Abstract
Cancer-associated fibroblasts (CAFs) actively contribute to the formation of tumor-supportive microenvironments, thereby promoting cancer progression and impacting therapeutic outcomes. This study utilized global microRNA (miRNA) expression profiling to identify specific miRNAs responsible for reprogramming normal lung fibroblasts (LFs) into CAFs. miR-224 demonstrates increased expression in CAFs, and its levels are elevated in lung tumors compared to those in normal tissues, according to data from public databases. Overexpression of miR-224 in LFs increases the overall expression of CAF activation markers. Furthermore, LFs overexpressing miR-224 enhanced the migration and invasion of lung cancer cells via direct cell-to-cell contact in a co-culture system. In a mouse orthotopic injection model, miR-224 overexpression in LFs increased lung cancer metastasis. Using target prediction tools and subsequent 3′-UTR luciferase assay, Akirin1 was validated as a direct target gene of miR-224. In addition, LFs depleted of Akirin1 by siRNAs stimulated the migration and invasion of lung cancer cells compared to control LFs. Overall, these findings indicate that miR-224 induces CAF activation and promotes the migration and invasion of lung cancer cells by targeting Akirin1 in co-culture systems.
Similar content being viewed by others
Introduction
Despite advances in targeted therapies and immunotherapies, lung cancer remains an important global health concern characterized by increased recurrence rates and unfavorable prognosis1,2. Recent cancer research has shifted focus from malignant cells to exploring the intricate interactions between cancer cells and their surrounding stroma, particularly cancer-associated fibroblasts (CAFs) residing within the tumor microenvironment3,4,5. CAFs stimulate the epithelial-mesenchymal transition of cancer cells6 and enhance the expression of matrix metalloproteinase-2 and vascular endothelial growth factor, thereby orchestrating the remodeling of the extracellular matrix7. Consequently, CAFs create a tumor-supportive microenvironment and facilitate the growth, invasion, and metastasis of various cancers, including lung cancer.
The initiation of CAF activation is prompted by cytokines, growth factors, and microRNAs (miRNAs)8,9,10. For instance, leukemia inhibitory factor acts as a pro-invasive fibroblast activator, independent of alpha-smooth muscle actin expression10. Platelet-derived growth factor, a pro-fibrotic growth factor released by cancer and stromal cells, triggers CAF activation11. Furthermore, CAF activation induced by hypoxia-inducible factor-1alpha (HIF-1α) can activate the NFκB signaling pathway and subsequently release chemokine (C-C motif) ligand 5, promoting tumor growth in lung cancer12. Lastly, miR-21 derived from lung cancer cells induces a CAF-like morphology in normal fibroblasts through the TGF-β pathway, thereby supporting the progression of lung cancer13. Previously, we reported that elevated levels of miR-196a in lung CAFs stimulate the migration and invasion of lung cancer cells by targeting Annexin A18. Increased expression of miR-196a is associated with unfavorable prognosis in patients with lung adenocarcinoma (LUAD); therefore, miR-196a has potential as a therapeutic target or biomarker for LUAD.
miR-224 is an oncogenic miRNA that promotes cancer progression through various mechanisms. In renal cell carcinoma, miR-224 facilitates cell proliferation, invasion, and migration by targeting Occludin14. In non-small cell lung cancer, miR-224 enhances cellular proliferation, migration, tumor growth, and metastasis by targeting TNFα-induced protein 1 and SMAD415. Additionally, miR-224-enriched exosomes promote epithelial-mesenchymal transition, accelerate cell-cycle progression, and inhibit apoptosis16. Herein, we demonstrated the noteworthy role of miR-224, which is significantly upregulated in lung CAFs, in influencing CAF activation. The activation of CAFs by miR-224 plays a crucial role in promoting the migration, invasion, and metastasis of lung cancer cells by modulating Akirin1 in the tumor microenvironment.
Results
miR-224 is elevated in CAFs and promotes their activation
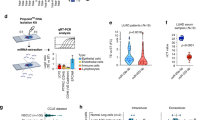
To explore the roles of miRNAs in lung CAFs derived from lung tumors in Kras-mutant (KrasLA1) mice17, we profiled the miRNA expression levels in CAFs and normal lung fibroblasts (LFs) using NanoString nCounter. The top differentially expressed miRNAs were selected and validated by quantitative reverse transcription-PCR (qRT-PCR), which was published previously18. Among the upregulated miRNAs, miR-224 exhibited the highest fold change, with a 7.68-fold increase in expression in CAFs compared to that in LFs (Fig. 1A). Moreover, miR-224 levels were found to be elevated in lung tumors compared to normal tissues, as indicated by dbDEMC (the database of differentially expressed miRNAs in human cancers; https://www.biosino.org/dbDEMC/index) (Fig. 1B). Analysis of The Cancer Genome Atlas (TCGA) LUAD data revealed upregulation of miR-224 in tumors compared to normal tissues (Fig. 1C). Subsequently, we assessed the effect of miR-224 on the survival of patients with LUAD using formalin-fixed, paraffin-embedded (FFPE) tumor samples. The Kaplan-Meier survival analysis demonstrated that increased miR-224 expression was associated with poor prognosis in patients (Fig. 1D). These data imply that elevated expression of miR-224 in CAFs participates in promoting lung cancer progression; therefore, we chose to explore the role of miR-224 in the interactions between CAFs and cancer cells within the tumor microenvironment.
miR-224 stimulates CAF activation. (A) qRT-PCR of miR-224 in LFs and CAFs. miR-224 expression levels were normalized to RNU6B snoRNA levels, and values relative to that of LFs (set at 1.0) are shown. Data are represented as mean ± standard deviation (SD; n = 3). P-value was assessed by two-tailed Student’s t-test. (B) Differentially expressed miR-224 levels in human cancers obtained from the dbDEMC3.0 database. The graph shows log2 fold-change values (cancer vs. normal). (C) miR-224 levels in normal and tumor tissues based on TCGA lung adenocarcinoma data. (D) Kaplan-Meier plot showing overall survival of patients with lung adenocarcinoma according to their miR-224 levels. miR-224 levels were measured by NanoString in FFPE tumor samples. P-value was assessed by log-rank test. (E) qRT-PCR of miR-224 in LFs transduced with empty (LF-vec) or miR-224 lentiviral vector (LF-224). Mean ± SD (n = 3). (F) Actin cytoskeleton staining of LF-vec and LF-224 cells. Cells were stained using Alexa Fluor® 594-conjugated phalloidin (red) and DAPI (blue), and cell circularity was then measured by ImageJ. Mean ± SD (LF-vec, n = 69; LF-224, n = 67). (G) Cell growth analysis in LF-vec and LF-224 cells. Cells were seeded in 12-well plates and counted on the indicated days after seeding. Mean ± SD (n = 4). *P < 0.05, **P < 0.01. (H) qRT-PCR of CAF activation markers in LF-vec and LF-224 cells. The levels of CAF activation markers are normalized to Rpl32. Mean ± SD (n = 3).
To elucidate the function of miR-224, we performed lentiviral transduction to overexpress miR-224 in LFs (Fig. 1E). Overexpression of miR-224 induced a shift in cell morphology, resulting in a slender and pointed shape accompanied by a reduction in cellular circularity (Fig. 1F). Moreover, miR-224 overexpression promoted cell growth (Fig. 1G) and increased the expression of markers associated with CAF activation, including Fap, Pdgfra, P4ha3, S100a4, Des, Ddr2, Vim, and Col1a2 (Fig. 1H). These findings suggest that miR-224 alters the characteristics of LFs to resemble those of CAFs, thereby triggering CAF activation.
miR-224 stimulates the migration and invasion of lung cancer cells in co-culture systems
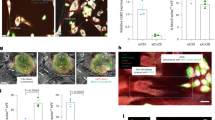
To examine how the overexpression of miR-224 in LFs influences the migration of lung cancer cells, we performed a transwell migration assay. When LFs overexpressing miR-224 (LF-224) were seeded in the bottom wells of transwell plates and 344SQ murine lung cancer cells were seeded in the upper inserts, the migration of 344SQ cells was unaffected by LF-224 cells compared to control LFs (LF-vec; Supplementary Fig. 1A). Next, LF-vec and LF-224 (labeled with enhanced green fluorescent protein) were directly co-cultured with 344SQ or 344LN cells (labeled with mCherry) in the upper inserts, and growth medium containing 10% fetal bovine serum (FBS) was added into the bottom wells to stimulate cell migration. Consequently, the migratory ability of lung cancer cells co-cultured with LF-224 cells was found to be higher than that of cancer cells co-cultured with LF-vec cells (Fig. 2A; Supplementary Fig. 1B). In spheroid invasion assays, 344SQ and 344LN cells exhibited greater invasive ability when co-cultured with LF-224 cells than when co-cultured with LF-vec cells (Fig. 2B; Supplementary Fig. 1C). These results suggest that LF-224 cells promote the migration and invasion of lung cancer cells through direct contact, rather than through a paracrine mechanism.
miR-224 promotes the migration and invasion of lung cancer cells in co-culture systems. (A) Transwell migration assay using 344SQ cells co-cultured with LF-vec or LF-224 cells. 344SQ cells were transduced with mCherry (red fluorescence), and LFs were transduced with GFP (green fluorescence). LFs and 344SQ cells were seeded on the upper chambers together, and migrated cells were imaged after 24 h using a fluorescence microscope. The migrated cells were then counted by using ImageJ. Mean ± SD (n = 3). P-values were assessed by two-tailed Student’s t-test. (B) Spheroid invasion assay in 344SQ cells (red) co-cultured with LF-vec or LF-224 (green) cells. Spheroid invasion ratios were measured using ImageJ after 96 h of incubation. Mean ± SD (LF-vec, n = 28; LF-224, n = 39). (C) qRT-PCR of miR-224 in LF-vec cells transiently transfected with miR-224 mimic. Mean ± SD (n = 3). (D) Transwell migration assay in 344SQ cells (red) directly co-cultured with control or LF-224 mimic cells (green). Mean ± SD (n = 3). (E) Spheroid invasion assay of 344SQ cells (red) co-cultured with control or LF-224 mimic cells (green). Mean ± SD (LF-con, n = 35; LF-224 mimic, n = 49). (F) qRT-PCR of miR-224 in human LFs (hLFs) transiently transfected with miR-224 mimic. Mean ± SD (n = 3). (G) Transwell migration assay of A549 cells (red) directly co-cultured with control or hLF-224 mimic cells. Mean ± SD (n = 3). (H) Spheroid invasion assay of A549 cells (red) co-cultured with control or hLF-224 mimic cells (green). ImageJ was used to calculate the invasion ratios after 96 h of incubation. Mean ± SD (hLF-con, n = 38; hLF-224 mimic, n = 38). (I) qRT-PCR of miR-224 in CAFs transiently transfected with miR-224 inhibitor. Mean ± SD (n = 3). (J) Transwell migration assay of 344SQ cells (red) directly co-cultured with control or CAF-224 inhibitor cells (green). Mean ± SD (n = 3). (K) Spheroid invasion assay of 344SQ cells (red) co-cultured with control or CAF-224 inhibitor cells (green). Mean ± SD (CAF-con, n = 54; CAF-224 inhibitor, n = 34).
To further investigate the direct effect of miR-224, LFs were transiently transfected with miR-224 mimic (LF-224 mimic; Fig. 2C). Subsequently, in the transwell migration assay, the migration of 344SQ cells increased significantly when directly co-cultured with LF-224 mimic cells (Fig. 2D). Moreover, spheroid invasion assays showed that LF-224 mimic cells promoted the invasion of 344SQ cells compared to control LFs (Fig. 2E). Subsequently, human LFs isolated from normal lung tissues adjacent to surgically resected lung tumors8 were transiently transfected with miR-224 mimic (Fig. 2F). When co-cultured with A549 human lung cancer cells, human LF-224 mimic cells significantly enhanced the migration and invasion of A549 cells (Fig. 2G, H). Conversely, miR-224 inhibitor was transfected into CAFs (CAF-224 inhibitor) to mitigate the effects of miR-224 (Fig. 2I). When co-cultured with CAF-224 inhibitor, the migration and invasion abilities of 344SQ cells were notably weaker than those cells directly co-cultured with control CAFs (Fig. 2J, K). Consequently, it can be inferred that elevated levels of miR-224 in LFs promote the migration and invasion of lung cancer cells in co-culture systems through direct contact.
miR-224 enhances the interaction between LFs and lung cancer cells, promoting metastasis in a mouse orthotopic model
Then, LFs were initially seeded as feeder cell layers in dishes, and 344SQ cell spheroids were placed on the LF layers after 24 h. In this setup, the expansion and invasion of the 344SQ spheroids into the LF layers were more pronounced in LF-224 cells compared to LF-vec cells (Fig. 3A). For the three-well migration assay, 344SQ, LF-vec, and LF-224 cells were seeded into individual wells of a three-well plate. After 24 h, the silicon wall separating each well was removed, and cell migration was observed under a fluorescence microscope daily for 5 d. The overlapping area of LF-224 and 344SQ cells was more extensive than that of LF-vec and 344SQ cells, indicating a more active interaction between 344SQ and LF-224 cells than that between 344SQ and LF-vec cells (Fig. 3B).
miR-224 enhances the interaction between LFs and lung cancer cells, promoting metastasis in a mouse orthotopic model. (A) Spheroid invasion assay of 344SQ cells on LF-vec or LF-224 feeder cell layers. Spheroids of 344SQ cells (red) were overlaid on the fibroblast layers. Spheroid invasion ratios were measured using ImageJ after 96 h of incubation. Mean ± SD (LF-vec, n = 39; LF-224, n = 37). P-value was assessed by two-tailed Student’s t-test. (B) Three-well migration assay of 344SQ cells (red) with LF-vec or LF-224 (green) cells. The overlapping migration areas were observed under a fluorescence microscope and measured using ImageJ on the indicated days after seeding. P-value was assessed by two-way analysis of variance with Tukey’s multiple comparison test. (C) Three-dimensional co-cultured spheroid invasion assay of 344SQ cells with LF-vec or LF-224 cells. Fibroblast projections were observed under a confocal microscope (left), and the percentage of projections with 344SQ cells was analyzed by using ImageJ. Mean ± SD (LF-vec, n = 19; LF-224, n = 16). (D) Orthotopic injection of 344SQ cells (red) with LF-vec or LF-224 cells in syngeneic mice. Cells were orthotopically injected into the left lungs of mice (LF-vec, n = 8; LF-224, n = 9). Then, metastatic tumor nodules on the right lungs and the mediastinal lymph nodes were counted.
CAFs guide lung cancer cells to form invasive projections from spheroids, a process that enhances cancer invasion19. In the three-dimensional spheroid invasion assay, LF-224 cells facilitated the invasion of 344SQ lung cancer cells, guiding them into distinct invasive projections, unlike LF-vec cells, which extended alone (Fig. 3C). These findings suggest that LF-224 cells promote lung cancer cell invasion and guide their invasion into other sites.
To investigate the effects of miR-224 on tumorigenesis and metastasis, orthotopic injection was performed in 129/Sv syngeneic mice. 344SQ cells (labeled with mCherry) along with LF-vec or LF-224 cells were injected directly into the left lung. After 10 d post-injection, the lungs were extracted and examined using a fluorescence stereomicroscope. The results revealed that LF-224 cells significantly enhanced the metastatic colonization of lung cancer cells on the opposite side of the lungs (Fig. 3D). This indicates that LF-224 cells actively promote the metastasis of lung cancer cells in an in vivo model.
Akirin1 is a direct target gene of miR-224 and attenuates the migration and invasion of lung cancer cells
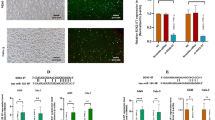
To identify the targets of miR-224, we utilized web-based tools (StarBase and TargetScan), along with RNA sequencing data obtained previously from LFs and CAFs8. Target genes were chosen based on the following criteria: (i) genes showing downregulation in CAFs according to RNA sequencing data of LFs and CAFs and (ii) genes predicted as target genes of miR-224 by StarBase and TargetScan. Subsequently, the expression levels of the predicted targets were assessed in LF-vec and LF-224 cells by qRT-PCR. Among the candidates, Akirin1, Cpne8, and Tial1 were downregulated in LF-224 cells compared to the control (Fig. 4A). Akirin1 and Cpne8 were also downregulated in CAFs compared to LFs (Fig. 4B). Additionally, Akirin1 protein levels were decreased in LF-224 cells (Fig. 4C). Thus, Akirin1 was identified as a promising target of miR-224, since it exhibited reduced expression in LF-224 cells and CAFs compared to LFs.
Akirin1 is a direct target gene of miR-224. (A,B) qRT-PCR of selected candidate target genes of miR-224 in LF-vec, LF-224 (A), LFs, and CAFs (B). Mean ± SD (n = 3). P-values were assessed by two-tailed Student’s t-test. *P < 0.05, **P < 0.01. (C) Western blotting of AKIRIN1 in LF-vec and LF-224 cells. β-Actin was used as a loading control. (D) 3′-UTR luciferase assay. Akirin1 3′-UTR region containing putative miR-224-binding sites was cloned into psiCHECK-2 vector and co-transfected with miR-224 mimic. Mean ± SD (n = 3). (E) Kaplan-Meier analysis showing overall survival of patients with lung adenocarcinoma based on their AKIRIN1 expression in TCGA data. P-value was assessed by log-rank test. (F) qRT-PCR of Akirin1 in LFs transiently transfected with Akirin1 siRNAs (#1, #2, and #3). Mean ± SD (n = 3). **P < 0.01. (G) Transwell migration assay of 344SQ cells (red) directly co-cultured with LFs (green) transfected with negative control or Akirin1 siRNAs (#1 and #3). Mean ± SD (n = 3). (H) Spheroid invasion assay of 344SQ cells (red) co-cultured with LFs (green) transfected with negative control or Akirin1 siRNAs. The invasion ratios were measured using ImageJ after 96 h of incubation. Mean ± SD (LF-con, n = 43; LF-Akirin1 siRNA_#1, n = 35; LF-Akirin1 siRNA_#3, n = 42). (I) qRT-PCR of Akirin1 in LF-224 cells transiently transfected with Akirin1_pcDNA3.1(-) vector. Mean ± SD (n = 3). (J) Western blot of AKIRIN1 in LF-224_control and LF-224_AKIRIN1 cells. β-actin was used as a loading control. (K) Transwell migration assay of 344SQ cells (red) directly co-cultured with LF-224 cells transfected with control and Akirin1 (green). Mean ± SD (n = 3). (L) Spheroid invasion assay of 344SQ-RFP cells (red) co-cultured with LF-224 cells transfected with control and Akirin1 (green). The invasion ratios were calculated using ImageJ after 48–96 h of incubation. Mean ± SD (LF-224_con, n = 51; LF-224_Akirin1, n = 57).
To examine whether Akirin1 is a direct target of miR-224, the 3′-UTR luciferase assay was performed. Co-transfection with miR-224 mimic led to the repression of luciferase activity in the Akirin1 3′-UTR (Fig. 4D). Furthermore, Kaplan-Meier analysis using data from KM plotter (https://kmplot.com/analysis/) revealed that LUAD patients with high AKIRIN1 expression exhibited a longer overall survival time than those with low AKIRIN1 expression (Fig. 4E). These findings suggest that Akirin1 is indeed a direct target of miR-224 and serves as a favorable prognostic marker for patients with LUAD.
Furthermore, Akirin1 knockdown by siRNAs (Fig. 4F) increased the migration and invasion capabilities of 344SQ cells (Fig. 4G, H). When Akirin1 was reintroduced into LF-224 cells (Fig. 4I, J), the migration and invasion of 344SQ cells were reduced in the co-culture system (Fig. 4K, L). Collectively, these findings indicate that Akirin1, as a direct target of miR-224, mediates the ability of LFs to facilitate the migration and invasion of lung cancer cells in a co-culture system.
Discussion
This study identified miR-224 as a driver of the conversion of LFs into CAFs, with higher expression in CAFs. miR-224 overexpression in LFs activated CAF traits and promoted lung cancer cell invasion and migration through direct contact. Mice showed increased cancer cell metastasis following injection of LF-224 cells. Akirin1 was confirmed to be a direct target of miR-224, and its knockdown in LFs increased lung cancer cell invasion and migration in co-culture experiments. Overall, miR-224-induced CAF activation promoted lung cancer cell migration and invasion in co-culture systems.
miR-224 is part of a miRNA cluster with miR-452 and is located within the GABRE gene20. miR-224 expression levels were increased in various tumor tissues, including non-small-cell lung cancer with lymphatic metastasis and pancreatic ductal adenocarcinoma21,22. miR-224 enhances the migration, invasion, and proliferation of lung cancer cells by regulating the tumor suppressors TNFα-induced protein 1 and SMAD415. miR-224 aggravates CAF-mediated effects that induce the proliferation, invasion, and epithelial-mesenchymal transition of lung cancer cells by forming a positive loop of the SIRT3/AMPK/mTOR/HIF-1α axis23. In addition, extracellular vesicles containing miR-224 derived from CAFs promote the migration, invasion, and anti-apoptosis of colorectal cancer cells by targeting SLC4A424. Here, we also demonstrated that CAFs activated by miR-224 promoted the migration and invasion of lung cancer cells when co-cultured directly with these cells.
miR-224 was clearly upregulated in CAFs compared to LFs, although the transcriptional mechanisms driving this upregulation in CAFs remain unclear. In cancer cells, HIF-1α has been shown to upregulate miR-224 expression at the transcriptional level25. In lung cancer cells, KRAS-mutant cell lines exhibit higher miR-224 expression than KRAS-wildtype cells, and active ERK signaling is known to induce miR-224 expression, implying that the RAS-ERK pathway regulates miR-224 transcription15. Additionally, hypomethylation on the miR-224 promoter region has been implicated in its higher expression15. It is possible that similar mechanisms could regulate miR-224 upregulation in CAFs, though this hypothesis requires further investigation.
CAFs directly interact with nearby cancer cells, promoting cancer cell invasion19,26. In squamous cell carcinoma (SCC), CAFs guide SCC cells through tracks in the extracellular matrix. Media or growth factors from CAFs could not enhance SCC cell invasion, emphasizing the need for direct contact between CAFs and SCC cells for invasion27. In colorectal cancer, CAFs induce colorectal cancer cell elongation, motility, and invasiveness via contact-dependent mechanisms. CAFs stimulate colorectal cancer cell migration and invasion via integrin-mediated cell adhesion28. Furthermore, in direct co-culture, CAFs enhance the invasion and proliferation of lung cancer cells by activating the β-catenin signaling pathway29. Additionally, through direct cell-to-cell contact, CAFs induce epithelial-mesenchymal transition and motility in H358 lung cancer cells by upregulating the hedgehog signaling pathway30. Conversely, CAFs promote the proliferation, invasion, and metastasis of lung cancer cells through indirect cell-to-cell interaction23. However, this study suggests that enhanced cancer cell motility by miR-224-activated CAFs primarily depends on direct cell-to-cell interaction rather than indirect interaction. Cancer cell motility was not significantly affected upon exposure to conditioned medium from LF-224 cells or when cultured separately with LF-224 cells as shown in the transwell migration assay. Additional research is required to elucidate the specific mechanisms by which miR-224-activated CAFs facilitate cancer cell motility.
In this study, Akirin1 was identified as a target of miR-224 in CAFs and it plays a direct role in mediating the enhancement of cancer cell migration by CAFs. Akirin1 serves as a co-transcription factor with a robust nuclear localization signal and protein-protein interaction features31. It directly interacts with the FoxO3 transcription factor, aiding its translocation to the nucleus and facilitating MuRF-1 transcription in the skeletal muscle31. Akirin1 overexpression in mouse myoblasts results in increased expression of MyoD, myogenin, and p21 proteins, thereby enhancing myogenic differentiation32. During muscle regeneration, β2-adrenoceptor contributes to regenerating muscle by activating TGF-β receptor I/SMAD2/3 and reducing the expression of Akirin133.
The role of Akirin1 in cancer development remains unclear. However, herein, knockdown of Akirin1 in LFs increased the motility of lung cancer cells in a co-culture system. Conversely, reintroduction of Akirin1 into LF-224 cells reduced the migration and invasion of lung cancer cells. Kaplan-Meier analysis indicated that low expression of AKIRIN1 correlated with poor prognosis in patients with LUAD. Collectively, these findings suggest that Akirin1 inhibits lung cancer progression. Contrastingly, Akirin2, another member of the Akirin family, has been identified as a promoter of migration, invasion, and tumorigenicity in hepatoma and lung adenocarcinoma cells34,35. Although Akirin1 and Akirin2 share a common N-terminal nuclear localization signal, Akirin2 possesses an additional nuclear localization signal36. Furthermore, Akirin2 contains two conserved helical regions that function as intrinsically disordered regions and contribute to protein-protein interactions37. Due to these differences, Akirin1 and Akirin2 may exert opposite effects on cancer cells.
The interaction between CAFs and cancer cells is mediated by direct heterotypic adhesion, involving N-cadherin on the CAF membrane and E-cadherin on the cancer cell membrane38. Cancer cells interact with fibronectin fibrils assembled on CAFs primarily through integrin-α5β1, migrating along the elongated protrusions of CAFs within the collagen matrix39. However, the precise mechanisms by which miR-224 and Akirin1 regulate the direct interaction between CAFs and cancer cells have not been elucidated yet. Further investigation into the roles of cadherins, integrins, and their downstream signaling pathways, such as β-catenin, in the regulation of CAF-cancer cell interactions by the miR-224/Akirin1 axis is needed.
In conclusion, miR-224, which is overexpressed in patients with lung cancer, activates CAFs and enhances direct contact-driven invasion and metastasis. Akirin1, as a target of miR-224, mediates at least part of these effects on cancer cell migration. miR-224 may have multiple targets beyond Akirin1; therefore, the observed promotive effects of miR-224 on cancer cell migration and invasion could be the combined result of the downregulation of several targets, including Akirin1. Low AKIRIN1 levels predicted poor prognosis in patients with LUAD, potentially serving as a prognostic marker. However, the precise mechanism of Akirin1 on lung cancer cell migration and invasion requires further investigation. Further research is crucial to uncover the mechanisms governing the direct interaction between miR-224-activated CAFs and lung cancer cells.
Methods
Cell culture
Normal murine LFs were extracted from the lungs of 129/Sv wild-type mice. CAFs were isolated from the lung tumors of KrasLA1 mice following previously established protocols17. Human LFs were isolated from normal lung tissues adjacent to surgically resected lung tumors. Subsequently, the fibroblasts were cultured in alpha-MEM (Welgene, Gyeongsan, Korea) supplemented with 10% FBS (Corning, Corning, NY, USA), penicillin/streptomycin (100 U/100 µg, Welgene), 2 mM L-glutamine (Welgene), and 1 mM sodium pyruvate (Welgene). LFs and CAFs were stably transfected with the TERT plasmid (pCDH-3xFLAG-TERT; a gift from Steven Artandi; Addgene plasmid #51631) using Lipofector-EXT (AptaBio, Yongin, Korea) for immortalization. The lung cancer cell lines 344SQ, 344LN (murine), and A549 (human) were cultured in RPMI 1640 (Welgene) supplemented with 10% FBS at 37 °C in a humidified atmosphere with 5% CO2. The A549 cell line was purchased from the Korean Cell Line Bank (Seoul, Korea). Fibroblasts were transduced with the pGIPZ control vector (green fluorescence) through lentiviral infection or were stained with the CellTracker Green CMFDA Dye (Invitrogen, Carlsbad, CA, USA) for fluorescent labeling. The 344SQ cells were transduced with the pLMP-mCherry retroviral vector (red fluorescence). To overexpress miR-224, the genomic DNA region including pre-miR-224 was amplified from TC-1 murine embryonic stem cell genomic DNA by PCR. This amplified DNA was then inserted into pLVX-Hyg, which is a modified version of the pLVX-Puro vector (Clontech, Mountain View, CA, USA). miR-224_pLVX-Hyg was transduced into LFs via lentiviral infection. Lentiviruses generated by 293T cells co-transfected with the lentiviral vectors pMD2.G and psPAX2 (gifts from Didier Trono; Addgene plasmids #12259 and #12260) were used to infect the cells for 2–3 d. The infected cells were selected using appropriate antibiotics for 4 weeks. The miR-224 mimic, miR-224 inhibitor, and Akirin1 siRNAs (Bioneer, Daejeon, Korea) were purchased and transiently transfected into fibroblasts through the TransIT-X2 Dynamic Delivery System (Mirus Bio, Madison, WI, USA). Mouse Akirin1 cDNA (pCMV-SPORT6_Akirin1, #mMU000542) was purchased from the Korean Human Gene Bank, Medical Genomics Research Center, KRIBB, Korea, and cloned into the pcDNA3.1(-) vector. For actin cytoskeleton staining, fibroblasts were seeded on collagen-coated (0.2 mg/mL) coverslips and stained with Alexa Fluor 594-conjugated phalloidin (Invitrogen), according to the manufacturer’s instructions. The stained cells were imaged using a fluorescence microscope (Leica DMi8; Wetzlar, Germany). For cell morphology, cell circularity was measured using the ImageJ software (https://imagej.net/ij).
miRNA expression profiling by NanoString
Total RNA from LFs and CAFs was isolated in triplicate using the AccuPrep Universal RNA Extraction Kit (Bioneer). After evaluating RNA quantity and quality, miRNA expression was profiled by PhileKorea (Seoul, Korea) using the nCounter Mouse v1.5 assay kit (NanoString Technologies, Seattle, WA, USA). miRNA probe sets were used to hybridize miRNAs containing oligonucleotide tags. miRNAs were then processed according to the manufacturer’s instructions. miRNA data were gathered, quantified, calculated using the nCounter Digital Analyzer, and analyzed using nSolver software.
Cell growth assay
Fibroblasts (9 × 103 cells per well) were seeded in 12-well plates and counted for 5 d after seeding. The cells were stained with 0.4% trypan blue solution (Sigma-Aldrich, St. Louis, MO, USA) and counted using the LUNA automated cell counter (Logos Biosystems, Anyang, Korea).
Cell migration assay
In the transwell migration assay for indirect co-culture, fibroblasts (5 × 104 cells per well) were seeded in the bottom wells of 24-well plates one day prior to the experiment. 344SQ cells (1 × 105 cells per chamber) were seeded in the upper chambers (Corning) which were coated with 0.1% gelatin in a serum-free medium and allowed to migrate towards the bottom wells containing 2 or 10% FBS. After 24 h, migrated 344SQ cells were stained with 0.1% crystal violet and photographed in four randomly selected microscopic fields per chamber. The cells were counted using the ImageJ Cell Counter plug-in. In the transwell migration assay for direct co-culture, fibroblasts (5 × 104 cells per well) and mCherry-labeled 344SQ cells (1 × 105 cells per well) were seeded in the upper chambers and allowed to migrate toward the bottom wells containing 10% FBS. Other conditions were similar as described previously.
Three-well migration assay
Fibroblasts (1.4 × 104 cells per well) and mCherry-labeled 344SQ cells (3.5 × 104 cells per well) were seeded in each well of Culture-Insert 3 Well in µ-Dish 35 mm (Ibidi, Martinsried, Germany). After appropriate cell attachment for 24 h, the Culture-Insert 3 Well was removed, and the dish was filled with cell culture medium. Migration was observed under a fluorescence microscope daily for 5 d. The overlapping area of fibroblasts and 344SQ cells was measured using the ImageJ software.
Spheroid invasion assay
To generate spheroids, green fluorescent protein (GFP)-labeled fibroblasts (1 × 105 cells) and mCherry-labeled 344SQ cells (5 × 104 cells) in 20% Methocel (Sigma-Aldrich) and 1% Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) were hung on the lids of 150-mm dishes for 2 d. The spheroids, which were mixed with 3 mg/mL type I collagen (Corning), 0.5× phosphate-buffered saline, and 0.01 N NaOH, were implanted in the center of each well of a 12-well plate. After polymerization, the wells were filled with complete cell culture medium. In another method, an agarose micromold was created using a 16 × 16 array in a three-dimensional petri dish (MicroTissues, Providence, RI, USA). To generate spheroids, mCherry-labeled 344SQ cells (3.57 × 104 cells) and GFP-labeled fibroblasts (7.13 × 104 cells) were seeded in the agarose micromold. After 2 d, the spheroids were harvested from the micromold. In addition, 344SQ-alone spheroids were overlaid on a confluent fibroblast layer. The invading spheroid patterns were imaged at the described times using phase contrast and fluorescence microscopy. The invasion ratio was calculated by dividing the total invaded area by the central spheroid area, which was measured using ImageJ. For the invasive projections of spheroids, three-dimensional petri dishes were used to produce spheroids with mCherry-labeled 344SQ cells and GFP-labeled fibroblasts (1:4 to 1:5 ratio, total 42–45 cells). The spheroids were mixed with 3 mg/mL type I collagen (Corning), 0.5× phosphate-buffered saline, and 0.01 N NaOH and were implanted in the center of confocal dishes. After 1 min of incubation, the dishes were turned over to place the spheroids in the middle of collagen. After 3 d, the invasive projections were observed under a confocal microscope.
qRT-PCR
Welprep Total RNA Isolation Reagent (Welgene) was used to isolate total RNA from the cells according to the manufacturer’s protocol. After reverse transcription using the ELPIS RT Prime Kit (Elpis-Biotech, Daejeon, Korea), the mRNA levels were measured using the BioFACT A-Star Real-Time PCR Kit with SFCgreen I (BioFACT, Daejeon, Korea). mRNA levels were normalized to Rpl32 mRNA and calculated using the ΔΔCT method. To quantify miRNA levels, the HB miR Multi Assay Kit (Heimbiotek, Seongnam, Korea) was used according to the manufacturer’s protocol. The miRNA levels were normalized to RNU6B snoRNA levels. The AriaMx real-time PCR system (Agilent Technologies, Santa Clara, CA, USA) was used for real-time PCR analysis. The primer sequences are listed in Supplementary Table 1.
3′-UTR luciferase assay
The Akirin1 3′-untranslated region (UTR, 1592 bp) was amplified by PCR from 344SQ cell genomic DNA and cloned into the psiCHECK-2 vector (Promega, Madison, WI, USA). One day prior to the experiment, 293T cells (1 × 105 cells per well) were seeded in 24-well plates. Cells were then transfected with the Akirin1 3′-UTR reporter (500 ng) and psiCHECK-2-control using Lipofector-EXT in the presence or absence of miR-224 mimic (20 nM; Bioneer). After 2 d, luciferase activity was measured using the Luc-Pair Duo-Luciferase Assay kit (GeneCopoeia, Rockville, MD, USA) on a microplate luminometer INNO-M (LTek, Seongnam, Korea).
Mouse experiment
Prior to the mouse experiment, all proposed mouse studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Ewha Womans University College of Medicine (EWHA MEDIACUC 23-030-t). The mice were cared for and euthanized according to the standards set forth by the IACUC. For the orthotopic injection, mCherry-labeled 344SQ cells and fibroblasts (each 5 × 105 cells in 50 µL of PBS) were directly injected into the left lungs of syngeneic mice (129/Sv, 8–10 weeks). Wildtype 129/Sv mice (129S4/SvJaeJ) were obtained from Jackson Laboratory (Bar Harbor, ME, USA) and bred in the animal facility following the approved protocol (EWHA MEDIACUC past-059-t). Mice were euthanized 10 d post-injection in an acrylic chamber using CO2 gas, with a filling rate of 30–70% per minute. After 5 min of exposure to 100% CO2, the gas flow continued for at least 1 min to confirm veterinary death, indicated by signs like respiratory arrest. The procedure was designed to be humane and effective. Euthanized mice were then necropsied to measure the metastatic tumors on the other side of the lungs. The extracted lungs from mice were observed under a fluorescence stereomicroscope. All the methods in this manuscript were carried out in accordance with the recommendations in the ARRIVE guidelines.
RNA extraction from FFPE tumor tissues
This study was approved by the Institutional Review Board of the Catholic Medical Centre (No. UC17SESI0073) and performed following the guidelines for human research. All methods were conducted in compliance with the guidelines and regulations set forth by the Institutional Review Board. Lung adenocarcinoma tumor specimens (n = 177) were acquired from patients who underwent surgical resection with a therapeutic aim. Informed consent was obtained from all participants. Total RNA was extracted from FFPE tumors using the miRNeasy FFPE Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The miRNA levels were measured using the NanoString nCounter.
Western blot analysis
Cells were washed with cold PBS and lysed using RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) containing protease inhibitors (Sigma-Aldrich). Protein samples were separated by SDS-PAGE and transferred onto PVDF membranes (Bio-Rad, Hercules, CA, USA). The membranes were then incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (Bio-Rad). The WestGlow PICO PLUS ECL chemiluminescent substrate (Biomax, Guri, Korea) was used to detect protein bands. The following antibodies were used: Akirin1 (dilution 1:500, #NBP2-81699, Novus Biologicals, CO, USA) and β-actin (dilution 1:10000, #BS6007M, Bioworld Technology, St. Louis Park, MN, USA).
Statistical analysis
Data were analyzed using Student’s t-test, analysis of variance test, and log-rank test using GraphPad Prism (GraphPad Software Inc., La Jolla, CA, USA). All methods were performed in accordance with the relevant guidelines and regulations.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files. Further inquiries can be directed to the corresponding author.
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249 (2021).
Lemjabbar-Alaoui, H., Hassan, O. U., Yang, Y. W. & Buchanan, P. Lung cancer: Biology and treatment options. Biochim. Biophys. Acta. 1856 (2), 189–210 (2015).
Baghban, R. et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell. Commun. Signal. 18 (1), 59 (2020).
Hanahan, D. & Coussens Lisa, M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell. 21 (3), 309–322 (2012).
Anderson, N. M. & Simon, M. C. The tumor microenvironment. Curr. Biol. 30 (16), R921–R925 (2020).
Wang, L. et al. Cancer-associated fibroblasts enhance metastatic potential of lung cancer cells through IL-6/STAT3 signaling pathway. Oncotarget. 8 (44), 76116–76128 (2017).
Xie, S., Wu, Z., Qi, Y., Wu, B. & Zhu, X. The metastasizing mechanisms of lung cancer: Recent advances and therapeutic challenges. Biomed. Pharmacother. 138, 111450 (2021).
Lee, S. et al. Cancer-associated fibroblasts activated by miR-196a promote the migration and invasion of lung cancer cells. Cancer Lett. 508, 92–103 (2021).
Ahn, Y. H. & Kim, J. S. Long non-coding RNAs as regulators of interactions between cancer-associated fibroblasts and cancer cells in the tumor microenvironment. Int. J. Mol. Sci. 21 (20), 7484 (2020).
Albrengues, J. et al. LIF mediates proinvasive activation of stromal fibroblasts in cancer. Cell. Rep. 7 (5), 1664–1678 (2014).
Wong, K. Y. et al. Cancer-associated fibroblasts in nonsmall cell lung cancer: From molecular mechanisms to clinical implications. Int. J. Cancer. 151 (8), 1195–1215 (2022).
Zhang, Y. et al. HIF-1alpha is necessary for activation and tumour-promotion effect of cancer-associated fibroblasts in lung cancer. J. Cell. Mol. Med. 25 (12), 5457–5469 (2021).
Kunita, A. et al. MicroRNA-21 in cancer-associated fibroblasts supports lung adenocarcinoma progression. Sci. Rep. 8 (1), 8838 (2018).
Liu, Y. et al. MiR-224-5p targeting OCLN promotes the proliferation, migration, and invasion of clear cell renal cell carcinoma cells. Urol. Int. 106 (11), 1185–1194 (2022).
Cui, R. et al. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc. Natl. Acad. Sci. U S A. 112 (31), E4288–E4297 (2015).
Zhou, J. et al. Mir-224-5p-enriched exosomes promote tumorigenesis by directly targeting androgen receptor in non-small cell lung cancer. Mol. Ther. Nucleic Acids. 23, 1217–1228 (2021).
Roybal, J. D. et al. miR-200 inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol. Cancer Res. 9 (1), 25–35 (2011).
Cheon, I., Lee, S., Oh, S. & Ahn, Y. H. Mir-200-mediated inactivation of cancer-associated fibroblasts via targeting of NRP2-VEGFR signaling attenuates lung cancer invasion and metastasis. Mol. Ther. Nucleic Acids. 35 (2), 102194 (2024).
Bota-Rabassedas, N. et al. Contextual cues from cancer cells govern cancer-associated fibroblast heterogeneity. Cell. Rep. 35 (3), 109009 (2021).
Gokhale, A. et al. Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J. Cancer Res. Ther. 6 (4), 521–529 (2010).
Han, H. et al. MiR-224 promotes lymphatic metastasis by targeting ANGPTL1 in non-small-cell lung carcinoma. Cancer Biomark. 34 (3), 431–441 (2022).
Zhu, G., Zhou, L., Liu, H., Shan, Y. & Zhang, X. MicroRNA-224 promotes pancreatic cancer cell proliferation and migration by targeting the TXNIP-mediated HIF1alpha pathway. Cell. Physiol. Biochem. 48 (4), 1735–1746 (2018).
Zhang, J., Han, L., Yu, J., Li, H. & Li, Q. miR-224 aggravates cancer-associated fibroblast-induced progression of non-small cell lung cancer by modulating a positive loop of the SIRT3/AMPK/mTOR/HIF-1α axis. Aging (Albany NY). 13 (7), 10431–10449 (2021).
Zheng, Y. et al. Extracellular vesicles derived from cancer-associated fibroblast carries Mir-224-5p targeting SLC4A4 to promote the proliferation, invasion and migration of colorectal cancer cells. Carcinogenesis 42 (9), 1143–1153 (2021).
He, C., Wang, L., Zhang, J. & Xu, H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol. Cancer. 16 (1), 35 (2017).
Yamaguchi, H. & Sakai, R. Direct interaction between carcinoma cells and cancer associated fibroblasts for the regulation of cancer invasion. Cancers (Basel). 7 (4), 2054–2062 (2015).
Gaggioli, C. et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell. Biol. 9 (12), 1392–1400 (2007).
Knuchel, S., Anderle, P., Werfelli, P., Diamantis, E. & Rüegg, C. Fibroblast surface-associated FGF-2 promotes contact-dependent colorectal cancer cell migration and invasion through FGFR-SRC signaling and integrin αvβ5-mediated adhesion. Oncotarget 6 (16), 14300–14317 (2015).
Otomo, R. et al. TSPAN12 is a critical factor for cancer-fibroblast cell contact-mediated cancer invasion. Proc. Natl. Acad. Sci. U S A. 111 (52), 18691–18696 (2014).
Choe, C. et al. Tumor-stromal interactions with direct cell contacts enhance motility of non-small cell lung cancer cells through the hedgehog signaling pathway. Anticancer Res. 33 (9), 3715–3723 (2013).
Rao, V. V., Sangiah, U., Mary, K. A., Akira, S. & Mohanty, A. Role of Akirin1 in the regulation of skeletal muscle fiber-type switch. J. Cell. Biochem. 120 (7), 11284–11304 (2019).
Marshall, A. et al. Mighty is a novel promyogenic factor in skeletal myogenesis. Exp. Cell. Res. 314 (5), 1013–1029 (2008).
Silva, M. T., Wensing, L. A., Brum, P. C., Camara, N. O. & Miyabara, E. H. Impaired structural and functional regeneration of skeletal muscles from beta2-adrenoceptor knockout mice. Acta Physiol. (Oxf). 211 (4), 617–633 (2014).
Komiya, Y. et al. A novel binding factor of 14-3-3beta functions as a transcriptional repressor and promotes anchorage-independent growth, tumorigenicity, and metastasis. J. Biol. Chem. 283 (27), 18753–18764 (2008).
Komiya, Y., Akiyama, H., Sakumoto, R. & Tashiro, F. FBI1/Akirin2 promotes tumorigenicity and metastasis of Lewis lung carcinoma cells. Biochem. Biophys. Res. Commun. 444 (3), 382–386 (2014).
Goto, A. et al. Akirins are highly conserved nuclear proteins required for NF-kappaB-dependent gene expression in drosophila and mice. Nat. Immunol. 9 (1), 97–104 (2008).
Tartey, S. et al. Akirin2 is critical for inducing inflammatory genes by bridging IkappaB-zeta and the SWI/SNF complex. EMBO J. 33 (20), 2332–2348 (2014).
Labernadie, A. et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell. Biol. 19 (3), 224–237 (2017).
Miyazaki, K. et al. Cancer cell migration on elongate protrusions of fibroblasts in collagen matrix. Sci. Rep. 9 (1), 292 (2019).
Acknowledgements
This work was supported by grants from the National Research Foundation of Korea (NRF), funded by the Korean government (MSIT) (NRF-2020R1A5A2019210 and NRF-2022R1A2C1003150).
Author information
Authors and Affiliations
Contributions
SO, SL, IC, and YHA conceived and designed this study. SO, SL, and IC performed the in vitro and in vivo experiments. SO, SL, IC, and YHA acquired and analyzed the data. SO and YHA drafted and revised the manuscript. All the authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Oh, S., Lee, S., Cheon, I. et al. miR-224 activates cancer-associated fibroblasts to enhance lung cancer cell migration and invasion by targeting Akirin1. Sci Rep 15, 3050 (2025). https://doi.org/10.1038/s41598-024-82189-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-82189-x