > 2, p < 0.05) in rd10 retinas at 5w and 8w compared to age-matched WT retinas50,51,52. (C) BP pathways by DAVID enrichment with DEPs (FC > 2, FDR < 0.25) in rd10 retinas at 5w and 8w. (D) (E) (F) Heatmaps showing proteins associated with top 10 BP pathways enriched in 5w or 8w. FC: fold change; 5w: 5 weeks; 8w: 8 weeks; DEP: differentially expressed protein; BP: biological process; FDR: false discovery rates.
As a result of GO-BP analysis, visual perception was the most influenced pathway at both 5 weeks and 8 weeks in rd10 mice (Fig. 4C). Besides, response to stimulus, detection of light stimulus involved in visual perception, phototransduction, photoreceptor cell maintenance, etc., nine pathways were also primarily affected pathways. Protein localization and glycolytic process pathways were affected solely at 8 weeks, while positive regulation of erythrocyte differentiation was affected exclusively at 5 weeks (Fig. 4C). GO-BP analysis revealed that the visual perception and photoreceptor relevant pathways were devastated badly in rd10 mice retinas.
Heatmaps were drawn using protein intensity ratios of rd10/WT. Figure 4D showed 41 proteins associated with 3 common pathways at 5w and 8w, including visual perception, response to stimulus, and detection of light stimulus involved in visual perception pathways. Among them, 36 proteins belonged to visual perception pathway. Heatmaps showed 32 enriched protein sets of 4 shared pathways at 5w and 8w, including phototransduction, photoreceptor cell maintenance, retina development in camera-type eye, and phototransduction, visible light pathways (Fig. 4E). Figure 4F showed the heatmaps of 56 proteins and the enriched 6 pathways, consisting of three shared enriched pathways (response to drug, negative regulation of neuron projection development, and cilium assembly), two solely enriched pathways (protein localization and glycolytic process) at 8w, and single enriched pathway (positive regulation of erythrocyte differentiation) at 5w. In addition, a horizontal analysis in time dimension was performed to identify the proteins and pathways affected by Pde6b mutation during growth and development (Supplementary Figure S4).
Heatmaps and PPI analysis of DEPs causing retinitis pigmentosa
Heatmaps of 25 DEPs (FC > 2, p < 0.05) causing autosomal recessive/dominant retinitis pigmentosa (ar/ad-RP) were drawn. Among 25 DEPs, Nr2e3, Sag and Rho could lead both ar-RP and ad-RP. We demonstrated heatmaps of 18 proteins causing ar-RP (Fig. 5A). Sixteen of them were identified from RetNet Database (https://sph.uth.edu, updated at June 9, 2022), and fifteen of them were ever reported by Daiger et al.3. Ten DEPs causing ad-RP were displayed with heatmaps (Fig. 5B). Protein-protein interaction networks of 25 DEPs causing ar/ad-RP were displayed in Fig. 5C. In the Pde6b-centered PPI networks, thirteen proteins including Reep6, Cnga1, Rho, Nr2e3, Rom1, Pde6a, Guca1b, Abca4, Tulp1, Sag, Rdh12, Prph2 and Rbp3 directly interact with Pde6b protein. Chord diagram (Fig. 5D) showed the most enriched 8 biological processes (GO terms) with the differentially expressed proteins contributing to RP. Among 25 DEPs causing ar/ad-RP, 11 DEPs belonged to visual perception pathway, and 10 overlapped DEPs related with response to stimulus, two proteins (Rho and Guca1b) associated with phototransduction pathway. Four DEPs (Arl3, Bbs2, Bbs7 and Bbs4) were relevant to cilium assembly pathway, three DEPs (Bbs2, Bbs7 and Bbs4) of which belonged to protein localization pathway.
Heatmaps and PPI analysis of DEPs causing RP in rd10 retinas (n = 3 retinas per group). (A) (B) Heatmaps showing significantly changed proteins (FC > 2, p < 0.05) causing autosomal recessive retinitis pigmentosa and autosomal dominant retinitis pigmentosa in rd10 retinas. (C) Protein-protein interaction networks of DEPs causing RP. (D) Chord diagram indicating biological processes and DEPs contributing to RP. In each chord, enriched GO biological processes are shown on the right, and DEPs are shown on the left. On the left side of the circle, each DEP is indicated by a rectangle which colour is correlated to the value of logFC. PPI: protein-protein interaction; DEP: differentially expressed protein; RP: retinitis pigmentosa; FC: fold change.
mRNA and protein levels confirmation of DEPs causing retinitis pigmentosa
qPCR was performed to verify the mRNA levels of DEPs causing ar/ad-RP at both 5w and 8w. The gene expressions of Rom1, Rho, Prph2, Cnga1 and Pde6a at 5w rd10 and 5w WT mice retinas were further examined (Fig. 6A). Figure 6B showed the mRNA levels of Rdh12, Rbp3, Abca4, Sag, Impdh1 at 5w in WT and rd10. The mRNA levels for Bbs2, Bbs4, Bbs7, Tulp1 and Arl6 were verified at 5w WT and rd10 retinas (Fig. 6C). Figure 6D showed the gene expression levels of Prom1, Nr2e3 and Guca1b at 5 weeks. The above qPCR results confirmed that the DEPs causing RP had correspondingly decreased gene expressions in mouse retinas.
Quantitative PCR verification of RP related DEPs in rd10 mice (n = 3 retinas per group). (A) (B) (C) (D) Q-PCR results of 18 RP genes in rd10 retinas at 5 weeks. (E) (F) (G) (H) Q-PCR results of 18 RP genes in rd10 retinas at 8 weeks. 3 duplicates were measured for each group. The results were mean ± SEM. significance of difference(**p < 0.01, ***p < 0.001, ****p < 0.0001) was determined by using two-way ANOVA with Bonferroni correction.
Figure 6E displayed the comparative mRNA levles of Rom1, Rho, Prph2, Cnga1 and Pde6a. Figure 6F illustrated the gene expressions of Rdh12, Rbp3, Abca4, Sag, Impdh1 at 8 weeks. Figure 6G showed the expression fold changes of Bbs2, Bbs4, Bbs7, Tulp1 and Arl6 at 8 weeks. The relative mRNA levels of Prom1, Nr2e3 and Guca1b at 8 weeks were confirmed (Fig. 6H). The qPCR results at 8 weeks presented the accordingly decrease for all DEPs except an unexpected increase for Arl6.
Western blots were conducted to confirm the protein expression levels of DEPs causing ar/ad-RP in rd10 retinas at 5/8 weeks. Figure 7A, E and F showed the proteins levels of Pde6a and Prph2 at 5w & 8w retinas. Protein levels of Pde6a and Prph2 at both 5w and 8w rd10 retinas were barely detectable. Figure 7B, G and I displayed Bbs2, Nr2e3 proteins reduced significantly, and Bbs7 had no significant decrease at rd10 retinas (Fig. 7H). Figure 7C, J and K indicated the down-regulation of Rho and Rom1 proteins at 5w and 8w rd10 retinas. The protein levels of Rho and Rom1 at 8w lower than those at 5w rd10 retinas. Figure 7D, L and N illustrated that the protein levels of Rbp3 and Bbs4 at rd10 retinas were significantly lower than WT retinas, in which protein expressions of 8w rd10 were lower than those of 5w rd10. Besides, Fig. 7D and M showed Tulp1 decreased significantly at 5w rd10 retinas, but had no significance at 8w rd10. Figure 7D and O presented the Arl6 protein decreased slightly at rd10 retinas, and had no significance. The Western blots results not only confirmed the decreased expressions of RP causative proteins in rd10 retinas, but also revealed the time course of photoreceptor degeneration. Compared to protein levels at 5 weeks in rd10 retinas, most proteins further decreased at 8 weeks. Besides, we checked the proteins expressions and locations in WT and rd10 retinas at 5 weeks with immunohistochemistry (Supplementary Figure S5).
Western blots validation of RP related DEPs in rd10 mice (n = 3 retinas per group). (A) (E) (F) Proteins levels of Pde6a and Prph2 in age-matched WT and rd10 retinas. (B) (G) (H) (I) Bbs2, Bbs7 and Nr2e3 expression in retinas at 5 or 8 weeks. (C) (J) (K) Proteins expression of Rho and Rom1 in WT and rd10 mice retinas. (D) (L) (M) (N) (O) Rbp3, Tulp1, Bbs4 and Arl6 protein levels in age-matched mice retinas. 3 repeated lanes were conducted for each group. The results were mean ± SEM. significance of difference(**p < 0.01, ***p < 0.001, ****p < 0.0001) was determined by using unpaired t-test.
Discussion
Retinitis pigmentosa is a blinding inherited retinal disease influencing millions of people in the world23,24. However, the concrete pathological mechanisms of retinitis pigmentosa still remain poorly understood. It is essential to explain the retinal degeneration by elucidating the molecular differences between rd10 and healthy retinas, which may meanwhile contribute to the development of novel therapies. We applied proteomic analysis to age-matched WT and rd10 retinas by using LC-MS/MS.
Rd10 mouse bearing Pde6b mutation is a naturally occurring mouse model of retinitis pigmentosa. The rd10 mouse carries a missense mutation (c.1678 C > T; p.R560C) in exon 13 of Pde6b gene, which was widely used for recessive retinal degeneration.
In rd10 mice, the 560th amino acid Arginine mutated to Cysteine. Arginine is a positively charged, polar amino acid, while Cysteine is an uncharged but polar amino acid25,26. The changes of primary amino acid sequences definitely affect the senior structures of Pde6b protein, leading to the decrease of protein structural stability. Besides, the molecular weight and other physicochemical properties between Arginine and Cysteine are different. The change of amino acid obviously influences the stability and function of Pde6b protein.
There are previous proteomics, secretom and metabonomics studies concerning retinas, vitreous and lens6,8,27,28,29,30. Nevertheless, rare study ever illustrated the alterations of retinitis pigmentosa associated proteins played a prominent role in retinal degeneration. We identified and quantified 3737 proteins in this study, and the largest ever protein quantity in this study assisted a comprehensive analysis. The number of differentially expressed proteins was also larger than previous reported correspondingly. Our results reconfirmed the up-regulation of Gfap protein in rd10 mice retinas, which was consistent with previous study8. Differently, the present study principally paid attention to decreased proteins in rd10 mouse retinas.
KEGG analysis showed 3 altered signal transduction pathways in common at 5 weeks and 8 weeks. They were Phototransduction, Purine metabolism and Metabolic pathways. Besides, GABAergic synapse, Glutamatergic synapse, SNARE interactions in vesicular transport, Fructose and mannose metabolism, Glycolysis / Gluconeogenesis, Carbon metabolism, Central carbon metabolism in cancer, Carbohydrate digestion and absorption, MAPK signaling pathway and HIF-1 signaling pathway changed in rd10 retinas at 8 weeks. These pathways were associated with the disorders of neural transduction, energy metabolism impairments and protein trafficking destruction31,32,33,34, which might be the research directions in the future.
GO-BP analysis revealed that protein transport and energy metabolism were messed up during retinal degeneration. Correspondingly, BBSome associated proteins Bbs1, Bbs2, Bbs4 and Bbs7 were responsible for protein transportation35,36,37,38. DEPs consisting of Arl3, Arl6, Bbs1, Bbs2, Bbs4 and Bbs7 were enriched for cilium assembly. Connecting cilium in photoreceptor was in charge of protein transport from inner segment to outer segment39,40,41. In addition, Tulp1 was reported to be associated with outer segment protein transport pathways in photoreceptors42. We verified the mRNA and protein levels of Bbs2, Bbs4, Tulp1 and Arl6 in WT and rd10 retinas. All of them decreased in rd10 retinas except Arl6. The deficiency of these proteins can aggravate retinitis pigmentosa in rd10 retinas.
Phototransduction was the most severely affected pathway at both 5 weeks and 8 weeks, which was enriched with DEPs like Pde6b, Pde6a, Gnat1, Rho, Guca1b, Cnga1, Sag and other 9 proteins. Guca1b gene, coding for Gcap2, regulated guanylyl cyclases activity in recovery of flash responses in rod and cone photoreceptors43. PPI network analysis showed strong and direct interactions between Guca1b and Pde6b. qPCR confirmed the decrease of Guca1b mRNA in rd10 retinas. Complex mechanisms and biological processes may lie behind this phenomenon. Besides, there is a network of proteins interacted with Pde6b, which demands further experimental confirmation.
Cep250, Bag1, Fat3 and Hp in rd10 mice did not increase with aging and retinal development as in WT mice. These proteins were associating with ciliogenesis, anti-apoptosis, amacrine cell development and anti-oxidation in retinas44,45,46,47. The abnormal proteins expression and retina developmental disorder may be results of Pde6b mutation.
GO-CC analysis exhibited the proteins subcellular localization (Supplementary Table 3). Rom1 abbreviated for rod outer segment membrane protein 1, played a role in the organization and maintenance of rod outer segment disks48. Rom1 decreased violently at both 5w and 8w in rd10 retinas, which was validated by qPCR and Western Blots. Physiological function of rods and cones is dependent on Peripherin 2 (Prph2), a tetraspanin protein specific to photoreceptors49. Prph2 expressions also decreased fiercely in rd10 retinas. Undoubtedly, the loss of these RP causative proteins was closely related with outer nuclear destruction.
In conclusion, our results demonstrated: (1) overall proteomic profile of rd10 mice retinas at 5 weeks and 8 weeks; (2) the alterations of RP causative proteins in retinal degeneration; (3) the pathways influenced by the RP causative proteins. This study focused on the roles of RP causative proteins in Pde6b mutated RP mouse model. The results sheds new light on the mechanisms underlying retinitis pigmentosa. An abundance of proteomic and experimental data in this research provided a foundation for novel therapeutic strategies in the future.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier IPX0004747000. This article includes data supporting the findings of this study.
References
Dias, M. F. et al. Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Prog. Retin. Eye Res.63, 107–131. https://doi.org/10.1016/j.preteyeres.2017.10.004 (2018).
Xu, W. et al. Integrative RNA-seq and ATAC-seq analyses of phosphodiesterase 6 mutation-induced retinitis pigmentosa. Int. Ophthalmol.https://doi.org/10.1007/s10792-022-02238-0 (2022).
Daiger, S. P., Sullivan, L. S. & Bowne, S. J. Genes and mutations causing retinitis pigmentosa. Clin. Genet.84, 132–141. https://doi.org/10.1111/cge.12203 (2013).
Hart, A. W. et al. Genotype-phenotype correlation of mouse pde6b mutations. Investig. Ophthalmol. Vis. Sci.46, 3443–3450. https://doi.org/10.1167/iovs.05-0254 (2005).
Han, J. et al. Review: The history and role of naturally occurring mouse models with Pde6b mutations. Mol. Vis.19, 2579–2589 (2013).
Murenu, E. et al. Metabolic Analysis of Vitreous/Lens and Retina in wild type and retinal degeneration mice. Int. J. Mol. Sci.22, 114. https://doi.org/10.3390/ijms22052345 (2021).
Power, M. et al. Cellular mechanisms of hereditary photoreceptor degeneration—focus on cGMP. Prog. Retin. Eye Res.74, 896. https://doi.org/10.1016/j.preteyeres.2019.07.005 (2020).
Ly, A. et al. Proteomic profiling suggests central role of STAT signaling during retinal degeneration in the rd10 mouse model. J. Proteome Res.15, 1350–1359. https://doi.org/10.1021/acs.jproteome.6b00111 (2016).
Barabas, P., Cutler Peck, C. & Krizaj, D. Do calcium channel blockers rescue dying photoreceptors in the Pde6b (rd1) mouse? Adv. Exp. Med. Biol.664, 491–499. https://doi.org/10.1007/978-1-4419-1399-9_56 (2010).
Chang, B. et al. Two mouse retinal degenerations caused by missense mutations in the beta-subunit of rod cGMP phosphodiesterase gene. Vision. Res.47, 624–633. https://doi.org/10.1016/j.visres.2006.11.020 (2007).
Scimone, C. et al. Germline Mutation Enrichment in pathways Controlling endothelial cell homeostasis in patients with brain arteriovenous malformation: Implication for molecular diagnosis. Int. J. Mol. Sci.21, 253. https://doi.org/10.3390/ijms21124321 (2020).
Donato, L., Scimone, C., Rinaldi, C., D’Angelo, R. & Sidoti, A. New evaluation methods of read mapping by 17 aligners on simulated and empirical NGS data: An updated comparison of DNA- and RNA-Seq data from Illumina and Ion Torrent technologies. Neural Comput. Appl.33, 15669–15692. https://doi.org/10.1007/s00521-021-06188-z (2021).
Qiu, R. et al. The Protective Effects of VVN001 on LPS-Induced Inflammatory Responses in Human RPE Cells and in a Mouse Model of EIU. Inflammation44, 780–794. https://doi.org/10.1007/s10753-020-01377-9 (2021).
Wang, W. et al. Intravitreal Injection of an Exosome-Associated Adeno-Associated viral vector enhances retinoschisin 1 Gene Transduction in the Mouse Retina. Hum. Gene Ther.32, 707–716. https://doi.org/10.1089/hum.2020.328 (2021).
Yang, M. et al. P2X7 Receptor Antagonist Attenuates Retinal Inflammation and Neovascularization Induced by Oxidized Low-Density Lipoprotein. Oxid. Med. Cell. lLngevity2021, 5520644. https://doi.org/10.1155/2021/5520644 (2021).
Guo, Q. et al. Phenotype heterogeneity and the Association between Visual Acuity and outer retinal structure in a cohort of Chinese X-Linked Juvenile Retinoschisis patients. Front. Genet.13, 832814. https://doi.org/10.3389/fgene.2022.832814 (2022).
Qiu, R. et al. AAV2-PDE6B restores retinal structure and function in the retinal degeneration 10 mouse model of retinitis pigmentosa by promoting phototransduction and inhibiting apoptosis. Neural Regener. Res.https://doi.org/10.4103/nrr.Nrr-d-23-01301 (2024).
Kemp, J., Ladwig, P. M. & Snyder, M. R. Alpha-1-Antitrypsin (A1AT) proteotyping by LC-MS/MS. Methods Mol. Biol. (Clifton N. J.)2750, 95–106. https://doi.org/10.1007/978-1-0716-3605-3_9 (2024).
Jin, X. et al. Identification of age-associated proteins and functional alterations in human retinal pigment epithelium. Genom. Proteom. Bioinform.https://doi.org/10.1016/j.gpb.2022.06.001 (2022).
Jin, X. et al. Retinal proteomic alterations and combined transcriptomic-proteomic analysis in the early stages of progression of a mouse model of X-Linked retinoschisis. Cells2022, 11. https://doi.org/10.3390/cells11142150 (2022).
Xue, M. et al. Proteomic analysis of aqueous humor in patients with pathologic myopia. J. Proteom.952021, 234. https://doi.org/10.1016/j.jprot.2020.104088 (2021).
Yan, B. et al. Data-driven tool for cross-run ion selection and peak-picking in quantitative proteomics with data-independent acquisition LC-MS/MS. Anal. Chem.95, 16558–16566. https://doi.org/10.1021/acs.analchem.3c02689 (2023).
Ducloyer, J. B., Le Meur, G., Cronin, T., Adjali, O. & Weber, M. [Gene therapy for retinitis pigmentosa]. Med. Sci.: M/S36, 607–615. https://doi.org/10.1051/medsci/2020095 (2020).
Iribarne, M. & Masai, I. Do cGMP levels drive the speed of photoreceptor degeneration? Adv. Exp. Med. Biol.1074, 327–333. https://doi.org/10.1007/978-3-319-75402-4_40 (2018).
Hamid, M. et al. Hb AHVAZ [α83(F4)Leu→Arg, CTG > CGG (α2); HBA2: c.251T > G],a new hemoglobin variant of the α2-globin gene. Hemoglobin37, 477–480. https://doi.org/10.3109/03630269.2013.792095 (2013).
LoPachin, R. M., Gavin, T., Petersen, D. R. & Barber, D. S. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: Nucleophilic targets and adduct formation. Chem. Res. Toxicol.22, 1499–1508. https://doi.org/10.1021/tx900147g (2009).
Carmy-Bennun, T., Myer, C., Bhattacharya, S. K. & Hackam, A. S. Quantitative proteomic analysis after neuroprotective MyD88 inhibition in the retinal degeneration 10 mouse. J. Cell. Mol. Med.25, 9533–9542. https://doi.org/10.1111/jcmm.16893(2021).
Cavusoglu, N. et al. Differential proteomic analysis of the mouse retina: The induction of crystallin proteins by retinal degeneration in the rd1 mouse. Mol. Cell. Proteom.: MCP2, 494–505. https://doi.org/10.1074/mcp.M300029-MCP200 (2003).
Rasmussen, M., Welinder, C., Schwede, F. & Ekström, P. The cGMP system in normal and degenerating mouse neuroretina: new proteins with cGMP interaction potential identified by a proteomics approach. J. Neurochem.157, 2173–2186. https://doi.org/10.1111/jnc.15251 (2021).
Donato, L. et al. Human retinal secretome: A cross-link between mesenchymal and retinal cells. World J. stem Cells15, 665–686. https://doi.org/10.4252/wjsc.v15.i7.665 (2023).
Grossman, G. H. et al. Protein partners of dynamin-1 in the retina. Vis. Neurosci.30, 129–139. https://doi.org/10.1017/s0952523813000138 (2013).
Hughes, E. G., Elmariah, S. B. & Balice-Gordon, R. J. Astrocyte secreted proteins selectively increase hippocampal GABAergic axon length, branching, and synaptogenesis. Mol. Cell. Neurosci.43, 136–145. https://doi.org/10.1016/j.mcn.2009.10.004 (2010).
Nishiwaki, Y. et al. The BH3-only SNARE BNip1 mediates photoreceptor apoptosis in response to vesicular fusion defects. Dev. Cell25, 374–387. https://doi.org/10.1016/j.devcel.2013.04.015 (2013).
Schramm, G. et al. Analyzing the regulation of metabolic pathways in human breast cancer. BMC Med. Genom.3, 45. https://doi.org/10.1186/1755-8794-3-39 (2010).
Landfear, S. M., Tran, K. D. & Sanchez, M. A. Flagellar membrane proteins in kinetoplastid parasites. IUBMB Life67, 668–676. https://doi.org/10.1002/iub.1411 (2015).
Lechtreck, K. F. et al. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol.187, 1117–1132. https://doi.org/10.1083/jcb.200909183 (2009).
Masek, M. et al. Loss of the Bardet-Biedl protein Bbs1 alters photoreceptor outer segment protein and lipid composition. Nat. Commun.13, 1282. https://doi.org/10.1038/s41467-022-28982-6 (2022).
Starks, R. D. et al. Regulation of insulin receptor trafficking by Bardet Biedl Syndrome proteins. PLoS Genet.11, e1005311. https://doi.org/10.1371/journal.pgen.1005311 (2015).
Anand, M. & Khanna, H. Ciliary transition zone (TZ) proteins RPGR and CEP290: Role in photoreceptor cilia and degenerative diseases. Expert Opin. Ther. Targets16, 541–551. https://doi.org/10.1517/14728222.2012.680956 (2012).
Armengot, M. et al. Nasal ciliary beat frequency and beat pattern in retinal ciliopathies. Investig. Ophthalmol. Vis. Sci.53, 2076–2079. https://doi.org/10.1167/iovs.11-8666 (2012).
Pulvermüller, A. et al. Calcium-dependent assembly of centrin-G-protein complex in photoreceptor cells. Mol. Cell. Biol.22, 2194–2203. https://doi.org/10.1128/mcb.22.7.2194-2203.2002 (2002).
Grossman, G. H., Watson, R. F., Pauer, G. J., Bollinger, K. & Hagstrom, S. A. Immunocytochemical evidence of Tulp1-dependent outer segment protein transport pathways in photoreceptor cells. Exp. Eye Res.93, 658–668. https://doi.org/10.1016/j.exer.2011.08.005 (2011).
Makino, C. L. et al. A role for GCAP2 in regulating the photoresponse. Guanylyl cyclase activation and rod electrophysiology in GUCA1B knock-out mice. J. Biol. Chem.283, 29135–29143. https://doi.org/10.1074/jbc.M804445200 (2008).
de Castro-Miró, M. et al. Novel candidate genes and a wide spectrum of structural and point mutations responsible for inherited retinal dystrophies revealed by exome sequencing. PloS One11, e0168966. https://doi.org/10.1371/journal.pone.0168966 (2016).
Hung, W. J., Roberson, R. S., Taft, J. & Wu, D. Y. Human BAG-1 proteins bind to the cellular stress response protein GADD34 and interfere with GADD34 functions. Mol. Cell. Biol.23, 3477–3486. https://doi.org/10.1128/mcb.23.10.3477-3486.2003 (2003).
Krol, A., Henle, S. J. & Goodrich, L. V. Fat3 and Ena/VASP proteins influence the emergence of asymmetric cell morphology in the developing retina. Dev. (Cambrid. Engl.)143, 2172–2182. https://doi.org/10.1242/dev.133678 (2016).
Fasano, A. Zonulin and its regulation of intestinal barrier function: The biological door to inflammation, autoimmunity, and cancer. Physiol. Rev.91, 151–175. https://doi.org/10.1152/physrev.00003.2008 (2011).
Clarke, G. et al. Rom-1 is required for rod photoreceptor viability and the regulation of disk morphogenesis. Nat. Genet.25, 67–73. https://doi.org/10.1038/75621 (2000).
Conley, S. M., Stuck, M. W., Watson, J. N. & Naash, M. I. Rom1 converts Y141C-Prph2-associated pattern dystrophy to retinitis pigmentosa. Hum. Mol. Genet.26, 509–518. https://doi.org/10.1093/hmg/ddw408 (2017).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587–D592 (2023).
Acknowledgements
This work was supported by Fundamental Research Program of Henan Eye Hospital (22JCQN010), Health Commission of Henan Province (LHGJ20220084), Key Technologies Research and Development Program of Henan Science and Technology Bureau (242102311068) and National Natural Science Foundation of China (82071008).
Author information
Authors and Affiliations
Contributions
Bo Lei designed and supported the experiments. Mingzhu Yang collected data and wrote the manuscript. Mingzhu Yang and Ruiqi Qiu conducted the experiments. Xiuxiu Jin supported the data analysis and figures drawing. Jingyang Liu and Weiping Wang helped the molecular experiments. Shun Yao and Guangming Liu helped the mouse raising and breeding. Jinfeng Han supported the animal experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
The ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines 2.0 and ARVO guidelines were followed for animal experiments. Procedures involving animals were permitted by the Ethics Committee of Henan Eye Hospital (Permit No: HNEECA-2021-10).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, M., Qiu, R., Jin, X. et al. Proteomics identifies multiple retinitis pigmentosa associated proteins involved in retinal degeneration in a mouse model bearing a Pde6b mutation. Sci Rep 14, 22090 (2024). https://doi.org/10.1038/s41598-024-72821-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-72821-1