Abstract
Hemolytic peptides are therapeutic peptides that damage red blood cells. However, therapeutic peptides used in medical treatment must exhibit low toxicity to red blood cells to achieve the desired therapeutic effect. Therefore, accurate prediction of the hemolytic activity of therapeutic peptides is essential for the development of peptide therapies. In this study, a multi-feature cross-fusion model, HemoFuse, for hemolytic peptide identification is proposed. The feature vectors of peptide sequences are transformed by word embedding technique and four hand-crafted feature extraction methods. We apply multi-head cross-attention mechanism to hemolytic peptide identification for the first time. It captures the interaction between word embedding features and hand-crafted features by calculating the attention of all positions in them, so that multiple features can be deeply fused. Moreover, we visualize the features obtained by this module to enhance its interpretability. On the comprehensive integrated dataset, HemoFuse achieves ideal results, with ACC, SP, SN, MCC, F1, AUC, and AP of 0.7575, 0.8814, 0.5793, 0.4909, 0.6620, 0.8387, and 0.7118, respectively. Compared with HemoDL proposed by Yang et al., it is 3.32%, 3.89%, 5.93%, 10.6%, 8.17%, 5.88%, and 2.72% higher. Other ablation experiments also prove that our model is reasonable and efficient. The codes and datasets are accessible at https://github.com/z11code/Hemo.
Similar content being viewed by others
Introduction
Therapeutic peptides are widely favored by the medical and pharmaceutical fields due to their advantages of high permeability and small side effects. However, some of their characteristics have two sides, and their therapeutic effect will be affected if these characteristics cannot be controlled within a certain range1,2. For example, the hemolytic activity of therapeutic peptides refers to their ability to bind to red blood cells, allowing water and other solute molecules to enter red blood cells, thereby increasing the osmotic pressure gradient inside red blood cells and causing them to swell or even rupture3. It can be seen that therapeutic peptides with high hemolytic activity have functions such as destroying cancerous cells and targeted delivery of drugs. But for other therapeutic tasks, the hemolytic activity of therapeutic peptides must be reduced so that them can stably express a specific function without damaging normal cells4. Therefore, accurate prediction of the hemolytic activity of therapeutic peptides is helpful to promote the development of peptide drugs. Hemolytik (http://crdd.osdd.net/raghava/hemolytik/) is a complete database of 3000 experimentally verified hemolytic peptides and non-hemolytic peptides5. The authors evaluated the hemolytic activity of peptide sequences on 17 different red blood cells and provided information related to each peptide sequence and its hemolytic activity. DBAASP is a constantly updated antimicrobial peptide database containing information on the bioactivity and toxicity of peptide sequences, which can be used for the study of hemolytic activity6. The establishment of these databases helps us to develop deep learning-based sequencing technologies with low cost and short time consumption compared to traditional biological experiments.
At present, the feature extraction methods used in identification models based on biological sequences can be broadly divided into two types: traditional hand-crafted feature extraction methods and feature extraction methods based on natural language processing technology. Hand-crafted feature extraction methods are designed by humans, offering low computational complexity and strong interpretability, such as binary encoding7, kme8, quasi-sequence-order (QSO)9. However, their effectiveness depends on the characteristics of the data itself, requiring the selection of appropriate descriptors based on the dataset’s properties. Feature extraction methods based on natural language can uncover structures and patterns that are difficult for humans to detect and are less influenced by the dataset’s inherent characteristics. Nevertheless, their drawback is that they struggle to learn effective features when the data quality is poor. Word embedding10 is one of the most basic feature extraction methods in natural language processing, and many models are built on it, such as Bert11,12. In addition, there are pre-trained language models such as ProtTrans13, evolutionary scale modeling (ESM)14. Of course, there are also many models that consider both types of methods in order to extract more comprehensive information15. Machine learning and deep learning algorithms are used for further feature mining and classification. Common machine learning methods include AdaBoost16, random forest (RF)17, hidden markov model (HMM)18, etc. While they are simple and easy to understand, they are not well-suited for handling large, high-dimensional data, resulting in lower model accuracy. In contrast, deep learning, with its ability to automatically learn features, has demonstrated exceptional performance, far surpassing traditional machine learning algorithms, such as convolutional neural network (CNN)19, capsule network20, recurrent neural network (RNN)21, transformer22,23. Machine learning and deep learning each have their own unique advantages, and sometimes work well when combined24.
Most of the existing identification models for hemolytic peptides use traditional hand-crafted features and machine learning algorithms, such as HemoPI25, HemoPred26, HLPpred-Fuse27, HAPPENN28, HemoPImod29. The involved feature extraction methods collect the information of hemolytic peptides from various aspects such as amino acid composition, peptide composition, physicochemical properties, and atomic descriptors. Classifiers cover almost all common machine learning algorithms. However, the methods and datasets used by these models are outdated. Moreover, HemoPImod is unable to identify hemolytic peptide sequences longer than 25 amino acids. Language model-based methods did not start to appear until 2021. HemoNet30 is the first to employ the SeqVec language model to capture the contextual features of amino acids, but it struggles to generalize well to unseen data. With the development of stronger transformer models, AMPDeep first used transformer-based pretrained model (PROT-ERT-BFD) to represent the features of peptide sequences in 202231. However, its fine-tuning process is extremely cumbersome, requiring the identification of a secondary distribution using specific keywords to fine-tune the model. In 2023, Sharma et al. tried to integrate multiple deep learning algorithms (Bi-LSTM, bi-directional temporal convolutional networks (Bi-TCN), CNN) to identify hemolytic peptides, and named it EnDL-HemoLyt32. EnDL-HemoLyt integrates hand-crafted features with deep learning features and also offers predictions for peptides with N/C-terminal modifications, though it may not be well-suited to the latest data. Yang et al. built the latest model, HemoDL, using both 7 hand-crafted features and 2 transformer-based language models (Prot-T5-XL-UniRef50 and ESM2)33. Multi-feature combination can collect more sequence information, but HemoDL only simply concatenates the two types of features and inputs them into LightGBM for classification, and there is no sufficient fusion and complementarity between the features.
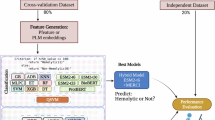
In this study, we continue the idea of multi-feature combination but make some improvements. We innovatively propose HemoFuse, an identification model of hemolytic peptides with multi-feature cross-fusion, as shown in Fig. 1. We use word embedding features and four hand-crafted features (BLOSUM62, dipeptide deviation from expected mean (DDE), dipeptide composition (DPC) and composition of k-spaced amino acid pairs (CKSAAP)) to represent peptide sequences. To be able to capture higher-order features, we add a transformer encoder layer after the embedding layer, similar to the structure of Bert. However, compared with Bert, this model has fewer parameters and is easy to transfer and use. In terms of hand-crafted feature methods, except DPC, the other three methods are applied to hemolytic peptides for the first time. Before feature fusion, we use bi-directional gated recurrent unit (Bi-GRU) to align the dimensions of hand-crafted features and embedding features. Then, multi-head cross-attention is used to deeply fuse the two features, which can strengthen the connection between them and complement each other’s advantages. Finally, CNN and multi-layer perceptron (MLP) are used as classifier to determine the hemolytic activity of peptide sequences.
Materials and methods
Datasets
In order to facilitate the comparison with the existing models, we directly use the datasets provided in the benchmark paper, and all positive samples are therapeutic peptides with hemolytic activity33. As shown in Fig. 1, there are four common datasets and an integrated dataset, and Table 1 shows the specific number of each dataset. Dataset 1 and dataset 2 are from Hemolytik and DBAASP v.2 databases, both collected and collated by Chaudhary et al.25, with 1014 and 1623 sequences, respectively. Dataset 3 was collected by Patrick et al.28 from Hemolytik and DBAASP databases with 3738 samples. Dataset 4 is derived from the DBAASP v3 database and has 4339 samples as the dataset used in EnDL-HemoLyt32. The integrated dataset is composed of these four datasets, and CD-HIT34 with a threshold of 0.7 is utilized for de-redundancy processing, resulting in 1993 sequences. These datasets are imbalanced, which better mimics the real-world situation where the positive and negative samples are not equal, so we do not apply an imbalanced treatment to enforce equality. In addition, these datasets are randomly divided into training datasets and independent test datasets according to 8:2.
Figures 2 and 3 show the sequence length distribution and amino acid composition of positive and negative samples from the training sets of datasets 1–4, respectively. As shown in Fig. 2, the lengths of positive and negative samples in dataset 1 are concentrated between 5 and 31 amino acids. In dataset 2, the lengths of positive and negative samples are roughly concentrated between 6 and 38 amino acids, with a few sequences having much longer lengths. The sequence length distribution is more uniform in datasets 3 and 4. Dataset 3 has sequences ranging from 7 to 35 amino acids, while dataset 4 ranges from 6 to 50 amino acids. We use the kpLogo tool35 to analyze the amino acid composition preferences between positive and negative samples, as shown in Fig. 3. In dataset 1, amino acids K and L are more abundant in positive samples, with no significant enrichment in negative samples. In dataset 2, amino acid K is more enriched in positive samples, whereas the negative samples are more varied. In dataset 3, amino acids K and A are highly expressed in positive samples, and amino acids K, R, and L are highly expressed in negative samples. In dataset 4, both positive and negative samples have higher frequencies of amino acids K, R, and L. These results indicate that using only peptide sequence composition for feature extraction is insufficient. Therefore, we will incorporate word embedding techniques into the feature extraction module to uncover the underlying patterns in the sequences.
Architecture of HemoFuse
As shown in Fig. 1, our model can be decomposed into three sub-modules: feature extraction and alignment module, feature cross-fusion module, and classification module. We use a combination of advanced word embedding features and traditional hand-crafted features to represent peptide sequences. Token embedding, position embedding, and transformer encoder together form a lightweight language model. BLOSUM62, DDE, DPC, and CKSAAP cover the evolutionary and compositional information of peptide sequences. Hand-crafted feature methods are specifically formulated based on the large number of available peptide sequences and still have great potential in representing peptide sequences. Bi-GRU can mine deeper context features and transform hand-crafted features to the appropriate dimensions to match embedding features. Then, we choose multi-head cross-attention to complete the deep fusion between different features, which is good at capturing the semantic relationship between two related but different sequences. This is the first application in the identification of hemolytic peptides. The classification module mainly consists of CNN and MLP.
Feature extraction and alignment
Word embedding feature
Word embedding technology follows the distributed semantics hypothesis, which uses the context around each word to express its semantic information36. In general, words with similar contexts will have similar semantic meanings. Compared with hand-crafted features, its biggest advantage is that its parameters can be continuously optimized throughout the training process, making it more suitable for the current data. Popular word embedding models include Word2Vec, GloVe, Bert, etc.
In this paper, we adopt token embedding and position embedding to initially represent the shallow information of protein sequences, and then use transformer encoder to capture the bidirectional relationship between amino acids in the sequence more thoroughly37. The architecture is more like a simplified Bert. Token embedding is a vector representation of the amino acid itself, and position embedding encodes the position information of the amino acid into a feature vector, just like RNN or LSTM can provide the position information of the sequence. The dimension of the two embeddings is 128. They are then combined and fed into transformer encoder layer. As shown in Fig. 1, transformer encoder is composed of multi-head attention, layer normalization, feed forword layer, and layer normalization in turn, with two residual connections. This structure does not have too many hidden layers, which can improve the computational efficiency. Self-attention mechanism can re-encode the target feature by the correlation between the target feature and other features, so that new features contain more interaction information without considering their distance in the sequence. The number of heads of self-attention in this module is 4.
Hand-crafted feature
BLOSUM62 is an amino acid substitution scoring matrix for protein sequence comparison, which stems from the conservation between the same amino acids38. It reflects the evolutionary information of the protein sequence. The score is essentially the logarithm of the ratio of the likelihood of the different amino acids being homologous and non-homologous, and the formula is as follows:
where \({p_{ab}}\) is the frequency of occurrence in the existing homologous sequences assuming that a and b are homologous. \({f_a}\) and \({f_b}\) are the frequencies of residues a and b occurring in either sequence, assuming that a and b are not homologous, respectively. \(\lambda\) is the scaling parameter. If residues a and b are homologous, then \({p_{ab}}>{f_a}{f_b}\) and the score is positive. If residues a and b are not homologous, then \({p_{ab}}<{f_a}{f_b}\) and the score is negative. In summary, the similarity between every two amino acids can be calculated. BLOSUM62 matrix can represent each amino acid as a 20-dimensional feature vector and the protein sequence as a L×20 feature matrix, where L is the sequence length.
DDE derives from the difference in dipeptide composition between epitopes and non-epitopes, and uses this to indicate the extent to which dipeptide frequencies deviate from the expected mean39. It is able to analyze the composition and distribution of amino acids in peptide sequences. The feature vector is constructed based on three parameters: dipeptide composition measure (\({D_c}\)), theoretical mean (\({T_m}\)), and theoretical variance (\({T_v}\)). \({T_m}\) does not depend on a specific peptide sequence, so \({T_{m\left( i \right)}}\) of 400 dipeptides is calculated first:
\({C_{i1}}\) is the codon number of the first amino acid in the dipeptide i and \({C_{i2}}\) is the codon number of the second amino acid. \({C_{L - 1}}\) is the total number of possible codons excluding stop codons. Given a peptide sequence of length L, \({D_{c\left( i \right)}}\) and \({T_{v\left( i \right)}}\) of the dipeptide i are calculated according to the following formula:
\({n_i}\) is the frequency of occurrence of the dipeptide i and \(L - 1\) is the number of dipeptides present in this sequence. Thus, DDE of the dipeptide i can be expressed as follows:
Finally, the peptide sequence can be represented as a 400-dimensional vector:
DPC is a common feature representation method based on amino acid composition information, which can provide detailed information about the arrangement of amino acids in peptide sequences40. It represents a protein sequence by counting the frequency of occurrence of all dipeptides in the protein sequence. The formula is as follows:
\({n_i}\) and \({{\text{f}}_i}\) are the number and frequency of occurrences of the dipeptide i in the sequence, respectively. There are a total of 400 dipeptides. Therefore, each protein sequence can be transformed into a 400-dimensional fixed-length feature vector.
CKSAAP converts a protein sequence into a feature vector by using the constituent ratio of k-spaced residue pairs in this protein sequence fragment41. It is of great significance to understand the function and structure of proteins. Given a protein sequence of length L and a value of k, two residues separated by a distance of k are extracted and considered as a residue pair, so a total of \(L - k - 1\) residue pairs can be extracted. The probability of occurrence of these residue pairs in the protein sequence is counted, resulting in a 400-dimensional feature vector:
\({L_{AA}}\), \({L_{AC}}\), and \({L_{YY}}\) is the number of occurrences of the corresponding residue pair. k can be set to 0, 1, 2, 3, 4, 5.
Bi-GRU can extract sequence information in proteins and deredundant hand-crafted features42. Both LSTM and GRU are common methods for processing long sequences, and the advantage of GRU is that it uses only two gates, which reduces the number of parameters by nearly a third and effectively avoids overfitting. Bi-GRU does not change its original internal structure, but only applies the model twice in different directions. This ensures that the forward and backward sequence features are captured, resulting in richer features. Notably, it can also perform the task of feature alignment. We set the number of neurons in the hidden layer to 64, resulting in a 128-dimensional feature vector, which is the same as the feature vector output by word embedding module.
Feature cross fusion
Word embedding features and hand-crafted features have collected a lot of information about the peptide sequence with different rules, so the next step is to consider how to make full use of these features to judge its hemolytic activity43,44,45. These two features contain their own information of interest, and their mutual relationship will be ignored if they are directly combined. In view of this, we adopt multi-head cross-attention mechanism to deeply fuse word embedding features and hand-crafted features, which is often used to deal with multi-modal features46,47. The fused new feature contains the interactive information of the two features. Compared with self-attention mechanism, the input of cross-attention mechanism has two parts: the “where” feature and the “what” feature. The “where” feature acts as the Query (Q), and the “what” feature is used to generate the Key and Value (K, V)48. In this study, we choose word embedding features \({X_{emb}}\) as the “where” feature, and hand-crafted features \({X_{hand}}\) as the “what” feature. The specific operations are as follows:
\({W_q}\), \({W_k}\), and \({W_v}\) are learnable parameter matrices. Then Q and K are used to calculate the correlation between each element of the two inputs to obtain attention weight. Next, update the feature vectors:
where D and h are the embedding dimensions and the number of heads, respectively. Finally, the fused features can be obtained by combining the outputs of multiple attention heads:
where N is the number of attention heads and \({W_c}\) is weight matrice. Obviously, in this process, cross-attention mechanism constantly updates the fusion features based on hand-crafted features with the information of word embedding features.
Classification
The final classifier consists of a CNN layer and four linear layers, and each layer corresponds to a set of a batch normalization layer and a dropout layer to prevent the model from overfitting. This structure can gradually reduce the dimension of the feature vector to avoid information loss.
Model evaluation
In this study, we train the model on each of the five datasets and evaluate the performance of the model on the corresponding independent test datasets using the following seven evaluation metrics: accuracy (ACC), specificity (SP), sensitivity (SN), Matthews correlation coefficient (MCC), F1 score, area under ROC curve (AUC) and average precision score (AP)33. The formulas are as follows:
Where TP, TN, FP, and FN represent correctly predicted positive samples, negative samples, and incorrectly predicted positive samples, negative samples, respectively. ACC represents the proportion of samples that are correctly predicted and reflects the global accuracy of the model. SP and SN are used to measure the ability of the model to correctly predict negative and positive samples, respectively. MCC combines true positives, true negatives, false positives, and false negatives, and is a more balanced index. F1 score is the average of precision and recall. When F1 is high, it also means that both precision and recall are high. The ROC curve can reflect the relationship between the true positive rate and the false positive rate of the model, the closer the curve is to the top left corner, the higher the accuracy of the model. The two ROC curves can be quantitatively compared by calculating AUC values. The precision-recall curve focuses on positive samples and is more suitable for imbalanced datasets. AP is the area under PR curve.
Results and discussion
Experimental settings
HemoFuse is built in Python under the PyTorch framework. In the model training process, the cross-entropy loss function and Adam optimizer are used to continuously optimize the model parameters and reduce the loss. The batch size is 15, the learning rate is 0.001, and the iteration epoch number is 100. After the shallow features are extracted, we use a transformer layer with built-in 8 attention heads and Bi-GRU with 64 neurons to extract the deep features. In the feature cross fusion module, the number of heads for cross attention is 8. In the classification, the number of convolution kernels in the CNN layer is 64, the kernel size is 3, and the dropout ratio is 0.5. The number of neurons in the linear layers decreases layer by layer to 512, 128, 64, 2 with a dropout ratio of 0.6. Finally, it is normalized by the softmax function.
Ablation experiments with different hand-crafted feature methods
Feature representation is the cornerstone of classification models, and given previous studies on hemolytic peptides, we believe that hand-crafted feature extraction methods are able to extract beneficial features of peptide sequences. This paper aims to find some hand-crafted feature methods that have not been used in hemolytic peptides, so as to provide more possibilities for the identification model. BLOSUM62, DDE, and CKSAAP are all applied to hemolytic peptides for the first time. In addition, the experimental results show that DPC helps to improve the performance of the proposed model, so we also use it. Figure 4 is for the results of the four methods of ablation experiments. It is clear that both BLOSUM62 and DDE achieve incredible results. BLOSUM62 is the best method on the dataset 1 and 4, and its ACC values are 0.8177 and 0.8517, respectively. And on the dataset 4, its SN value is slightly higher than the SN value of combined features, which indicates that it is good at identifying positive samples. On the dataset 2 and 3, DDE has the best overall performance, with ACC values of 0.7846 and 0.8743, respectively, which are less than 1% different from the ACC values of combined features. The overall performance of CKSAAP and DPC is inferior, but they can produce better results when combined with the first two feature methods. On the dataset 1, the addition of CKSAAP and DPC improves the ACC, SN, MCC, and F1 values of the model by 2.47%, 4.51%, 5.02%, and 2.53%, respectively, compared with the best single method. Figure 5 shows the corresponding ROC and PR curves. CKSAAP and DPC have greater improvement on the datasets 1 and 4. Compared with the best single method, the AUC values are increased by 2.31% and 3.49%, and the AP values are increased by 1.72% and 4.71%, respectively.
The effectiveness of cross-attention mechanism
In this paper, multi-head cross-attention mechanism is used to establish the connection between word embedding features and hand-crafted features, focusing on the key information of sequences. To illustrate the advantages of cross-attention mechanism, we compare it with common self-attention mechanism. Compared with cross-attention mechanism, self-attention mechanism has higher requirements on the input features, and all dimensions need to be equal. So we add a linear layer in front of multi-head self-attention module to align the dimensions of the two features. As shown in Fig. 6, the overall performance of cross-attention is better than that of self-attention. The ACC values on the four datasets are increased by 2.96%, 2.16%, 3.88%, and 4.14%, and the MCC values are increased by 3.68%, 4.02%, 8.31%, and 8.77%, respectively. To sum up, multi-head cross-attention can mine more discriminative features, thus improving the identification ability of the model. In addition, cross-attention mechanism abandons the feature concatenation operation, which greatly reduces the feature dimension and accelerates the training speed of the model.
Visualization of features before and after fusion
We use t-distributed stochastic neighbor embedding technique (t-SNE) to visualize the features obtained from word embedding feature module, hand-crafted feature module, and multi-head cross-attention module on a two-dimensional plane. As shown in Fig. 7, the four rows represent feature visualizations of the three modules on four independent test datasets, respectively. It can be seen that the positive and negative samples after embedding feature encoding are mixed states. However, relatively speaking, the negative samples at this time have a slight tendency to aggregate, which is obvious on the independent test dataset 3 and 4, as shown in Fig. 7 (C-D). The second column is the feature distribution obtained by hand-crafted feature module. Because the built-in Bi-GRU is used for depth feature extraction, there is a clear boundary between the positive and negative samples. After multi-head cross-attention module and CNN layer, two obvious clusters are formed, and only a small number of samples are not separated correctly. The interval between the positive and negative samples is also further compared to the previous module. Thus, each of the submodules of HemoFuse can extract beneficial information to achieve the desired effect.
Comparison with other state-of-the-art methods
The model in this paper converts peptide sequences into word embedding feature vectors and hand-crafted feature vectors respectively, and innovatively uses multi-head cross-attention mechanism to complete further fusion. This operation can effectively integrate and fuse information from multiple modalities. In order to show that the structure of our model is feasible and effective, it is compared with nine existing models on four independent test datasets, and the results are shown in Tables 2, 3, 4 and 5. From the results of the dataset 1, we can see that our model is of great potential in identifying hemolytic peptides, second only to the best model HemoDL. Although the performance on the dataset 2 is unsatisfactory, it still outperforms most other models. On the dataset 3 and 4, the overall performance of HemoFuse is improved to some extent. It is worth noting that on the dataset 3, the SP and SN values are 0.8884 and 0.8706, respectively, and the MCC value is also increased by 2.17% compared with HemoDL. This indicating that the model can correctly distinguish the positive and negative samples on imbalanced datasets.
To test the comprehensive performance of the model, we combined the dataset 1–4 and controlled the similarity between sequences within 0.7. The model is also trained and tested in the same way on the integrated dataset, and the results are shown in Table 6. HemoFuse achieves surprising results, with improvements in all metrics. ACC, SP, SN, MCC, F1, AUC, AP are increased by 3.32%, 3.89%, 5.93%, 10.6%, 8.17%, 5.88%, 2.72%, respectively. These results show that our model has strong identification and generalization ability, so it can be used to predict hemolytic activity.
Case study
To conduct the case study, we incorporate 10 peptides with varying degrees of hemolytic activity and 10 non-hemolytic peptides. These data come from Adiba et al.30, collected from the DBAASP and Hemolytik databases, which contain detailed information on experimentally verified cell toxicity/hemolytic activity of peptides. By using these unseen data, we more accurately simulate real-world prediction scenarios, thereby effectively assessing the model’s practical utility and generalizability. The experimental results, as shown in Tables 7 and 8, demonstrate a total accuracy of 90% for identifying both positive (hemolytic) and negative (non-hemolytic) samples. This result indicates that our model is capable of accurately recognizing different levels of hemolytic activity, particularly for peptides with lower hemolytic activity, which represents a more challenging task. Moreover, the model performs excellently in distinguishing between hemolytic and non-hemolytic peptides, proving its ability to discern subtle differences between the two categories. Misclassifying a hemolytic peptide as non-hemolytic in vivo could potentially lead to underestimating the degree of cellular damage, affecting disease diagnosis. Therefore, the model’s actual performance is crucial for understanding the occurrence and development of hemolysis in various diseases.
Discussion
Based on the experimental results, the proposed model, HemoFuse, demonstrates strong recognition capability by effectively integrating sequence features extracted from multiple aspects and performing classification tasks. As shown in Figs. 4 and 5, the four hand-crafted features are feasible and effective for identifying hemolytic peptide sequences, and their combination further enhances the model’s recognition ability. Figure 6 illustrates that the performance of cross-fusing features from two modalities is superior to simply concatenating them before inputting them into the self-attention mechanism. Word embedding features and hand-crafted features extract peptide sequence information using different rules: word embeddings utilize contextual information, while hand-crafted features focus on compositional information. The cross-attention mechanism captures the relationship between these two types of information, leading to improved model performance.
However, several issues warrant further investigation. As shown in Tables 2, 3, 4 and 5, the model exhibits higher sensitivity (SN) values on datasets 1 and 2, while the specificity (SP) value is higher on dataset 4. We hypothesize that these results are related to dataset balance. Datasets 1 and 2 have a higher proportion of positive samples, whereas dataset 4 has more negative samples. The model’s ability to recognize certain sample types is significantly influenced by the quality of the data. To address this, we conduct experimental analysis on an integrated dataset derived from four datasets, but even after CD-HIT processing, the dataset remained imbalanced. Therefore, future research will involve collecting more hemolytic peptide sequences to train the model, aiming to reduce the model’s dependency on dataset balance. Additionally, the model’s training time is relatively long, which may be due to the model’s complexity or the computing resources used. In the next steps, we will attempt to streamline the model to improve training efficiency without compromising performance.
Conclusions
The hemolytic activity of therapeutic peptides is one of the key factors restricting their entry into clinical trials, so it is urgent to find more available low hemolytic peptides. In view of the tedious and time-consuming biological experiments, the classification model of deep learning is the best scheme to identify hemolytic peptides. Following the principle that multiple features can obtain richer sequence information, we use word embedding technique, BLOSUM62, DDE, DPC and CKSAAP to extract features. Since these methods are based on two different ideas, multi-head cross-attention mechanism is used to integrate the information from the two modalities. Cross-attention mechanism improves the accuracy and robustness of the model by effectively exploiting the interaction information between two input features. Experimental results show that the proposed model is competitive in identifying hemolytic peptides compared with the baseline model. However, it is worth considering that although HemoFuse performs better than other existing models on the integrated dataset, it does not significantly improve on the four basic datasets. Therefore, we will continue to explore appropriate methods to improve the model to promote the development of peptide drugs.
Data availability
Data is provided within the manuscript.
References
Zhao, J., Zhao, C., Liang, G. Z., Zhang, M. Z. & Zheng, J. Engineering Antimicrobial peptides with Improved Antimicrobial and hemolytic activities. J. Chem. Inf. Model.53 (12), 3280–3296 (2013).
Orlov, N., Geraskina, O. & Feofanov, A. Study of membrane defects induced by antimicrobial and hemolytic peptide Ltc1 in erythrocyte membrane. Microsc. Microanal.27 (S1), 1728–1729 (2021).
Wang, T. R. et al. The effect of structural modification of antimicrobial peptides on their antimicrobial activity, hemolytic activity, and plasma stability. J. Pept. Sci.27 (5), e3306–e3306 (2021).
Vinod, K., Rajesh, K., Piyush, A., Sumeet, P. & Gajendra, P. S. R. A Method for Predicting Hemolytic Potency of chemically modified peptides from its structure. Front. Pharmacol.11 (54), 1–8 (2020).
Ankur, G. et al. Hemolytik: a database of experimentally determined hemolytic and non-hemolytic peptides. Nucleic Acids Res.42 (Database), D444–D449 (2014).
Malak, P. et al. DBAASP v3: database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res.49 (D1), D288–D297 (2021).
Zhang, S. L. & Li, X. J. Pep-CNN: an improved convolutional neural network for predicting therapeutic peptides. Chemometr. Intell. Lab. Syst.221, 104490 (2022).
Shi, H. Y. & Zhang, S. L. Accurate prediction of anti-hypertensive peptides based on Convolutional Neural Network and gated recurrent unit. Interdiscip Sci. Comput. Life Sci.14, 879–894 (2022).
Mir, T. H., Hilal, T. & Kil, T. C. Meta-IL4: an Ensemble Learning Approach for IL-4-Inducing peptide prediction. Methods. 217, 49–56 (2023).
Jing, Y. Y., Zhang, S. L. & Wang, H. Q. DapNet-HLA: adaptive dual-attention mechanism network based on deep learning to predict non-classical HLA binding sites. Anal. Biochem.666, 115075 (2023).
Wang, R. H. et al. MVIL6: Accurate identiffcation of IL-6-induced peptides using multi-view feature learning. Int. J. Biol. Macromol.246, 125412 (2023).
Xing, W. X., Zhang, J., Li, C., Huo, Y. J. & Dong, G. F. iAMP-Attenpred: a novel antimicrobial peptide predictor based on BERT feature extraction method and CNN-BiLSTM-Attention combination model. Brief. Bioinform. 25 (1), 1–9 (2024).
Zhu, Y. H., Liu, Z., Liu, Y., Ji, Z. W. & Yu, D. J. ULDNA: integrating unsupervised multi-source language models with LSTM-attention network for high-accuracy protein-DNA binding site prediction. Brief. Bioinform. 25 (2), 1–10 (2024).
Du, Z. J., Xu, Y. X., Liu, C. Q. & Li, Y. H. pLM4Alg: protein Language Model-based predictors for allergenic proteins and peptides. J. Agric. Food Chem.72 (1), 752–760 (2024).
Nhat, T. P., Rajan, R., Jongsun, P., Adeel, M. & Balachandran, M. H2Opred: a robust and efficient hybrid deep learning model for predicting 2’-O-methylation sites in human RNA. Brief. Bioinform. 25 (1), 1–13 (2024).
Zhou, C. M., Peng, D. J., Liao, B., Jia, R. R. & Wu, F. X. ACP_MS: prediction of anticancer peptides based on feature extraction. Brief. Bioinform. 23 (6), 1–10 (2022).
Beltrán, J. F. et al. VirusHound-I: prediction of viral proteins involved in the evasion of host adaptive immune response using the random forest algorithm and generative adversarial network for data augmentation. Brief. Bioinform. 25 (1), 1–8 (2024).
Chen, Y. G. et al. Quantitative model for genome-wide cyclic AMP receptor protein binding site identification and characteristic analysis. Brief. Bioinform. 24 (3), 1–12 (2023).
Yu, Y. T. et al. Cooperation of local features and global representations by a dual-branch network for transcription factor binding sites prediction. Brief. Bioinform. 24 (2), 1–9 (2023).
Ma, C. W. & Wolfinger, R. A prediction model for blood-brain barrier penetrating peptides based on masked peptide transformers with dynamic routing. Brief. Bioinform. 24 (6), 1–12 (2023).
Wang, L. Y. et al. ncRFP: a novel end-to-end method for non-coding RNAs Family Prediction based on deep learning. IEEE/ACM Trans. Comput. Biol. Bioinf.18 (2), 784–789 (2021).
Pang, Y. X., Yao, L. T., Xu, J. Y., Wang, Z. & Lee, T. Y. Integrating transformer and imbalanced multi-label learning to identify antimicrobial peptides and their functional activities. Bioinf. (Oxford England). 38 (24), 1–7 (2022).
Chang, K. L. et al. Short human eccDNAs are predictable from sequences. Brief. Bioinform. 24 (3), 1–11 (2023).
Wang, X. Y. et al. ASPIRER: a new computational approach for identifying non-classical secreted proteins based on deep learning. Brief. Bioinform. 23 (2), 1–12 (2022).
Kumardeep, C. et al. R Gajendra P.S. A web server and Mobile App for Computing hemolytic potency of peptides. Sci. Rep.6 (1), 22843 (2016).
Su, W. T. et al. HemoPred: a web server for predicting the hemolytic activity of peptides. Future Med. Chem.9 (3), 275–291 (2017).
Hasan, M. M. et al. HLPpred-Fuse: improved and robust prediction of hemolytic peptide and its activity by fusing multiple feature representation. Bioinformatics. 36 (11), 3350–3356 (2020).
Timmons, P. B. & Hewage, C. M. HAPPENN is a novel tool for hemolytic activity prediction for therapeutic peptides which employs neural networks. Sci. Rep.10 (1), 10869 (2020).
Kumar, V., Kumar, R., Agrawal, P., Patiyal, S. & Raghava, G. P. S. A Method for Predicting Hemolytic Potency of chemically modified peptides from its structure. Front. Pharmacol.11, 1–8 (2020).
Adiba, Y., Sadaf, G., Naeem, A., Imran, A. & Fayyaz, M. HemoNet: Predicting hemolytic activity of peptides with integrated feature learning. Journal of bioinformatics and computational biology. ;19(5):2150021. (2021).
Milad, S., Arash, A. K. & Shiun, J. Y. AMPDeep: hemolytic activity prediction of antimicrobial peptides using transfer learning. BMC Bioinform.23 (1), 389 (2022).
Ritesh, S. et al. EnDL-HemoLyt: Ensemble Deep Learning-based Tool for identifying therapeutic peptides with low hemolytic activity. IEEE J. Biomedical Health Inf.28 (4), 1896–1905 (2023).
Yang, S. & Xu, P. Hemolytic peptides prediction by double ensemble engines from Rich sequence-derived and transformer-enhanced information. Anal. Biochem.690, 115523 (2024).
Li, W. Z. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22 (13), 1658–1659 (2006).
Wu, X. B. & Bartel, D. P. kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res.45 (W1), W534–W538 (2017).
Li, Z. T. et al. Adapt-Kcr: a novel deep learning framework for accurate prediction of lysine crotonylation sites based on learning embedding features and attention architecture. Briefings Bioinf.23 (2), bbac037 (2022).
Zhang, Y., Lin, J. Y., Zhao, L. M., Zeng, X. X. & Liu, X. R. A novel antibacterial peptide recognition algorithm based on BERT. Briefings Bioinf.22 (6), bbab200 (2021).
Li, A. et al. Phosphorylation site prediction with a modified k-nearest neighbor algorithm and BLOSUM62 matrix. Conf. Proceedings: Annual Int. Conf. IEEE Eng. Med. Biology Soc.2005, 6075–6078 (2005).
Vijayakumar, S. & Namasivayam, G. Harnessing Computational Biology for exact Linear B-Cell Epitope Prediction: a novel amino acid composition-based feature descriptor. Omics: J. Integr. Biology. 19 (10), 648–658 (2015).
Manoj, B. & Raghava, G. P. S. ESLpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucleic Acids Res.32, W414–W419 (2004).
Chen, Z. et al. Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs. PLoS ONE. 6 (7), e22930 (2017).
Wang, M., Lei, C. Q., Wang, J. X., Li, Y. H. & Li, M. TripHLApan: predicting HLA molecules binding peptides based on triple coding matrix and transfer learning. Brief. Bioinform. 25 (3), bbae154 (2024).
Fang, Y. T. et al. AFP-MFL: accurate identification of antifungal peptides using multi-view feature learning. Brief. Bioinform. 24 (1), bbac606 (2023).
Guan, J. H. et al. A two-stage computational framework for identifying antiviral peptides and their functional types based on contrastive learning and multi-feature fusion strategy. Brief. Bioinform. 25 (3), bbae208 (2024).
Yao, L. T. et al. AMPActiPred: a three-stage framework for predicting antibacterial peptides and activity levels with deep forest. Protein Sci.33 (6), e5006 (2024).
Zhang, W. T., Xu, Y. C., Wang, A. W., Chen, G. & Zhao, J. B. Fuse feeds as one: cross-modal framework for general identification of AMPs. Brief. Bioinform. 24 (6), 1–14 (2023).
Huang, Z. J., Zhang, P. & Deng, L. DeepCoVDR: deep transfer learning with graph transformer and cross-attention for predicting COVID-19 drug response. Bioinformatics(Oxford England). 39 (Supplement_1), i475–i483 (2023).
Nguyen, N. Q., Park, S., Gim, M. & Kang, J. MulinforCPI: enhancing precision of compound-protein interaction prediction through novel perspectives on multi-level information integration. Brief. Bioinform. 25 (1), 1–11 (2024).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No.12101480), the Natural Science Basic Research Program of Shaanxi (No.2024JC-YBMS-004), and Xidian University Specially Funded Project for Interdisciplinary Exploration (No.TZJH2024028).
Author information
Authors and Affiliations
Contributions
Ya Zhao and Shengli Zhang wrote the main manuscript text and Yunyun Liang prepared Figs. 1, 2, 3, 4 and 5. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, Y., Zhang, S. & Liang, Y. HemoFuse: multi-feature fusion based on multi-head cross-attention for identification of hemolytic peptides. Sci Rep 14, 22518 (2024). https://doi.org/10.1038/s41598-024-74326-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-74326-3