Abstract
Rapid and accurate diagnosis of acute myeloid leukemia (AML) remains a significant challenge, particularly in the context of myelodysplastic syndrome (MDS) or MDS/myeloproliferative neoplasm with NPM1 mutations. This study introduces an innovative approach using holotomography (HT), a 3D label-free quantitative phase imaging technique, to detect NPM1 mutations. We analyzed a dataset of 2073 HT myeloblast images from 48 individuals, including both NPM1 wild-type and mutated samples, to distinguish subcellular morphological changes associated with NPM1 mutations. Employing a convolutional neural network, we analyzed 3D cell morphology, focusing on refractive index distributions. The machine learning model showed high accuracy, with an area under the receiver operating characteristic curve of 0.9375 and a validation accuracy of 76.0%. Our findings reveal distinct morphological differences between the NPM1 wild-type and mutation at the subcellular level. This study demonstrates the potential of HT combined with deep learning for early, efficient, and cost-effective diagnosis of AML, offering a promising alternative to traditional stepwise genetic testing methods and providing additional assistance in morphological myeloblast discrimination. This approach may revolutionize the diagnostic process in leukemia, facilitating early detection and potentially reducing the reliance on extensive genetic testing.
Similar content being viewed by others
Introduction
The classification of hematologic neoplasms has continually evolved1,2,3,4 since the first global consensus in 19975, transitioning from a morphology-centric approach to one incorporating genomic data. This evolution reflects a paradigm shift in our understanding of these diseases. Notably, the 2022 revision of acute myeloid leukemia (AML) diagnosis de-emphasizes the traditional 20% leukemic blast cutoff in the presence of driver mutations3,4, highlighting the need for comprehensive, yet costly, diagnostic methods for effective treatment planning and risk assessment.
Nucleophosmin (NPM1), a member of the nucleophosmin/nucleoplasmin family, was discovered around 40 years ago6,7. Functioning as a nucleo-cytoplasmic shuttling protein, NPM1’s activity is closely linked to its structure, particularly mutations in its C-terminal region, which are crucial in leukemogenesis8,9. About 80% of these mutations are type A, characterized by a TCTG duplication in the C-terminal. NPM1 mutations, found in 30–35% of AML cases10,11, were initially identified as a provisional entity in 20081, but were included in the 2017 WHO diagnostic criteria2. Their detection methods have evolved from immunohistochemistry to real-time PCR and next-generation sequencing12, offering a broader understanding of coexisting genetic alterations.
The mutational status of NPM1 is vital in guiding treatment decisions, including bone marrow transplantation and specific chemotherapy regimens4,10,13,14,15. The 2024 WHO classification highlight the rapid progression of myelodysplastic syndrome (MDS) or MDS/myeloproliferative neoplasm (MPN) with NPM1 mutations to AML16,17, underscoring the importance of early detection, especially in cases with less than 20% leukemia cells or evidence of multilineage dysplasia. However, identifying NPM1-mutated leukemic blasts remains challenging due to the lack of distinct morphological features.
Holotomography (HT), a 3D quantitative phase imaging technique, offers high-resolution, label-free imaging by leveraging refractive index (RI) distributions18,19,20. Functioning as an optical analog to X-ray computed tomography, HT constructs 3D tomograms from multiple 2D holographic images21,22,23. Its advantages include label-free detection, fast assessment, user-friendliness, and the feasibility of objective measurement. HT’s efficacy in differentiating cell types and its integration into deep learning models for advanced screening holds significant promise.
Prior studies have shown the effectiveness of HT in analyzing diverse pathophysiological samples. This includes distinguishing mature cells from blasts24, analyzing CD8 + T cells as sepsis biomarkers23, and studying biomolecular condensates in bone metastasis25. Combined with machine learning approaches26, HT has also been effective in classifying lymphocyte subtypes27, a task challenging for manual methods. The incorporation of HT data into deep learning models has shown potential in enhancing screening accuracy beyond traditional visual inspection, especially as recent deep learning approaches based solely on image data have demonstrated high sensitivity in classifying bone marrow cells28. This study aimed to develop a deep learning model using HT data to predict NPM1 mutations in leukemic blasts, potentially revolutionizing the diagnostic process for AML.
Results
Morphological features of leukemic blasts revealed by holotomography
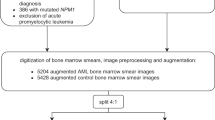
In our investigation, we analyzed 2,073 HT images of leukemic blasts from 48 specimens. This dataset included 1,173 images from 22 NPM1MUT specimens and 900 images from 26 NPM1WT specimens (Fig. 1). The demographic characteristics of the patient groups were similar, as detailed in Table 1. Traditional Wright stain slides did not reveal significant morphological differences between the groups (Fig. 2a,c). However, HT facilitated an in-depth 3D analysis of the leukemic blasts, revealing intricate intracellular RI contrasts (Fig. 2b,d).
Representative images of myeloblasts from bone marrow (BM) of acute myeloblastic leukemia (AML) patients. (a) Wright-stained BM smear image of an NPM1 wild-type AML patient. (b) Myeloblast holographic tomography (HT) image from the same patient, showing central sections in the xy, zy, and xz planes, a line profile of the refractive index through a nucleolus (right), and a three-dimensional rendering of the full volumetric image. (c) Wright-stained BM smear image of an NPM1 mutant AML patient. (d) Myeloblast HT image from the same patient, with similar features as in (B).
The nuclei of these cells displayed a pronounced nuclear to cytoplasmic ratio (N/C ratio) and were faintly contoured by changes in RI. Within the nuclei, the nucleoli were distinctly visible with defined outlines and typically exhibited a higher RI. The nucleoplasm region showed spatial variations in RI values, possibly reflecting chromatin distribution heterogeneity.
Notably, differences in RI intensity were associated with NPM1 mutation status. Cells from the NPM1WT group showed increased RI in the nucleolar regions, while those from the NPM1MUT group demonstrated comparatively lower RI in these areas. A striking observation was the more defined delineation between the nucleoli and the surrounding nucleoplasm in the NPM1MUT group. This suggests a potential disruption of the nucleolus structure due to NPM1 protein dislocation, a well-documented consequence of NPM1 mutations29.
In the cytoplasm, cells with NPM1 mutations exhibited higher RI overall. However, droplets of high RI density were also observed in some NPM1WT specimens. Additionally, certain HT images of leukemic blasts presented particulate structures with high RI, indicative of a lipid-based composition30.
Numerical parameters of leukemic blasts
Our study identified 39 distinct numerical parameters associated with leukemic blasts, previously undocumented and detailed in Supplementary Table 1. Comparative analysis of these parameters revealed significant differences in cell and nucleus sphericity, nucleolus volume, N/C ratio, and RI values of the cytoplasm and nucleolus between the NPM1WT and NPM1MUT groups (Fig. 3).
Statistical comparison of myeloblast properties obtained from holotomography (HT) of individual myeloblasts. Each data point represents an individual myeloblast HT image. The average ± standard deviation is displayed on the right of each swarm chart. Data points beyond the 95% confidence interval are not shown to visualize the typical value range better. *, **, and *** denote P < 0.01, P < 0.001, and P < 0.0001 in the Mann–Whitney U test. The absence of any asterisk indicates no significance.
Regarding cell volume, NPM1MUT blasts demonstrated a slightly larger average volume (462.18 ± 187.43 fL) compared to NPM1WT blasts (446.24 ± 181.18 fL), although this difference was not statistically significant. Similar marginal distinctions were observed in nucleolus volume (NPM1WT: 14.48 ± 12.54 fL, NPM1MUT: 14.99 ± 13.09 fL), cytoplasm volume (NPM1WT: 259.28 ± 84.73 fL, NPM1MUT: 260.90 ± 80.70 fL), and nucleoplasm RI (NPM1WT: 1.3692 ± 0.0039, NPM1MUT: 1.3692 ± 0.0031).
Notably, NPM1MUT blasts exhibited significantly reduced cell sphericity (NPM1WT: 0.804 ± 0.052, NPM1MUT: 0.797 ± 0.057) and lower cytoplasm RI (NPM1WT: 1.3635 ± 0.0032, NPM1MUT: 1.3632 ± 0.0029). The nucleus volume in NPM1MUT blasts was also larger (NPM1WT: 186.71 ± 101.13 fL, NPM1MUT: 201.07 ± 109.97 fL). More pronounced differences were evident in nucleus sphericity (NPM1WT: 0.501 ± 0.065, NPM1MUT: 0.538 ± 0.057), nucleoplasm volume (NPM1WT: 172.22 ± 90.94 fL, NPM1MUT: 186.08 ± 98.36 fL), nucleolus RI (NPM1WT: 1.3764 ± 0.0044, NPM1MUT: 1.3759 ± 0.0034), and NC volume ratio (NPM1WT: 0.400 ± 0.059, NPM1MUT: 0.417 ± 0.054). Intriguingly, the N/C ratio difference was only appreciable in the 3D analyses, as opposed to the 2D projected area (NPM1WT: 0.633 ± 0.053, NPM1MUT: 0.629 ± 0.047). These observed morphological changes, particularly the spherical inflation of the nucleus, disruption of nucleolar aggregation, and alteration in cytoplasmic roundness, are hypothesized to correlate with the dislocation of the mutated NPM1 protein.
Artificial-intelligence-assisted prediction model of NPM1 mutation: classifier development and validation
We leveraged HT combined with deep learning to identify mutation-related alterations in leukemic blasts. Our deep learning classifier was trained on 1,740 HT images of blasts from 40 patients and validated using an independent dataset of 333 HT images from eight patients. The training dataset consisted of 984 images from 18 NPM1MUT patients and 756 images from 22 NPM1WT patients. The validation dataset included 189 images from four NPM1MUT patients and 144 images from four NPM1WT patients (Fig. 1).
In the validation phase, the classifier accuracy for single blast HT images was 76.0%. Misclassification rates were 20.4% for NPM1WT and 25.8% for NPM1MUT blasts (Fig. 4a,b). The t-SNE analysis demonstrated a clear separation of latent features learned by the classifier between NPM1MUT and NPM1WT (Fig. 5). Notably, the classifier exhibited strong correlation between the training and validation datasets within both the NPM1MUT and NPM1WT groups, underscoring its robustness. Importantly, this correlation is significant because the training and validation data were independently acquired from distinct patient cohorts, highlighting the classifier’s ability to generalize across different datasets by focusing on HT features relevant to NPM1 mutations.
Deep learning classification of myeloblast holotomography (HT) images for NPM1 mutation status. (a) Validation result of the deep learning classifier on myeloblast HT images of four NPM1 wild-type patients. (b) Validation result of the deep learning classifier on myeloblast HT images of four NPM1 mutant patients. (c) Receiver operating characteristic curve representing the performance of patient-wise NPM1 mutation screening based on different threshold positive prediction ratios. (d) Plot of deep learning output values corresponding to the two classes (wild-type and mutant). Each data point represents an individual myeloblast HT image. The dashed diagonal line indicates the boundary where the two output values are equal.
Latency of 264 dimensions visualized in 2D using t-SNE. (a) Validation data marked based on prediction accuracy, depicting consistent feature extraction and error distribution. Central sections of error cases shown in various regions of latent feature space. Scale bar = 5 µm. (b) NPM1 mutant data marked with size dependent on variant allele frequency, showing moderate correlation with mutation degree. (c) All data marked with size dependent on cell volume, illustrating size-dependent clustering of myeloblast data.
However, some misclassifications occurred, particularly in cells with atypical sizes or poorly defined nuclear boundaries (Fig. 5a). Additional analyses revealed key insights into the classifier’s decision-making process, including spatial feature mapping for each class (Supplementary Fig. S1). While the current network design outperformed lighter variants in our trials (Supplementary Fig. S2), these alternatives may offer benefits such as faster training times and reduced overfitting. Moreover, the high 3D resolution provided by HT was found to enhance single-blast accuracy, as reducing data resolution led to diminished performance (Supplementary Fig. S3).
Insights from t-SNE mapping and ROC analysis
Further t-SNE mapping correlated misclassifications with variant allele frequency (VAF) (Fig. 5b) and cell volume (Fig. 5c). Misclassified cases and those outside the NPM1MUT prediction lobe typically showed low VAF, and cells with abnormally large volumes were more common in the apical zones of both the NPM1WT and NPM1MUT lobes.
To evaluate the classifier’s effectiveness in determining the NPM1 genotype in the validation patients, we employed the receiver operating characteristic (ROC) curve by varying the threshold of the NPM1MUT prediction rate (Fig. 4c). The optimal true positive rate (TPR) with a zero false positive rate (FPR) was 0.75 at a threshold of 93.3%. Conversely, the lowest FPR with a TPR of 1.0 was 0.25 at a threshold of 39.2%. The area under the ROC curve was 0.9375.
An analysis of classifier outputs revealed that variations in prediction across patients resulted from overall shifts in classifier outputs rather than individual outliers (Fig. 4d). The distribution of outputs for both genotypes showed trends toward the predicted genotype, with the extent of the shift correlating with the prediction rate.
Discussion
The morphological overlap among leukemic blasts often hinders differentiation using conventional optical microscopy. However, HT and AI have shown promise in identifying NPM1 mutations, with an accuracy of 76.0% in single-cell mutation determination and an AUC of 0.9375 for patient-wise detection. HT’s unique capability in 3D label-free imaging, coupled with deep learning image classification, provides a rapid and cost-effective method for detecting NPM1 mutations in AML patients. The entire process, from cell imaging to AI-driven final analysis, was completed within a mere 20 min with no requirements for the use of experts or exogenous labeling agents (Fig. 6).
Schematic illustration of the proposed NPM1 mutation screening method in comparison to the gold standard test. The gold standard test provides the confirmatory genetic profile based on DNA next-generation sequencing, yet the typical turnaround time may stretch to 2 weeks. The proposed method can provide an early screening based on the three-dimensional morphology of myeloblasts using holotomography (HT) imaging aided by statistical and artificial intelligence analyses.
HT’s key advantages include minimal examination costs and the ability to confirm cellular RI images while calculating numerical data. The RI features of the nucleoli and cytoplasm are particularly pronounced in NPM1MUT blasts, facilitating their identification. This observation is supported by IHC staining, which confirms protein dislocation associated with NPM1 mutations10. HT’s sensitivity in detecting minute protein changes (as subtle as 5.4 fg/fL with a noticeable RI difference of 0.001)31 and its linear correlation between the RI value and protein concentration32,33 allow for precise quantification of protein and cellular alterations due to genetic mutations.
Our study’s numerical parameter analysis indicates that NPM1 mutations cause spherical nuclear inflation, nucleolar aggregation disruption, and cytoplasmic morphology deformation11. These alterations, previously observed with anti-N-terminal antibody staining, are more pronounced in three-dimensional views. In two-dimensional optical microscopy, these changes mimic monoblast-like morphology, suggesting characteristics of FAB M4 or M5 subtypes in AML with mutated NPM12.
The integration of AI with an HT database focused on detecting various genetic alterations in leukemic blasts is crucial for objective and precise cell classification. As molecular genetic alterations play an increasingly important role in predicting patient outcomes, especially as the traditional 20% blast threshold for leukemia diagnosis becomes less definitive, this approach has significant potential34,35,36. In resource-limited settings, where advanced molecular tests like NGS may not be routinely performed, crucial diagnoses may be missed. For instance, if the blast count exceeds 10%, the absence of molecular testing could lead to incorrect or delayed treatment decisions. Our approach addresses this gap by enabling early mutation detection in hematologic malignancies, facilitating timely and accurate classification. This not only expedites the identification of cases requiring further genetic testing but also ensures that patients receive the appropriate treatment for their specific disease state, optimizing the allocation of medical resources.
The limitations of our study include the relatively small validation dataset, which may affect the generalizability of our findings. Future studies with larger patient cohorts are necessary to enhance the method’s robustness and reliability, as indicated in our comparative study with reduced training data (Supplementary Fig. S4). Additionally, the genetic heterogeneity of AML introduces significant challenges related to inter-patient variability. Even within individual patients, somatic mutation changes can vary depending on the specific clone producing each cell37,38,39. This diversity is well-established and is associated with various prognoses based on coexisting gene mutations, particularly among AML patients with mutated NPM14,40. While a dominant clone within a one patient may exhibit similar characteristics, as demonstrated in our study, challenges in achieving higher accuracy may arise due to associated morphological changes among patients.
To improve early screening of leukemic blasts that require urgent assessment, it is critical to train AI models on a wider variety of AML blasts encompassing diverse diagnostic classifications. This will enhance the effectiveness of screening tools. Moreover, as data volume increases, future studies will require improvements in both software and hardware. With our current setup—comprising the network design, input pipeline, and hardware—200 training iterations take 36 h and utilize 30.96 GB of video memory, while each set of network parameters requires 925.30 MB of storage. Although feasible on a high-performance personal computer, larger databases in future studies may necessitate more efficient software or advanced computational resources. Software enhancements, such as refining the loss function and incorporating data augmentation, offer promising strategies to better leverage large-scale data. Additionally, future research should focus on more finely stratified genetic subgroups to improve the differentiation of morphological features and mitigate variability within the AML patient population.
In conclusion, our work underscores the potential of combining HT and AI for early NPM1 mutation detection, offering a complementary tool to traditional diagnostic methods. The integration of this approach with genetic confirmation and prognostic analysis will bolster its utility in AML diagnosis. Despite current limitations, our promising results lay the groundwork for further research aimed at refining and expanding this methodology, contributing to more effective AML diagnostic strategies.
Methods
Specimen selection
Between March 2021 and July 2023, our research focused specifically on utilizing residual specimens obtained from patients who underwent bone marrow (BM) examinations. The process of specimen selection for analysis unfolded in three distinct stages.
In the first stage, residual BM specimens were characterized by a leukemic blast percentage that exceeded 50%. Utilizing HT-1H (Tomocube Inc., Daejeon, Republic of Korea), we captured images and numerical indices of leukemic blasts within 24 h of specimen collection.
Moving to the second stage, the procedure involved filtering specimens based on the results of NGS. Specifically, individuals with confirmed NPM1 mutations identified in the NGS results were designated as the experimental group (Mutated NPM1, NPM1MUT). Specimens lacking molecular abnormalities associated with AML defined by genetic abnormalities or AML with recurrent genetic abnormalities were selected as potential specimens for the control group (Wild-type NPM1, NPM1WT).
In the third stage, two pathologists conducted a meticulous examination of the Wright-stained slides of the selected specimens for the final selection of the NPM1WT group. By comparing the morphology of leukemic blasts in the NPM1MUT group specimens, those with visually indistinguishable features were selected for the NPM1WT group.
HT imaging system and numerical parameter extraction
We utilized a laser-based holotomography system (HT-1H, Tomocube Inc., Daejeon, Republic of Korea). To capture individual images of leukemic blasts, residual BM specimens were diluted in a phosphate-buffered saline (PBS) solution, adjusted to the physiological pH of the living organism41,42. Then, we place 20 μL of the diluted specimen, on a 25 × 50 mm cover slide (C025501, Matsunami Glass Ind., Japan), covered it with another cover slide, and conducted the measurements.
During cell measurements, the HT-1H microscope not only provides visualization of the cells but also enables the measurement of the RIs of various intracellular structures (For more information, refer to the Supplementary Text.). Through this, it allows for the generation of numerical data pertaining to spatial information and physical characteristics. The image reconstruction was performed with TomoStudio 2.7.3 (Tomocube Inc., Daejeon, Republic of Korea), specialized software for the measurement and processing of the HT-1H data.
The cellular properties were calculated from each HT image using the linear relationship between the biomolecule concentration and the RI contrast, along with image segmentation for subcellular regions. The properties of the entire cell or an intracellular region include volume, surface area, sphericity, average RI, and dry mass.
The inter-group difference in the distribution of properties was assessed using the Mann–Whitney U test due to uneven sample sizes and the necessity for a non-parametric approach to the distributions. For subcellular region segmentation, we utilized ilastik 1.4.043, a machine learning pixel classification software, followed by numerical post-processing based on priors about cellular morphologies. The numerical processing and analyses were performed using MATLAB 2022a (MathWorks, Natick, MA).
Deep learning classifier and optimization
For deep learning image classification, a convolutional neural network (CNN) designed for multi-resolution feature preservation was implemented. This CNN architecture, inspired by FishNet44, comprises three primary components—the tail, body, and head—in the feed-forward order. Additionally, it includes initial convolutional layers, tail-body bridging layers, as well as classifying layers. The tail, body, and head components extract features while downsampling, upsampling, and downsampling, respectively. A 2 × 2 × 2 max-pooling operation downsamples the feature map, whereas a 2 × 2 × 2 nearest-neighbor resizing operation upsamples the feature map. A distinct mechanism of this architecture is the recollection of equal-sampling features from the shallower layers. For instance, the feature maps after each upsampling operation in the body layer are concatenated with the first equal-sampling feature maps of the tail layer.
For a more quantitative description of the CNN, it includes a total of 80,839,106 adjustable parameters, mostly accounted for by the 247 convolutional layers. Residual bottleneck blocks45 precede every downsampling or upsampling operation. Each residual block results in a summation of two feature maps: one through a single 1 × 1 × 1 convolutional layer and the other through a series of 1 × 1 × 1, 3 × 3 × 3, and 1 × 1 × 1 convolutional layer that reduce the number of feature map channels. Every convolutional layer is preceded by instanceNorm46 and leaky ReLU pre-activation47. A more graphical description of the CNN classifier can be found in Supplementary Fig. S5.
The parameters of the CNN were optimized through gradient-based reduction of the cross-entropy loss function. The cross-entropy loss was weighted inversely proportional to the class-specific size of the training dataset to evenly penalize errors despite the class imbalance in the training dataset. The Adam optimizer was employed to adjust the parameters with the following hyperparameters: an initial learning rate of 0.0005, learning rate decay of 0.0001, gradient moment decay of 0.5, and second-order gradient moment decay of 0.999. The learning rate was further modulated through cosine annealing with a period of 32 to facilitate the exploration of a wide range of parameters. The batch size was 16, and each HT image of the training dataset was fed twice—once in its original form and once in a randomly augmented form—during each iteration through the dataset. Augmentation included the addition of Gaussian noise, random translation, horizontal flipping, and rotation. The size of each input HT image was 160 × 160 × 72.
The optimization process ran on a computational workstation, utilizing four graphics processing units of GeForce GTX 1080 Ti (NVIDIA Corporation, Santa Clara, CA) and eight workers from an Intel Xeon E5-2630 processor (Intel Corporation, Santa Clara, CA). In this hardware setting, each iteration through the training dataset consumed approximately 6 min. The training accuracy of the CNN saturated within 200 iterations. The CNN and its optimization process were implemented in a Python 3.7.16 environment that included the deep learning library Torch 1.0.0.
The final parameters of the CNN classifier were chosen based on the performance in the training dataset. To be specific, a portion of the training dataset with the size comparable to the validation dataset was used to assess the generalization accuracy instead of directly reflected during optimization. The quantities of optimization data is 637 and 574 for NPM1WT and NPM1MUT respectively. The quantities of generalization data is 144 and 410 for NPM1WT and NPM1MUT respectively. Among different sets of the CNN parameters yielded from the 200 iterations, the one with the highest sum of training accuracy and generalization accuracy was selected.
For the investigation of deep learning features, a 264-dimensional feature was extracted from each HT image prior to the final output layer of the CNN. After reducing the dimensions through t-distributed stochastic neighbor embedding (t-SNE), a 2D visualization was performed. The optimization process of t-SNE was conducted under a learning rate of 200 and a perplexity of 200, after standardizing each feature dimension to have 0 average and 1 standard deviation. The embedding and visualization were carried out using MATLAB 2022a (MathWorks, Natick, MA, USA).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Swerdlow, S. H. et al. WHO classification of tumours of haematopoietic and lymphoid tissues Vol. 2 (International agency for research on cancer Lyon, 2008).
Swerdlow, S. H. et al. WHO classification of tumours of haematopoietic and lymphoid tissues (International agency for research on cancer Lyon, 2017).
Khoury, J. D. et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: Myeloid and histiocytic/dendritic neoplasms. Leukemia 36, 1703–1719 (2022).
Döhner, H. et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood J. Am. Soc. Hematol. 140, 1345–1377 (2022).
Jaffe, E. S., Harris, N. L., Diebold, J. & Muller-Hermelink, H.-K. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: A progress report. Am. J. Clin. Pathol. 111, S8-12 (1999).
Kang, Y.-J., Olson, M. O., Jones, C. & Busch, H. Nucleolar phosphoproteins of normal rat liver and Novikoff hepatoma ascites cells. Cancer Res. 35, 1470–1475 (1975).
Schmidt-Zachmann, M., Hügle-Dörr, B. & Franke, W. A constitutive nucleolar protein identified as a member of the nucleoplasmin family. EMBO J. 6, 1881–1890 (1987).
Heath, E. et al. Biological and clinical consequences of NPM1 mutations in AML. Leukemia 31, 798–807 (2017).
Federici, L. & Falini, B. Nucleophosmin mutations in acute myeloid leukemia: A tale of protein unfolding and mislocalization. Prot. Sci. 22, 545–556 (2013).
Falini, B., Brunetti, L. & Martelli, M. P. How I diagnose and treat NPM1-mutated AML. Blood J. Am. Soc. Hematol. 137, 589–599 (2021).
Falini, B., Brunetti, L., Sportoletti, P. & Martelli, M. P. NPM1-mutated acute myeloid leukemia: From bench to bedside. Blood J. Am. Soc. Hematol. 136, 1707–1721 (2020).
Lopez, A. et al. Comparison of multiple clinical testing modalities for assessment of NPM1-mutant AML. Front. Oncol. 11, 701318 (2021).
Lachowiez, C. A. et al. Outcomes of older patients with NPM1-mutated AML: Current treatments and the promise of venetoclax-based regimens. Blood Adv. 4, 1311–1320 (2020).
Tiong, I. S. et al. Venetoclax induces rapid elimination of NPM1 mutant measurable residual disease in combination with low-intensity chemotherapy in acute myeloid leukaemia. Br. J. Haematol. 192, 1026–1030 (2021).
Sharma, N. & Liesveld, J. L. NPM 1 Mutations in AML—The Landscape in 2023. Cancers 15, 1177 (2023).
Forghieri, F. et al. NPM1-mutated myeloid neoplasms with< 20% blasts: A really distinct clinico-pathologic entity?. Int. J. Mol. Sci. 21, 8975 (2020).
Board, W. C. O. T. E. Haematolymphoid Tumours, WHO Classification of Tumours Vol. 11 (International Agency for Research on Cancer (IARC), 2024).
Kim, D. et al. in Advanced Imaging and Bio Techniques for Convergence Science 211–238 (Springer, 2021).
Park, Y., Depeursinge, C. & Popescu, G. Quantitative phase imaging in biomedicine. Nat Photon. 12, 578–589 (2018).
Kim, G. et al. Holotomography. Nat. Rev. Methods Prim. 4, 51 (2024).
Wolf, E. Three-dimensional structure determination of semi-transparent objects from holographic data. Opt. Commun. 1, 153–156 (1969).
Kim, K. et al. Optical diffraction tomography techniques for the study of cell pathophysiology. J. Biomed. Photon. Eng. 2, 020201 (2016).
Jin, D., Zhou, R., Yaqoob, Z. & So, P. T. Tomographic phase microscopy: Principles and applications in bioimaging. JOSA B 34, B64–B77 (2017).
Park, N. et al. Characteristics of leukemia blasts to differentiate from mature cells by optical diffraction tomography. Int. J. Lab. Hematol. 44, e258–e261 (2022).
Esposito, M. et al. TGF-β-induced DACT1 biomolecular condensates repress Wnt signalling to promote bone metastasis. Nat. Cell Biol. 23, 257–267 (2021).
Park, J. et al. Artificial intelligence-enabled quantitative phase imaging methods for life sciences. Nat. Methods 20, 1–16 (2023).
Yoon, J. et al. Identification of non-activated lymphocytes using three-dimensional refractive index tomography and machine learning. Sci. Rep. 7, 6654 (2017).
Matek, C., Krappe, S., Münzenmayer, C., Haferlach, T. & Marr, C. Highly accurate differentiation of bone marrow cell morphologies using deep neural networks on a large image data set. Blood J. Am. Soc. Hematol. 138, 1917–1927 (2021).
Mitrea, D. M. et al. Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat. Commun. 9, 842 (2018).
Kim, K. et al. Three-dimensional label-free imaging and quantification of lipid droplets in live hepatocytes. Sci. Rep. 6, 36815 (2016).
Jung, J. et al. Label-free non-invasive quantitative measurement of lipid contents in individual microalgal cells using refractive index tomography. Sci. Rep. 8, 6524 (2018).
Barer, R. Determination of dry mass, thickness, solid and water concentration in living cells. Nature 172, 1097–1098 (1953).
Popescu, G. et al. Optical imaging of cell mass and growth dynamics. Am. J. Physiol.-Cell Physiol. 295, C538–C544 (2008).
Montalban-Bravo, G. et al. NPM1 mutations define a specific subgroup of MDS and MDS/MPN patients with favorable outcomes with intensive chemotherapy. Blood Adv. 3, 922–933 (2019).
Rogers, H. J. et al. Complex or monosomal karyotype and not blast percentage is associated with poor survival in acute myeloid leukemia and myelodysplastic syndrome patients with inv (3)(q21q26. 2)/t (3; 3)(q21; q26. 2): A Bone Marrow Pathology Group study. Haematologica 99, 821 (2014).
Döhner, H. et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood J. Am. Soc. Hematol. 129, 424–447 (2017).
Cocciardi, S. et al. Clonal evolution patterns in acute myeloid leukemia with NPM1 mutation. Nat. Commun. 10, 2031 (2019).
Vosberg, S. & Greif, P. A. Clonal evolution of acute myeloid leukemia from diagnosis to relapse. Genes Chromosomes Cancer 58, 839–849 (2019).
Li, S. et al. Somatic mutations drive specific, but reversible, epigenetic heterogeneity states in AML. Cancer Discov. 10, 1934–1949 (2020).
Shimony, S., Stahl, M. & Stone, R. M. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 98, 502–526 (2023).
Hoang, V. T. et al. Optical properties of buffers and cell culture media for optofluidic and sensing applications. Appl. Sci. 9, 1145 (2019).
Phillips, J. R. & Cadwallader, D. E. Behavior of erythrocytes in phosphate buffer systems. J. Pharm. Sci. 60, 1033–1035 (1971).
Berg, S. et al. Ilastik: Interactive machine learning for (bio) image analysis. Nat. Methods 16, 1226–1232 (2019).
Sun, S., Pang, J., Shi, J., Yi, S. & Ouyang, W. Fishnet: A versatile backbone for image, region, and pixel level prediction. Adv. Neural Inf. Process. Syst, 31 (2018).
He, K., Zhang, X., Ren, S. & Sun, J. in Proceedings of the IEEE conference on computer vision and pattern recognition. 770–778.
Ulyanov, D., Vedaldi, A. & Lempitsky, V. Instance normalization: The missing ingredient for fast stylization. https://arxiv.org/abs/1607.08022 (2016).
Xu, B., Wang, N., Chen, T. & Li, M. Empirical evaluation of rectified activations in convolutional network. https://arxiv.org/abs/1505.00853 (2015).
Acknowledgements
This work was supported by the research fund of the Korea Society of Hematology (No. ICKSH-2023-07), National Research Foundation of Korea (2015R1A3A2066550, RS-2024-00442348), and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (RS-2023-00241278).
Author information
Authors and Affiliations
Contributions
H.K. and H.P. performed the research; H.K. and S.J. designed the research study; Y.P. contributed essential reagents or tools; H.K., G.K., M.L. analysed the data; H.K. and G.K. wrote the paper; Y.P. and S.J. contributed to critical revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Geon Kim, Manjae Lee, YongKeun Park, and Seongsoo Jang have financial interests in Tomocube Inc., a company that commercializes holotomography instruments. The other authors declare no competing interests.
Ethical approval
The study protocol was approved by the Institutional Review Board of Asan Medical Center (no. S2020-2538-0001) and Seoul National University Bundang Hospital (no. X-2309-853-901). This study was conducted using residual samples, and therefore, confirming that the need for informed consent was waived by Asan Medical Center. All methods of this study were conducted in accordance with the guidelines and regulations approved by the IRB.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, H., Kim, G., Park, H. et al. Integrating holotomography and deep learning for rapid detection of NPM1 mutations in AML. Sci Rep 14, 23780 (2024). https://doi.org/10.1038/s41598-024-75168-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-75168-9
Keywords
This article is cited by
-
From genotype to phenotype: decoding mutations in blasts by holo-tomographic flow cytometry
Light: Science & Applications (2025)