Abstract
Histopathologically, oral squamous cell carcinoma (OSCC) consists of well-defined interfaces with adjacent non-cancerous epithelium. Previously, we found that SCC tissues expressed higher levels of specific proteins at this interface. Ladinin-1 (LAD1) is one of the specific molecules that has increased expressions in cancer fronts; however, its function in OSCC is unknown. Therefore, this study aimed to elucidate the function of LAD1 in human OSCC cells. LAD1 was localized on the actin arc at the distal periphery of cell clusters in the OSCC cell lines HSC-2, HSC-3, and HSC-4. When LAD1 was knocked down, cellular migration was repressed in wound scratch assays but was reversed in three-dimensional collagen gel invasion assays. Characteristic LAD1 localization along actin arcs forming the leading edge of migrating cells was diminished with loss of filopodia formation and ruffling in knockdown cells, in which the expression levels of cell motility-related genes—p21-activated kinase 1 (PAK1) and caveolin-1 (CAV1)—were upregulated and downregulated, respectively. LAD1 expression was also associated with the downregulation of vimentin and increased histological differentiation of OSCC. These results suggest that LAD1 is involved in actin dynamics during filopodia and lamellipodia formation, and in maintaining the epithelial phenotype of OSCC cells.
Similar content being viewed by others
Introduction
Oral squamous cell carcinoma (OSCC) frequently forms an evident lateral interface with the surrounding non-cancerous epithelium, wherein cancer cells and non-cancerous epithelial cells come into direct contact with each other, harboring different gene alterations, protein expressions, and other molecular phenotypes1. Using histopathological tissue specimens of OSCC, we previously characterized interface morphologies. In addition, using liquid chromatography–tandem mass spectrometry (LC–MS/MS)-based proteomic analysis, we showed dynamic molecular expression profiles between the cancerous and non-cancerous sides1. As a result, several proteins that were specifically enhanced on the cancerous side piqued our interest. Among these, we first focused on ladinin-1 (LAD1), whose function in OSCC cells still remains unknown.
LAD1 was first described as a protein comprising the basement membrane and as an autoantigen of a cutaneous blistering autoimmune disease, known as linear IgA bullous dermatosis, in mice2,3; however, we previously suggested that it functions within the cytoplasm because it was immunohistochemically localized in the periphery of OSCC cells1. More recently, it has been shown to function in the migration of mammary4 and colorectal5 cancer cells, and its higher expression levels are related to the poor prognosis of patients with these cancer types. In addition, LAD1 expression is reportedly higher in laryngeal SCCs with metastasis than in those without metastasis6. These reports suggest that LAD1 may play an important role in the motility of either adenocarcinoma or SCC.
The association between LAD1 expression and cancer biology has been reported in various cancer types, such as mammary invasive ductal carcinoma4, colorectal adenocarcinoma5, gastric adenocarcinoma7, lung adenocarcinoma8, and prostatic adenocarcinoma9. Li et al. reported that high LAD1 expression in prostate cancer was involved in drug resistance9. Among the various types of adenocarcinomas, abundant expression of LAD1 has been linked to cancer cell progression, and its expression is possibly regulated by miR-331 in lung adenocarcinoma10. Moreover, in our previous study, we focused on the peculiar site-specific higher expression of LAD1 in OSCC cells laterally facing non-cancer epithelia1. The expressional imbalance of LAD1 potentially arise the concept of cancer heterogeneity in OSCC.
In the context of the association of LAD1 and cell motility, Roth et al. reported that intracellular LAD1 in breast cancer cells binds to actin fibers via filamin-A4. The actin filament consists of a cytoskeleton supporting cell shape and is involved in cellular motility. Among the specific actin structures in the cytoplasm, cell processes such as filopodia, lamellipodia, and lamella, and their associates, known as actin arcs are pivotal factors in generating cell crawling force via myosin II contraction11.
Therefore, in this study, we hypothesized that LAD1 is involved in the tumor heterogenic phenotype and the lateral invasion of human OSCC cells via regulation of cell motility with background associations with actin molecules, which promoted their superficial spread.
Results
LAD1 was mainly localized in actin arcs and associated with actin filament formation
We first evaluated LAD1 expression at both the protein and mRNA levels in three OSCC cell lines: HSC-2, HSC-3, and HSC-4, because these three cell lines commonly express LAD1. Immunofluorescence revealed linear localization of LAD1 at the periphery of the cytoplasm of HSC-2 HSC-3, and HSC-4 (Fig. 1a). The linear pattern for LAD1 was most pronounced along the outermost zones of small-cell clusters, which were composed of fewer than 10 cells. The cell–cell attachment surface was not strongly positive. The mRNA and protein expression levels of LAD1 were confirmed to be similar in the three cell lines, using real-time polymerase chain reaction (real-time PCR; Fig. 1b) and western blotting (Fig. 1c), respectively.
Expressions of ladinin-1 (LAD1) in oral squamous cell carcinoma (OSCC) cell lines, HSC-2, HSC-3, and HSC-4. (a) Immunofluorescence for LAD1 (green) and DAPI (blue); scale bars: 50 μm. (b) LAD1 mRNA expression via real-time polymerase chain reaction (real-time PCR). (c) Protein expression using western blotting. LAD1 was localized in a linear pattern in the periphery of HSC-2, HSC-3, and HSC-4 cells, and such peripheral linear signals were most enhanced when they formed round clusters in HSC-2 and HSC-3 cells (a). The three SCC cell lines showed similar gene (b) and protein (c) expression levels for LAD1.
High-resolution morphological analysis of HSC-4 cells using structured illumination microscopy (SIM) revealed that fine granular LAD1 signals were localized on arch-shaped actin bundles (Fig. 2); these are considered to be actin arcs12. Actin arcs were enhanced and formed in the cell periphery, facing the direction of cell migration. Actin arcs were arranged perpendicularly at the innermost ends of the filopodia or along the basal part of the lamellipodia (Fig. 2a). The reconstructed three-dimensional (3D) images in both orthogonal projection (Fig. 2b) and pseudo-color images (Fig. 2c) revealed that LAD1 and actin filaments were localized within the actin arcs formed at the cell bottom, from which filopodia and lamellipodia were projected. The colocalization of LAD1 and actin filaments was not clearly observed on the inner side of the actin arcs or in the lamellipodia zone, including filopodial spikes, although punctuated LAD1 signals, which were independent of actin filaments, were diffusely scattered in the central zone of the cells (Fig. 2a).
Comparative three-dimensional (3D) localization of actin versus LAD1 in HSC-4 cells. (a) Maximum intensity projection images: phalloidin-rhodamine fluorescence for F-actin, immunofluorescence for LAD1, and their merges; scale bars: 20 μm. (b) Orthogonal projection image of F-actin (red) and LAD1 (green); scale bar: 5 μm. (c) Pseudo-color image; scale bars: 4 μm. (d) Immunofluorescent images of F-actin (red) and LAD1 (green) of 3D on-gel culture section and intensity plots of the LAD1; scale bars: 20 μm (left panel) and 10 μm (a and b). Arch-shaped actin bundles, called actin arcs (white arrowheads), are arranged along the lamellipodia base, represented by fine actin meshwork, and filopodial protrusions represented by actin filament spikes. Dot-like signals inside the cell may represent cross-sectional actin fibers that are not associated with the actin arc, and LAD1 granular signals co-localized with actin filaments within the actin arc range (a). The orthogonal projection image (b) and its pseudo-color images (c) at an identical height at the cellular base revealed that LAD1 exclusively colocalized with actin filaments forming the actin arc; however, LAD1 signals scattered inside the cell did not always colocalize with short actin filaments (a–c). In the immunofluorescence of the on-gel culture section (d), peripheral accumulation of LAD1 was observed in the stacking cells on the gel layer (d-i, white arrowheads), but not in the cells invading the collagen gel (d-ii). Quantitatively, the intensity plots of LAD1 indicate peripheral accentuation (black arrowheads).
To visualize LAD1 localization in stacked and invading cells simultaneously, we used confocal microscopy on sections of the collagen gel invasion assays with immunofluorescence (Fig. 2d). HSC-4 cells were stacked on the collagen gel layer, and some cells began to invade the collagen gels after incubation. In the vertical sections of the collagen gel assay with immunofluorescence, we simultaneously observed both stacked cells on the collagen layer (Fig. 2d-i) and cells invading the collagen (Fig. 2d-ii). Small cancer-cell-clusters were observed in the collagen layer, which were isolated from stacked cells on the collagen surface. HSC-4 cells stacking on the collagen layer showed a wider cytoplasm and polygonal shape, and LAD1 linear positivity was retained at the cell border (Fig. 2d-i, white arrowheads); however, infiltrating HSC-4 cells in the collagen gel layer showed shrunken cytoplasm and irregular elongation of the cell process (Fig. 2d-ii). In addition, LAD1 linear positivity in the cell periphery was completely diminished in irregularly shaped cells (Fig. 2d-ii). Observational findings were visualized using intensity plots from the initial to terminal points of the yellow arrows. Black arrowheads in the figure indicate peripheral accentuation of the LAD1.
LAD1 promotes horizontal, but not vertical SCC cell migration
To evaluate the effects of LAD1 on cell migration, gel invasion and transwell migration assays were performed via siRNA transfection against LAD1. The siRNA efficacy in the three cell lines was analyzed using real-time PCR (Supplementary Fig. S1), and western blotting (Supplementary Fig. S2). Thus, LAD1 mRNA expression was successfully knocked down. The cell survival analysis of the knockdown cells were determined using the MTS assay (Fig. 3a).
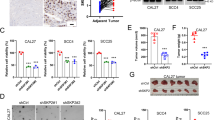
Functional and morphological effects of LAD1 knockdown in OSCC cell lines. (a) Cell survival analysis by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2 H-tetrazolium, inner salt (MTS) assay. (b) Collagen gel invasion assay. (c) Transwell migration assay. (d) Scratch wound-healing assay. (e) Cell-tracking plots of timelapse imaging for 24 h and comparisons of velocity and distance/frame; scale bars: 100 μm (b, c). Bars in bar charts, means ± s.e.; *p < 0.05, **p < 0.01. In MTS assay (a), three cell lines showed suppression of cell survival rates in si#1 treatments. HSC-4 showed the common suppressive effect of LAD1 knockdown on cell survival. In the collagen gel invasion assay, depths of cell infiltration, i.e., absolute length from the uppermost collagen surface to the deeper cancer cells in captured foci were measured in the vertical sections of paraffin-embedded culture samples. In si#1 LAD1knockdown of the three cell lines, the cancer cells showed considerable deep infiltration (b). HSC-2 and HSC-4 with si#2 and HSC-2 and HSC-3 with si#3 also showed increased depth of infiltration. In transwell migration assays, si#1 and si#3 effectively suppressed cell migration 24 h after seeding, whereas si#2 did not (c). In the wound scratch assay with serum-free media, LAD1-knockdown cells showed wider gaps than controls 48 h after scratching only in HSC-2 and HSC-4 with si#1 (d). The time-lapse analysis was performed to analyze cellular planer motility (e). The LAD1-knockdown cells showed decreased average velocity and moving distance between the time frames.
Cell survivals of the knocked-down cells were determined by MTS assays. Both si#1 and si#3 suppressed cell survival rates in HSC-3 and HSC-4 significantly (p < 0.05) at days 7 after transfection (Fig. 3a). si#2 showed a suppressive effect in HSC-4 but not in HSC-2 and HSC-3. For HSC-4, three siRNAs commonly showed suppressive effects on cell survival.
In the collagen gel invasion assays, we measured the depth to which cancer cells infiltrated the underlying collagen layer as the absolute length from the uppermost surface of the collagen layer to the lower end of the deeper infiltrating cells in hematoxylin and eosin-stained sections. Among all cell lines, the cell infiltration depth was significantly increased (p < 0.05) in at least two siRNAs (Fig. 3b). In transwell migration assays, si#1 effectively enhanced the migrating cells number 24 h after seeding in the three cell lines (Fig. 3c), whereas si#2 enhanced those of HSC-4, but not of HSC-2 and HSC-3, although the differences were not statistically significant.
Conversely, in the wound-scratch assays, LAD1-knockdown HSC-2 and HSC-4 showed larger remaining cell-free areas than controls 48 h after scratching (Fig. 3d). Scratched surface closure was most effectively inhibited by si#1 and not si#2 or si#3. Although this was a common tendency among the three cell lines, only si#1 showed statistical significance in HSC-2 and HCS-4 cells (Fig. 3d).
Such contrasting alterations between vertical motility, as shown in transwell migration and collagen gel invasion assays, and horizontal motility, as shown in the wound scratch assay, and time-lapse analysis comparing control and si#1, which had more phenotypic effect, suggest that LAD1 is involved in cancer cell motility, but its functional mode may differ slightly in two-dimensional (2D) cell migration or 3D cell invasion.
Furthermore, we evaluated the morphological effects of LAD1 knockdown using siRNA (Fig. 4a). Control HSC-4 cells showed elongated filopodial spikes and widely spread lamellipodia in the cell migration direction, in which LAD1 was mainly localized in the actin arcs. LAD1-knockdown cells showed shrunken or irregularly shaped filopodia and lamellipodia. These results indicate that LAD1 is associated with filopodial and lamellipodial formation in the actin arc. We performed the fractal dimension analysis using the box-counting method to quantify the cell shape outline complexity12. The fractal dimension significantly increased (p < 0.05) in HSC-3 and HSC-4 with si#1 and HSC-2 and HSC-4 with si#3. These results showed that LAD1 expression level was associated with cellular shape complexity.
Cell morphological effects of knockdown of LAD1. (a) Representative immunofluorescence images in HSC-4 cells on day three after plating; siRNAs against LAD1 (si#1) and negative control (cont). (b) The fractal dimension was obtained by the fractal analysis of immunofluorescence images using FracLac; scale bars, 50 μm (a). In immunofluorescence, the HSC-4 cells (cont) form radially extended filopodia and widely spread lamellae (asterisk), which are composed of thick and elongated actin filaments (white arrows). At the base of the filopodia and lamella, LAD1 is localized along the actin arc. The cells form polygonal and paved shapes when in contact with each other. In contrast, LAD1-knockdown cells appear shrunken with irregularly formed filopodia (white arrowheads) and a lack of lamellar expansion, resulting in a further irregular ruffled cell border shape, in which LAD1 is not localized. The fractal analysis by the box-counting method on auto-segmentation images showed HSC-3 and HSC-4 indicated significantly (p < 0.05) higher average values of fractal dimension in LAD1 knockdown cells by si#1. Inversely, HSC-2 showed a decrease in fractal dimension in si#2.
Transcriptome analysis revealed that LAD1 expression level was associated with cell motility
To examine the effect of alterations in LAD1 expression on the transcriptome profile, we conducted a comprehensive gene expression analysis using RNA sequencing (RNA-seq). Triplicate samples of total RNA isolated from LAD1-knockdown cells (si#1) and controls (cont) were analyzed. The control and siRNA sample groups were clearly separated in tSNE (Fig. 5a). Differentially expressed gene analysis revealed 1059 upregulated and 694 downregulated genes in LAD1-knockdown cells, as indicated in the MA-plot (Fig. 5b). Furthermore, bicluster analysis showed that sample groups were clustered into control and suppressed groups; “cell motility” and “localization of cell” were listed in the specific cluster of genes (Fig. 5c). Although biclustering analysis is typically useful for large sample sizes, our results support the hypothesis that LAD1 expression is associated with cell motility (Supplementary Table S1).
Transcriptome analysis using high-throughput RNA sequencing (RNA-seq). (a) The t-distributed stochastic neighbor embedding (t-SNE) plot. (b) MA-plot of expressing genes. (c) Heatmap of the selected clusters from the biclustering analysis. (d) Gene ontology analysis according to biological processes in the selected cluster. Transformed RNA-seq data were analyzed using iDEP. Principal component analysis using t-SNE revealed clear differences between control and LAD1-knockdown samples (a). Differential expression gene analysis revealed 1059 upregulated and 694 downregulated genes (b). Further biclustering analysis using the BCCC method revealed five clusters, and the selected cluster No. 1 enriched several biological processes including “cell motility” and “cell localization” (c).
LAD1 expression was linked with cell motility-related molecules
To further explore the expression of genes associated with actin regulation, the expression levels of 84 cell motility-related genes in HSC-4 cells were compared with those in si#1 treated cells using PCR arrays. The results were plotted on a volcano plot based on fold regulations of each gene (horizontal axis) against their p-values: in the vertical axis (Fig. 6a) si#1 upregulated genes with > two-fold regulations and p-values < 0.05 were plotted with red dots, and downregulated ones with < two-fold regulations and p-values < 0.05 were plotted with green dots. As shown in Fig. 6b, the gene expression levels of p21-activated kinase 1 (PAK1), vimentin (VIM), epithelial growth facter receptor (EGFR), phospholipoase D1 (PLD1), phosphatase and tensin homolog deleted from chromosome 10 (PTEN), and protein kinase C alpha (PRKCA) were significantly enhanced (p < 0.05), whereas those of caveolin-1 (CAV1), fibroblast growth factor 2 (FGF2), matrix metalloprotainase 2 (MMP2), myosin light chain kinase (MYLK), paxillin (PXN), actinin 4 (ACTN4), and breast cancer anti-estrogen resistance protein 1 (BCAR1) were suppressed upon LAD1 knockdown.
Cell motility-related gene expression profiles in LAD1-knockdown HSC-4 cells. (a) Listing of representative upregulated or downregulated genes. Red dots, upregulated genes; green, downregulated genes; black, stable genes; gray, genes with no significant differences. (b) Volcano plot of listed genes. In LAD1-knockdown HSC-4 cells (si#1), the mRNA expression levels of CAV1, FGF2, and the other five genes were significantly repressed (p < 0.05) (a) among the 84 cell-motility-related genes in the PCR array kit used, whereas those of PAK1, VIM, and the other four genes were significantly enhanced (b).
LAD1-expressing cells showed low vimentin expression
Among the overexpressed genes in the PCR array, vimentin (VIM; Fig. 6b) is a major marker of the mesenchymal phenotype in cancer cells during epithelial–mesenchymal transition13. In the immunofluorescence study, LAD1-knockdown cells by si#1 showed a significantly increased (p < 0.05) vimentin-positive cell ratio when cultured on a plastic plate (Fig. 7a). HSC-3 showed increased vimentin positivity in all siRNA condition. Immunofluorescent staining for LAD1 and vimentin in the collagen gel culture section revealed that LAD1 positivity was predominantly observed in the upper layer of stacked cells; however, vimentin positivity was enhanced in the deeper part, particularly in the invading cells in the collagen layer (Fig. 7b). This reciprocal expression pattern is shown in the intensity plot (Fig. 7c) of the line from the surface to the deep layer (yellow arrow) in Fig. 7b. In the upper stacked cells on the collagen gel (asterisks in Fig. 7b), cancer cells expressed higher LAD1 levels and lower vimentin levels. In contrast, in the deeper part of the gel (daggers in Fig. 7b), the intensity of LAD1 decreased; however, the intensity peak of vimentin was frequently detected. These results indicate that LAD1 and vimentin expression is regulated reciprocally.
Expression levels of vimentin and LAD1 and histological differentiation of the public cancer dataset. (a) Representative immunofluorescent images of HSC-3 and dot plots of vimentin-positive cell ratio in HSC-2, HSC-3, and HSC-4 cells. (b) A confocal immunofluorescence image of the vertical section of HSC-4 cells cultured on the collagen gel for LAD1 (red) and vimentin (green) with nuclear staining (Hoechst, blue). (c) Fluorescent intensity plot of vimentin (green) and LAD1 (red) on the yellow arrow in panel (b). (d) The histological grades of LAD1-mRNA high and low expression groups in the head and neck squamous cell carcinoma dataset of the cancer genome atlas; scale bars: 200 μm (a) and 20 μm (b). Bars in the chart, means ± s.e.; *p < 0.05, **p < 0.01. G1, well-differentiated; G2, moderately-differentiated; G3, poorly-differentiated; G4, undifferentiated; GX, not assessed. In LAD1-knockdown cells, the ratio of vimentin-positive cells significantly increased, and HSC-3 cells showed a high vimentin-positive percentage in LAD1-knockdown conditions (a). In the section of HSC-4 cells cultured on collagen gel, stacked cells on the gel were positive for LAD1 (red); however, invading cells in the gel showed strong positivity for vimentin (green) (b). In the measurement of fluorescence intensity (c), the vimentin intensity (green line) gradually increased from the surface (asterisks) to the deeper parts (daggers). Reciprocally, the LAD1 intensity (red line) was higher in surface cells (asterisks) but lower in deeper cells (daggers). Public database analysis via cBioportal revealed a significant increased rate (p < 0.05) of well-differentiated histology (G1) in the LAD1-mRNA high-expressing squamous cell carcinoma group.
LAD1 expression was related to differentiation in the clinical samples
Analysis of the cancer genome atlas database on cBioPortal (https://www.cbioportal.org/) showed that well-differentiated histology was considerably predominant in the LAD1-high expression group of head and neck SCC, compared with the low expression group (Fig. 7d). These results indicate the histopathological differentiation, i.e. degree of epithelial differentiation, is associated with LAD1 expression level in the clinical cancer tissue samples.
Discussion
LAD1 localization on the actin arc emphasizes its potential involvement in the structural modulation underlying cell motility. In OSCC cells, LAD1 was localized in the outermost cell periphery of small-cell clusters and was colocalized with actin filaments in the actin arc. The outermost cells had more space to expand their cell processes and migrate than the cells in the central part of the cluster. Moreover, the actin arc is composed of actin filaments aligned with the leading edge of the cell14, and myosin II proteins are involved in its contraction11. During each edge retraction, these arc-shaped actomyosin structures are formed in the lamellipodium15. As the edge switches from protrusion to retraction, lamellipodial actin is converted into an actin arc15. During epithelial cell crawling, the positional advance of the new-actin-arc addition to the lamellar front is balanced by the positional advance of the previous-actin-arc removal site at the lamellar rear15. The continuous linear positivity of LAD1 at the outermost edge of small cell clusters of cancer cells (Fig. 1a) is identical to the localization at the base of the leading edge in individual cells (Fig. 2a), suggesting that actin arc formation and LAD1 localization on the actin arc may be essential in the direction in which the cells can migrate. In addition, the actin arc is reportedly involved in vimentin localization in the U2OS osteosarcoma cell line16. Actin arc-depleted osteosarcoma cells show loss of perinuclear localization of the vimentin network, and vimentin is diffusely distributed in the cytoplasm of the cells16.
Regarding morphological changes and their effects on cell migration, elevated PAK1 expression is an important finding related to actin dynamics. The immunofluorescent images and the fractal analysis showed cell shape complexity increased in HSC-3 and HSC-4 by LAD1 downregulation (Fig. 4 and Supplementary Fig. S3). These results suggested that the cell process, an essential factor in determining cell shape, is modulated by the LAD1 expression level. Morphologically, cell membrane ruffling suggests local activation of Rac and Cdc4217, including the requirement of RhoA activation17. We detected PAK1 upregulation in LAD1-knockdown cells. PAK1 links Rac-Cdc42 activation18 and is critical in forming lamella and lamellipodia19. Thus, the altered expression of PAK1 under LAD1 knockdown treatment suggested the novel possibility of pathway links between LAD1 and the Rac-Cdc42 axis; this molecular background potentially explains the LAD1-mediating promotion of cell invasion.
Molecular regulation of the actin molecules by LAD1 may be related to the regulation between lateral progression, vertical invasion to the stroma, and cell migration. Regulation of directional cell migration is associated with planar cell polarity, which is regulated via actin fiber arrangement, a critical mechanism determining the direction of cell migration20. Within planar cell polarity, Wnt non-canonical pathways mediate actin molecule rearrangement21 and have been implicated in cancer cell migration across various cell types20, including head and neck SCCs22. The reciprocity of LAD1 suppression in planar and 3D cell migration led us to hypothesize that LAD1 is also associated with cell polarity. Although investigations of LAD1 function in mammary adenocarcinoma by Roth et al.4 and in colorectal adenocarcinoma by Moon et al.5 suggested that LAD1 knockdown was associated with a lower migration potential of adenocarcinoma cells in transwell migration/invasion assays, OSCC cells showed opposite tendencies in this study; for example, LAD1 knockdown was associated with a higher potential for 3D migration. This discrepancy may be attributed to the differences in cell differentiation between adenocarcinoma and SCC, and the complex disparities in cell polarity based on cell differentiation.
Among the genes that showed altered expression upon LAD1 suppression, the decreased expression of caveolin-1 could explain the conflict between 3D and planar migration. Caveolin-1 was the most downregulated gene, with less than 1/17th of the mRNA expression compared to the control. Caveolin-1 is a major structural protein in the caveolae23. Caveolin-1 is located at the “rear” of migrating cells, and the loss of caveolin-1 by siRNA prevents cell polarization and directional movement in endothelial cells24. In OSCC cells, caveolin-1 is associated with cellular polarity and 2D cell migration25. Moreover, Caveolin-1 accumulates on the trailing edge of migrating cells during 2D migration; however, it is polarized at the migrating cell front during 3D migration26. These findings suggest that downregulation of caveolin-1 is essential for modulating cell polarity and migration in LAD1-knockdown cells.
LAD1 expression affected epithelial morphology. LAD1-knockdown cells showed shrinking and ruffling characteristics in non-epithelial cells with increased proportions of vimentin-positive cells due to the LAD1 knockdown, and the tissue differentiation grade of the SCC case histology was related to LAD1 expression levels. Lu et al. reported that downregulation of caveolin-1 was necessary and sufficient for epidermal growth factor (EGF)-induced upregulation of Snail, and that EGF-induced upregulation of Snail transcription played an essential role in the transcriptional downregulation of E-cadherin27. Simultaneous Snail upregulation and E-cadherin downregulation induce epithelial–mesenchymal transition28. Although our study did not clarify the underlying molecular mechanisms, LAD1 may be involved in maintaining the epithelial phenotype of cancer cells.
The in vivo results, which were reported previously1, indicated intra-cancer tissue difference in LAD1 protein amount. An integrated interpretation of the previous results and the ones in this study would suggest that LAD1 reflects intratumoral heterogeneity. The results show that LAD1 expression positively regulates planar cell motility and negatively regulates 3D cell migration. LAD1 expression is higher in the periphery and lower in the center of OSCC tissues1. OSCC cells with high LAD1 expression can be characterized by intraepithelial planar migration against non-cancerous cells, whereas those with low LAD1 expression can progress to stromal invasion. However, throughout the experiments, the present data had a discrepancy of effect depending on the differences in siRNA sequencing. And this study had technical limitations in monitoring LAD1 protein level in individual cells and in simulating the in vivo conditions of cancer heterogeneity under controlled LAD1 expression. In addition, it is possible that changes in LAD1 expression levels are strictly regulated at the cellular level in a small number of cells, and therefore quantification of expression levels using bulk samples may not elucidate the real picture. Further studies are required to unveil the protein interactions involved in actin fiber dynamics, cell polarity associated with LAD1 using single cell level analysis, and the clinical specificity of LAD1 to evaluate the biological behavior of in vivo cancerous and non-cancerous cells.
Conclusions
In this study, we demonstrated the intracellular localization of LAD1 in OSCC cells and provided evidence for its possible functions, presumably via influencing actin structure through its specific subcellular localization within the actin arc. Modulation of LAD1 expression could potentially be a critical determinant of 3D stromal invasion and an incomplete modulator of lateral intraepithelial expansion. The reciprocal effect observed in the direction of invasion suggests that LAD1 may be pivotal in maintaining the epithelial phenotype of OSCC cells. Thus, further investigations on signaling pathways associated with LAD1 can provide new perspectives on cancer cell biology, and LAD1 may have the potential to become a novel target for cancer therapy that focuses on cell motility.
Materials and methods
Cells
Human SCC cell lines HSC-2, HSC-3, and HSC-4, established from the metastatic lymph nodes of patients with OSCC29, were obtained from the RIKEN BRC Cell Bank (Tsukuba, Japan). The experimental use of these cells is exempt from Ethics Committee/Ethics Committee on Genetic Analysis of Niigata University; all experiments in this study were conducted in accordance with the principles of Declaration of Helsinki. HSC-2 and HSC-3 were maintained in minimum essential medium (MEM; Gibco™; Thermo Fisher Scientific Inc., MA, USA) and HSC-4 in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco™) containing 10% fetal calf serum (FCS, Gibco™) under a humidified 5% CO2/95% air atmosphere at 37 °C.
Antibody
A rabbit polyclonal antibody against human LAD1 (HPA028732) was purchased from Sigma-Aldrich (MO, USA). Mouse monoclonal antibodies against actin (mAbcam 8226, IgG1) and vimentin (V9, IgG1) were purchased from Abcam (Cambridge, UK).
Immunofluorescence
HSC-2, HSC-3, and HSC-4 cells were plated onto a 35-mm plastic dish on which a piece of cover glass with no coating was placed. The cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 10 min. Permeabilization was performed by adding 0.2% Triton X-100 to the fixative for 5 min. After overnight treatment at 4 °C with 5% milk protein in PBS, the membranes were incubated with primary and secondary antibodies for 1 h and 30 min, respectively, at room temperature (range 20–25 °C, RT). Alexa Fluor 488-conjugated goat antibodies against mouse or rabbit IgG (H + L; Life Technologies, Thermo Fisher Scientific) were used as secondary antibodies. Rhodamine-phalloidin (Invitrogen™, Thermo Fisher Scientific) was used with a secondary antibody to visualize actin filaments. The cells were then mounted using ProLong™ Diamond Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen™).
Structured illumination microscopy (SIM)
Immunofluorescence for SIM observation was performed as previously described30. Briefly, HSC-4 cells were fixed with 1% glutaraldehyde directly added to the culture medium for 5 min at room temperature (20–25 °C). After fixation, the cells were treated with 1% sodium tetrahydroborate in cytoskeletal stabilizing buffer31 for 10 min at room temperature (20–25 °C). The cells were incubated with a primary, and then secondary antibody solution containing rhodamine-phalloidin. The labeled cells were then covered with ProLong™ Diamond Antifade Mountant without DAPI. Stained samples were analyzed using a Zeiss ELYRA S.1 microscope (Carl Zeiss Microimaging, Jena, Germany). The Z-stack images were acquired in 0.11 μm stacking heights. Orthogonal and pseudo-color 3D images were processed using ZEN software (release version 8.01; Carl Zeiss).
Real-time polymerase chain reaction (real-time PCR)
Total RNA was isolated from HSC-2, HSC-3, and HSC-4 cells using the ISOGEN system (Nippon Gene Co., Ltd., Tokyo, Japan) or NucleoSpin® RNA (Macherey-Nagel, Düren, Germany), and reverse-transcribed to cDNA using the SuperScript III First-Strand Synthesis System (Life Technologies, Thermo Fisher Scientific). Quantitative RT-PCR was performed with specific primers and SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad Laboratories, Inc., CA, USA) using a Light cycler® 96 (F. Hoffmann-La Roche, Ltd., Basel, Switzerland) and a QuantStudio1 (Thermo Fisher Scientific). Target gene amplification was monitored in real-time, and gene expression levels were quantified using the lightcycler® 96 software (Roche) and Design & analysis software v2.8.0 (Thermo Fisher Scientific). The expression of peptidylprolyl isomerase A (PPIA) was used for variance normalization, and the expression levels of specific genes were represented as ratios to those of PPIA from the same master reaction. The PCR primers used for each gene were as follows: LAD1, Prime PCR™ (qHsaCED0004063, Bio-Rad); PPIA, 5′-GCAGTAATGGGTTACTTCTGAAAC-3′; and 5′-TGCCTCAGGTAATACATTACAGAC-3′.
Western blotting
HSC-2, HSC-3, and HSC-4 cells, which were cultured in 60 mm dishes, were lysed using 100 µl of lysis buffer, 50 mM HEPES (pH 7.4), 150 mM sodium chloride, and 1% Triton X-100, containing a protease inhibitor cocktail (Nakalai Tesque, Kyoto, Japan). The protein concentrations of the lysate samples were determined using a standard Bradford assay. A 10 µg aliquot of proteins from lysed cells was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, and the proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad). After incubation with 2% ECL™ blocking agent (GE Healthcare, Little Chalfont, UK) (Fig. 1) or 5% skim milk (Meg milk Snow Brand Co Ltd, Tokyo, Japan) (Supplementary) in 50 mM Tris-buffered saline (pH 7.4) containing 0.1% Tween-20 (TTBS) for 1 h at RT, the membranes were incubated overnight at 4 °C with primary antibodies diluted in TTBS. After washing with TTBS, the membranes were incubated with Envision Plus (diluted 1:500 in TTBS) for 1 h at RT. Target protein bands were visualized using ECL Prime western blotting detection reagent (GE Healthcare) (Fig. 1) or Western BLoT Quant HRP Substrate (Takara Bio Inc., Tokyo, Japan) (Supplementary).
Small interference RNA
Three different sequences of Stealth Select RNAi™ siRNAs against LAD1 (Invitrogen), HSS105926 (si#1), HSS105927 (si#2), and HSS105925 (si#3); Stealth RNAi™ siRNA Negative Control High GC Duplex (cont) (Invitrogen), and sterile water (vehicle) were transfected into HSC-2, HSC-3, and HSC-4 cells using Lipofectamine™ RNAiMAX reagents (Life Technologies) via reverse transfection, as per manufacturer’s instructions. After 48 h of transfection, cells were used for subsequent analyses. The effects of RNAi were evaluated using real-time PCR and western blotting.
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2 H-tetrazolium, inner salt (MTS) assay
HSC-2, HSC-3, and HSC-4 cells transfected with siRNA were seeded in a concentration of 0.02 × 105 with 150 µl of FCS-containing medium in each well of a 96-well microplate. At 24 h, 48 h, 72 h, and 120 h after re-plating, 30 µl of CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega Corporation, WI, USA) was added into each well and incubated for 1.5 h under a humidified 5% CO2/95% air atmosphere at 37 °C. Absorbance at 490 nm was measured with a reference absorbance of 600 nm using the GloMax® Discover System (Promega).
Scratch wound-healing assay
At 48 h after siRNA-transfection, artificial wounds were inflicted on the confluent monolayers of HSC-2 cells in 24-well culture plates using a 200 µl pipette tip. At 48 h incubation with serum-free media after scratch, cell migration into the wound area was monitored using a phase-contrast microscope equipped with a digital camera. The wound surface area was measured from the captured images of the three fields using Photoshop CS5.5 Extended (Adobe Systems Inc., CA, USA). The percent (%) closure of the wound surface area was calculated as the empty surface area/total surface area (surface area at 0 h) × 100. Cells were incubated through the experiment with no inhibition reagent of cell proliferation. Experiments were performed three times independently, and we analyzed three different foci in each replicate.
Transwell migration assay
Cell migration was determined using a BD Falcon cell culture insert 12-well companion tissue culture plate system with an 8 μm pore size polyethylene terephthalate membrane (BD Biosciences, NJ, USA). HSC-2, HSC-3, and HSC-4 cells at a concentration of 2.0 × 105 were seeded in 1 ml of FCS-free MEM or RPMI 1640 in the insert chamber, and the lower chamber was filled with 2 ml of MEM or RPMI 1640 containing 10% FCS. After 24 h of incubation, the cells on the upper surface of the insert chamber were carefully removed with cotton swabs and the bottom surface of the membrane was fixed and stained using a Hemacolor staining kit (Merck KGaA, Darmstadt, Germany). Images of cells traversing the membrane pores were captured and counted using ImageJ ver. 1.52a (National Institutes of Health). Three different foci were analyzed in each stained membrane. Mean values were calculated from triplicate experiments.
Collagen gel invasion assay
Cell invasion into the collagen matrix was determined using a cell culture insert 6-well companion tissue culture plate system with a 3 μm pore size polyethylene terephthalate membrane (Cellster®; Greiner Bio-One GmbH, Kremsmünster, Austria). The membrane was covered with 3 ml of type I collagen solution, including 2.1 ml of Cell matrix type IA (final concentration of collagen, 2.1 mg/ml), 0.6 ml 5 × condensed Dulbecco’s modified Eagle’s medium (DMEM), and 0.3 ml of reconstruction buffer (Nitta Gelatin, Tokyo, Japan). The collagen solution was incubated for 30 min under a humidified 5% CO2/95% air atmosphere at 37 °C for polymerization. Cells at a concentration of 5.0 × 105 were seeded in 2 ml of 10% FCS-containing MEM or RPMI 1640 onto the polymerized collagen layer. After one week of incubation, the cells and collagen layer were removed from the insert and fixed with 4% PFA for 1 h. Paraffin-embedded tissue sections (2.5 μm) were stained with hematoxylin and eosin. The cell infiltration depth from the uppermost surface of the collagen layer was measured in an image captured using ImageJ ver. 1.52a. Cell infiltration depths were measured in five different foci from each sample, and the assays were independently repeated three times.
To evaluate the morphology of migrating cells in the collagen layer, HSC-4 cells (5.0 × 105) were seeded onto a type IA collagen gel layer. After a seven days of period, the cells and collagen gel were fixed with 4% PFA-containing PBS. Paraffin-embedded tissue sections (20 μm) were prepared, and immunofluorescence for LAD1, vimentin, and β-actin was performed. Z-stacking immunofluorescence images were acquired using an FV-1200 confocal laser scanning microscope (Olympus Co., Ltd., Tokyo, Japan). The acquired images were analyzed using ImageJ ver. 1.52a.
Cell tracking analysis
HSC-2, HSC-3, and HSC-4 cells were incubated with siRNA treatments and incubated for 48 h. Cells were suspended and seeded in 0.34 × 105 to a 35 mm dish. After 24 h from seeding, aspiration of the old medium and serum-free medium was added, and timelapse images were taken by a CytoWatcher instrument (WSL-1800-B, ATTO Corporation, Tokyo, Japan) at 5-minute intervals for 24 h. Then, the image sequences were analyzed using ImageJ/Fiji ver. 1.54f with TrackMate (version 7.1.1.)32, and the cell continuously tracked for more than 60 frames (5 h) were statistically analyzed by using R (4.2.1; R Core Team, Vienna, Austria). Tracking plots were depicted by the Chemotaxis and Migration tool 2.0 (ibidi GmbH, Gräfelfing, Germany). The cell tracks were depicted on the relative X-Y coordinates plots (Fig. 3e).
Cell morphological analysis of immunofluorescent images
Immunofluorescent images using phalloidin (Invitrogen™) for F-actin and Hoechst 33,342 (1:200; Dojindo Laboratories Co., Ltd., Kumamoto, Japan) for nucleus on HSC-2, HSC-3, and HSC-4 were taken by BZ-9000 system (Keyence, Osaka, Japan). Obtained images were analyzed by cellpose 2.133 for cell segmentation with the “cyto” model. Subsequent fractal analysis was done by the Fraclac plugin (2016Apr120248a502) with the box-counting method in default parameters in ImageJ/Fiji ver.1.54 f.
Transcriptome analysis
Total RNA samples from HSC-4 with siRNA control treatment or si#1 were isolated with RNeasy® Plus Mini kit (Qiagen NV, Hilden, Germany). The quality of the extracted RNA was assessed using the Bioanalyzer 2100 system (Agilent Technologies, Inc., CA, USA), and all samples were of sufficient quality. NEBNext® Poly(A) mRNA Magnetic Isolation Module and NEBNext® Ultra™ ll Directional RNA Library Prep Kit (New England Biolabs Inc., MA, USA) were used for poly-A selection and library preparation for RNA sequencing (RNA-seq), respectively. RNA-seq was performed on a NovaSeq 6000 (Illumina, Inc., San Diego, CA, USA) with 150 bp paired-end reads, as per the manufacturer’s instructions. Base-calling data were processed using Trimmomatic (version 0.39)34 for adaptor trimming, STAR (version 2.7.10b)35 for alignment, and RSEM (version 1.3.3)36 for read counting with default parameters. Read count data were analyzed using iDEP 1.137 run on R studio (2023.03.0 + 386) (RStudio, PBC, MA, USA) and R (4.2.1) in a local environment.
PCR array
A total of 1 µg isolated RNA was used for cDNA synthesis with RT2 First Strand Kit (Qiagen). The analyses with RT² Profiler™ PCR Array Human Cell Motility (PAHS-128ZF, Qiagen) and RT2 SYBR Green Master Mix (Qiagen) were performed as per the manufacturer’s instructions. Real-time PCR was performed using a Light cycler® 96 (Roche). The array reaction was repeated thrice with independently isolated RNA-based cDNA. Measurements were analyzed using the GeneGrobe data analysis center (Qiagen; https://geneglobe.qiagen.com). Data sets were normalized based on the arithmetic means of the Ct values of housekeeping genes, such as β-actin (ACTB), β2-microglobulin (B2M), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hypoxanthine phosphoribosyltransferase 1 (HPRT1), and ribosomal protein lateral stalk subunit P0 (RPLP0), included in the array plates, as per manufacturer’s instructions.
Public database analysis
We explored the effects of LAD1 mRNA expression levels in clinical samples of head and neck SCC from The Cancer Genome Atlas (TCGA) dataset (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) on cBioPortal38,39 (https://www.cbioportal.org/, accessed on 2023/01/24). We selected a dataset of head and neck SCCs (Pancancer Atlas; sample size 523). In genomic profiles, mRNA expression z-scores relative to diploid samples (RNA Seq V2 RSEM) were selected, and the z-score threshold was set to ± 2.0. Of the dataset, 515 were included in the analysis.
Statistical analysis
Experimental data were analyzed using a one-way analysis of variance and Dunnett’s post-hoc comparisons, and Welch’s t-test. Statistical analyses were performed using R (4.2.1 and 4.4.0). A p-value < 0.05 was considered statistically significant. In the public database analysis, the chi-square test was performed using cBioPortal (https://www.cbioportal.org/), and secondary residual analysis was performed using GraphPad Prism 6 (GraphPad Software, Inc., CA, USA).
Data availability
The experimental and analyzed data of the current study are available from the corresponding author upon reasonable request. The RNA-seq data of the cell lines are available in the DNA Data Bank of Japan (DDBJ; https://www.ddbj.nig.ac.jp)40 under the BioProject accession number PRJDB16201.
References
Abé, T. et al. Proteomic and histopathological characterization of the interface between oral squamous cell carcinoma invasion fronts and non-cancerous epithelia. Exp. Mol. Pathol. 102, 327–336 (2017).
Ishiko, A. et al. 97-kDa linear IgA bullous dermatosis (LAD) antigen localizes to the Lamina lucida of the epidermal basement membrane. J. Investig. Dermatol. 106, 739–743 (1996).
Marinkovich, M. P., Taylor, T. B., Keene, D. R., Burgeson, R. E. & Zone, J. J. LAD-1, the linear IgA bullous dermatosis autoantigen, is a novel 120-kDa anchoring filament protein synthesized by epidermal cells. J. Investig. Dermatol. 106, 734–738 (1996).
Roth, L. et al. SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Sci. Signal. 11, eaan0949 (2018).
Moon, B. et al. LAD1 expression is associated with the metastatic potential of colorectal cancer cells. BMC Cancer 20, 1180 (2020).
Klobučar, M. et al. Basement membrane protein ladinin-1 and the MIF-CD44-β1 integrin signaling axis are implicated in laryngeal cancer metastasis. Biochim. Biophys. Acta 1862, 1938–1954 (2016).
Jiang, Y. et al. LAD1 promotes malignant progression by diminishing ubiquitin-dependent degradation of vimentin in gastric cancer. J. Transl. Med. 21, 632 (2023).
Chang, C. Y. et al. Ladinin 1 shortens survival via promoting proliferation and enhancing invasiveness in lung adenocarcinoma. Int. J. Mol. Sci. 24, 431 (2022).
Li, J., Wang, Z. & Tie, C. High expression of ladinin-1 (LAD1) predicts adverse outcomes: a new candidate docetaxel resistance gene for prostatic cancer (PCa). Bioengineered 12, 5749–5759 (2021).
Wang, Y. circ-ANXA7 facilitates lung adenocarcinoma progression via miR-331/LAD1 axis. Cancer Cell Int. 21, 85 (2021).
Yi, J., Wu, X. S., Crites, T. & Hammer, J. A. Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in Jurkat T cells. Mol. Biol. Cell 23, 834–852 (2012).
Jiang, C., Cui, C., Li, L. & Shao, Y. The anomalous diffusion of a tumor invading with different surrounding tissues. PLoS One 9, e109784 (2014).
Thompson, E. W., Newgreen, D. F. & Tarin, D. Carcinoma invasion and metastasis: a role for epithelial-mesenchymal transition? Cancer Res. 65, 5991–5995 (2005).
Heath, J. P. Behaviour and structure of the leading lamella in moving fibroblasts. I. Occurrence and centripetal movement of arc-shaped microfilament bundles beneath the dorsal cell surface. J. Cell Sci. 60, 331–354 (1983).
Burnette, D. T. et al. A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol. 13, 371–381 (2011).
Jiu, Y. et al. Bidirectional interplay between vimentin intermediate filaments and contractile actin stress fibers. Cell Rep. 11, 1511–1518 (2015).
Kurokawa, K. & Matsuda, M. Localized RhoA activation as a requirement for the induction of membrane ruffling. Mol. Biol. Cell 16, 4294–4303 (2005).
Edwards, D. C., Sanders, L. C., Bokoch, G. M. & Gill, G. N. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1, 253–259 (1999).
Delorme-Walker, V. D. et al. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J. Cell Biol. 193, 1289–1303 (2011).
VanderVorst, K., Hatakeyama, J., Berg, A., Lee, H. & Carraway, K. L. Cellular and molecular mechanisms underlying planar cell polarity pathway contributions to cancer malignancy. Semin. Cell Dev. Biol. 81, 78–87 (2018).
Kikuchi, A., Yamamoto, H., Sato, A. & Matsumoto, S. Wnt5a: its signalling, functions and implication in diseases. Acta Physiol. (Oxf) 204, 17–33 (2012).
Lee, J. K. et al. KITENIN increases invasion and migration of mouse squamous cancer cells and promotes pulmonary metastasis in a mouse squamous tumor model. FEBS Lett. 583, 711–717 (2009).
Okamoto, T., Schlegel, A., Scherer, P. E. & Lisanti, M. P. Caveolins, a family of scaffolding proteins for organizing preassembled signaling complexes at the plasma membrane. J. Biol. Chem. 273, 5419–5422 (1998).
Beardsley, A. et al. Loss of caveolin-1 polarity impedes endothelial cell polarization and directional movement. J. Biol. Chem. 280, 3541–3547 (2005).
Chan, N. N. et al. Cholesterol is a regulator of CAV1 localization and cell migration in oral squamous cell carcinoma. Int. J. Mol. Sci. 24, 6035 (2023).
Parat, M. O., Anand-Apte, B. & Fox, P. L. Differential Caveolin-1 polarization in endothelial cells during migration in two and three dimensions. Mol. Biol. Cell 14, 3156–3168 (2003).
Lu, Z., Ghosh, S., Wang, Z. & Hunter, T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of β-catenin, and enhanced tumor cell invasion. Cancer Cell 4, 499–515 (2003).
Pastushenko, I. & Blanpain, C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29, 212–226 (2019).
Momose, F. et al. Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J. Oral Pathol. Med. 18, 391–395 (1989).
Nozumi, M. et al. Identification of functional marker proteins in the mammalian growth cone. Proc. Natl Acad. Sci. USA 106, 17211–17216 (2009).
Conrad, P. A., Nederlof, M. A., Herman, I. M. & Taylor, D. L. Correlated distribution of actin, myosin, and microtubules at the leading edge of migrating Swiss 3T3 fibroblasts. Cell. Motil. Cytoskelet. 14, 527–543 (1989).
Ershov, D. et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–832 (2022).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Ge, S. X., Son, E. W. & Yao, R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 19, 534 (2018).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Kodama, Y., Shumway, M., Leinonen, R & International Nucleotide Sequence Database Collaboration. The sequence read Archive: explosive growth of sequencing data. Nucleic Acids Res. 40, D54–D56 (2012).
Acknowledgements
This work was supported by the Center for the Coordination of Research Facilities, Institute for Research Promotion (CCRF), Niigata University, Japan.
The authors wish to acknowledge Dr. Takashi Saku, the emeritus professor, Niigata University, Japan, for his helpful advice on this study manuscript.
Funding
This work was supported by Grants-in-Aid for Scientific Research (C) (18K09550 and 21K09841 to TA) from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Contributions
T.A.: Project administration, Conceptualization, Methodology, Investigation, Formal analysis, Writing–Original Draft, Review & Editing; M.Y., M.N., S.M., K.T., and R.O.: Investigation, Writing–Review & Editing; Y.A. and J-I.T.: Supervision, Writing–Review & Editing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Abé, T., Yamazaki, M., Nozumi, M. et al. Ladinin-1 in actin arcs of oral squamous cell carcinoma is involved in cell migration and epithelial phenotype. Sci Rep 14, 22778 (2024). https://doi.org/10.1038/s41598-024-74041-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74041-z
Keywords
This article is cited by
-
Visualization of nanostructures in neuronal growth cones and axons using super-resolution structured illumination microscopy
Anatomical Science International (2025)