Abstract
PANoptosis is closely associated with tumorigenesis and therapeutic response, yet its role in multiple myeloma (MM) remains unclear. This study analyzed bulk transcriptomic and clinical data from the TCGA and GEO databases to identify seven PANoptosis-related genes (PRGs) using machine learning (LASSO regression and random forest models) and univariate Cox analysis, and constructed a prognostic risk model. The model demonstrated robust predictive performance across three external validation cohorts. High-risk patients exhibited higher tumor purity, increased tumor mutational burden, and distinct immune cell infiltration patterns. Drug sensitivity analysis revealed heightened sensitivity to cyclophosphamide, Sinularin, Wee1 inhibitor, osimertinib, JQ1, VE-822, and AZD6738 in high-risk patients. Single-cell transcriptomic analysis revealed significant enrichment of PARP1, ZBP1, LY96, and CASP3 in plasma cells. Quantitative PCR (qPCR) further validated differential expression patterns of the seven core PRGs between MM patients and healthy controls. Immunohistochemical analysis demonstrated distinct expression profiles of PARP1, ZBP1, LY96, and CASP3 in high-risk versus standard-risk MM patients. Furthermore, CCK-8 assays and Wright-Giemsa staining confirmed the crucial role of PARP1 in regulating MM cell viability. This PANoptosis-based prognostic model provides a valuable tool for predicting MM prognosis and guiding personalized treatment.
Similar content being viewed by others
Introduction
Multiple myeloma (MM) is a hematological malignancy caused by the aberrant proliferation of malignant plasma cells, accounting for 10–15% of hematological malignancies. The median age of onset typically ranges from 65 to 75 years. With the intensification of global population aging, its incidence shows an upward trend1. Clinical manifestations such as refractory anemia, pathological fractures, hypercalcemia, renal impairment, and immune dysfunction markedly degrade quality of life and ultimately culminate in the fatality of some patients2. Breakthroughs in therapeutic approaches, such as the development of proteasome inhibitors, immunomodulatory drugs, monoclonal antibodies, and cellular immunotherapies, have substantially improved the prognosis of multiple myeloma. The National Cancer Institute reports that the 5-year overall survival (OS) rate for MM patients increased from 32% (1995–1997) to 58% (2012–2018)3,4. However, prognosis varies significantly depending on clinical staging and cytogenetic risk stratification, while relapse and drug resistance remain formidable challenges5. Therefore, the identification of novel biomarkers is crucial for optimizing clinical management and accurately predicting patient outcomes.
PANoptosis plays a critical role in tumor initiation, progression and metastasis6. Anti-cancer therapies, such as radiotherapy, chemotherapy, and immunotherapy, aim to induce regulated cell death (RCD). Programmed cell death (PCD), a specific form of RCD under strict physiological conditions, encompasses apoptosis, pyroptosis, autophagy, necroptosis, ferroptosis, and other modalities7,8. Apoptosis, pyroptosis, and necroptosis were initially considered independent processes; however, recent studies have revealed extensive crosstalk and cross-regulation among them. In 2019, Dr. Kanneganti’s team proposed “PANoptosis”, an inflammatory form of PCD mediated by the PANoptosome complex. PANoptosis integrates core features of pyroptosis, apoptosis, and necroptosis but cannot be simply classified as any single type of PCD9.
Currently, the implication of PANoptosis in tumors has attracted significant attention, although its precise mechanisms remain incompletely understood. In colorectal cancer, inducing PANoptosis significantly enhances cancer cell sensitivity to oxaliplatin10. In diffuse large B-cell lymphoma, SAMHD1 silencing induces PANoptosis by activating MLKL, Caspase 3, and Gasdermin E, and concurrently enhances the efficacy of PD-L1 inhibitors11. Additionally, TNF-α and IFN-γ synergistically induce PANoptosis in over 10 tumor cell types, with leukemia cells included, resulting in tumor growth suppression12.
Gene expression profiles have been demonstrated to be deeply involved in the pathogenesis of various tumors13,14,15. Investigating the expression patterns of PANoptosis-related genes (PRGs) in multiple myeloma could provide new insights into disease prognosis and treatment; however, no relevant studies have been reported to date. In this study, we comprehensively analyzed the differences in gene expression profiles between multiple myeloma patients and healthy individuals, identified PRGs, and developed a novel risk scoring system to predict prognosis. We further characterized the relationships between the risk score and the tumor microenvironment (TME), tumor mutational burden, and immune checkpoint genes, and predicted the sensitivity of different risk groups to 198 drugs. The reliability of the aforementioned model was validated in three independent cohorts. Moreover, we systematically investigated the expression profiles of core PRGs through single-cell RNA sequencing, quantitative PCR, and immunohistochemical analysis, with functional validation performed using CCK-8 assays and Wright-Giemsa staining. These studies elucidate novel insights into the advancement of precision medicine strategies and the optimization of clinical outcomes in patients with MM.
Materials and methods
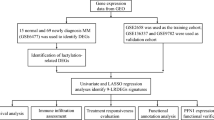
Data collection and processing
Transcriptomic profiles and associated clinical data from MM patients were retrieved from The Cancer Genome Atlas (TCGA) database (GDC MMRF-COMMPASS, https://portal.gdc.cancer.gov/) and the Gene Expression Omnibus (GEO) database (GSE39754, GSE4581, GSE57317, GSE136337; https://www.ncbi.nlm.nih.gov/geo/). Following rigorous data preprocessing and quality control, the samples within these datasets were validated to exhibit high-purity bone marrow plasma cell populations. Comparative analysis of differentially expressed genes (DEGs) between MM patients and healthy donors was conducted utilizing the GSE39754 dataset. The subsequent four datasets, encompassing comprehensive survival information, were employed to elucidate the prognostic significance of PRGs. The MMRF-COMMPASS cohort was designated as the training set for the development of a prognostic risk model, and in parallel, GSE4581, GSE57317, and GSE136337 were utilized as independent validation cohorts to evaluate the robustness and generalizability of the prognostic model. Detailed sample sizes and platform specifications for each dataset are tabulated (Supplementary Table S1). Furthermore, the PRG set was curated by combining genes identified from the GeneCards database (https://www.genecards.org/Search/Keyword?queryString = panoptosis, accessed on November 13, 2024) with a relevance score greater than 0.8, along with PRGs reported in previous studies16,17,18,19.
This integration yielded 114 PRGs for subsequent analyses.
Differential expression analysis and functional enrichment
DEGs between the disease and healthy groups were identified by applying the “limma” package, with a significance threshold of p < 0.05 and |log2 fold change| > 1. Volcano plots and heatmaps were generated through the “ggplot2” and “pheatmap” packages, respectively, to visualize the DEGs. Functional and pathway enrichment analyses were conducted with the “clusterProfiler” R package, including Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis20,21,22. Additionally, Gene Set Enrichment Analysis (GSEA) was employed to delineate the enrichment of different gene sets.
Identification of PANoptosis-related DEGs
The intersection between the previously identified DEGs and the PRG set was visualized with the “VennDiagram” package. Protein-protein interaction (PPI) analysis of the PANoptosis-related DEGs was performed via the STRING database (https://cn.stringdb.org/cgi/input?sessionId=bd3Y1ShAIfn8&input_page_show_search=on), and the results were depicted through Cytoscape software.
Construction of a prognostic risk model
Least Absolute Shrinkage and Selection Operator (LASSO) regression effectively addresses multicollinearity issues and enhances the model’s generalizability, thereby preventing overfitting. To implement this method, the “glmnet” R package was utilized to perform lasso regression and cross-validation for screening key PRGs. Random Forest (RF) is another robust clinical prediction model that improves accuracy and stability by constructing multiple decision trees and aggregating their predictions. For the implementation of RF, the “randomForestSRC” R package was employed with parameters set to ntree = 1000 and an importance threshold of 0.01, to identify key genes. Subsequently, the intersection of key PRGs identified by both machine learning methods was further analyzed through univariate Cox regression to screen core PRGs. The risk score was calculated with the following formula: Riskscore = h0(t)×exp (β1X1+β2X2+…+βnXn), where h0(t) represents the baseline hazard function (a constant), β denotes the regression coefficients, and X corresponds to mRNA expression levels. MM patients were stratified into high-risk and low-risk groups based on the median risk score, and the results displayed in scatter plots. Kaplan-Meier (KM) analysis was performed to assess the impact of the risk score on OS. To further evaluate the prognostic accuracy of the risk model, Time-dependent Receiver Operating Characteristic (ROC) curves and corresponding Area Under Curve (AUC) values were generated using the “survivalROC” R package. In addition, the chromosomal locations of the core PRGs were illustrated through the “RCircos” package.
Analysis of independent prognostic factors and construction of a nomogram
Gender, age, disease stage, and risk score were analyzed by performing univariate Cox regression to identify significant prognostic factors, followed by multivariate Cox regression to determine independent prognostic factors. A nomogram and calibration curves were developed by means of the “rms” and “survival” R packages to assess predictive accuracy.
Tumor microenvironment analysis
The CIBERSORT algorithm was leveraged to profile immune cell infiltration in tumor samples and explore the associations among the risk score, core PRGs, and 22 immune cell subsets. Spearman correlation analysis was further utilized to unravel the link between immune checkpoint gene expression and the risk score.
Tumor mutational burden analysis
Tumor mutation burden (TMB) is a known predictor of immunotherapy response. Somatic mutation data from the MMRF-COMMPASS cohort were retrieved via the GDCquery function in the “TCGAbiolinks” R package. The “maftools” R package was used to generate waterfall plots visualizing tumor mutational profiles across risk groups.
Drug sensitivity prediction analysis
Drug sensitivity in MM patients across risk groups was predicted via the “oncoPredict” R package, with training datasets obtained from https://osf.io/c6tfx/. Differences between high-risk and low-risk groups were compared using the Wilcoxon test, and results were visualized with the “ggplot2” R package.
Single-cell RNA sequencing (scRNA-seq) analysis
The single-cell RNA sequencing dataset (GSE175385) was retrieved from the Tumor Immune Single-cell Hub (TISCH) database, which provides preprocessed datasets filtered through standardized quality control criteria (mitochondrial gene content < 6%, 500–2,500 detected genes per cell). Cells from 15 multiple myeloma samples were extracted for downstream analysis. Data processing was conducted using the Seurat package following this workflow: Raw counts underwent normalization through the NormalizeData method followed by scaling with the ScaleData function. Highly variable genes (HVGs) were computationally identified (the top 2,000 genes) using the FindVariableFeatures algorithm. Cellular clustering was achieved through shared nearest neighbor (SNN) graph construction (FindNeighbors function) followed by community partitioning at a resolution parameter of 0.5 (FindClusters function). The t-distributed stochastic neighbor embedding (t-SNE) algorithm was implemented for non-linear dimensionality reduction. Cluster-specific differentially expressed genes (DEGs) were determined via the FindAllMarkers function. Cellular populations were annotated according to established lineage-defining markers: plasma cells (CD27, PRDM1, CD38), B lymphocytes (CD79A, CD79B), dendritic cells (HLA-DQB2, HLA-DPB1), and macrophages (BIRC3, MAF).
Sample collection and eligibility criteria
All specimens (including paraffin-embedded bone marrow biopsy tissues, bone marrow aspirates, and peripheral blood samples) were obtained from eligible multiple myeloma (MM) patients or healthy volunteers. MM patients were diagnosed according to the International Myeloma Working Group (IMWG) updated diagnostic criteria. Healthy volunteers had no history of hematological disorders or organ failure-related diseases. The age range for both groups was 10–80 years. This study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University (Approval No. RG2023-306-02), and written informed consent was obtained from all participants.
Real-Time quantitative polymerase chain Reaction(qPCR)
Bone marrow aspirates were collected from six newly diagnosed multiple myeloma (MM) patients, and peripheral blood samples were obtained from five healthy volunteers; all samples were anticoagulated with EDTA. Mononuclear cells were isolated using lymphocyte separation medium (Goonie Biotech, Guangzhou, China) via density gradient centrifugation. Total RNA was extracted from cell samples using the RNAsimple Total RNA Kit (Tiangen Biotech, Beijing, China; Cat. #4992858) according to the manufacturer’s protocol. RNA concentration and purity were measured with a NanoDrop One spectrophotometer (Gene Company Limited, Shanghai, China). Reverse transcription was performed with the Reverse Transcription Kit (Lanjeke Biotech, Beijing, China; Cat. #BL696A) on a GeneAmp 9700 PCR System (Applied Biosystems, Foster City, CA, USA). cDNA was then amplified by qPCR on a LightCycler 480 System (Roche Diagnostics, Indianapolis, IN, USA) with Universal SYBR qPCR Master Mix (Lanjeke Biotech, Beijing, China; Cat. #BL697A). GAPDH served as the housekeeping gene for normalization, and gene expression levels were calculated using the 2−ΔCT method(△CT = CT target gene – CTGAPDH). Primer sequences are provided in Supplementary Table S2.
Immunohistochemical staining (IHC)
Paraffin-embedded bone marrow biopsy tissues from 4 high-risk and 5 standard-risk MM patients (stratified according to mSMART 3.0 criteria) were sectioned to a thickness of 4 μm using a microtome. After deparaffinization and rehydration through graded alcohols, antigen retrieval was performed by heating sections in Tris-EDTA buffer (pH 9.0) to boiling for 10 min, followed by gradual cooling to room temperature. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide at room temperature for 15 min. Non-specific binding sites were blocked by incubating sections with 5% bovine serum albumin (BSA) in a humidified chamber at 37 °C for 1 h. Following overnight incubation with primary antibodies at 4 °C, species-matched secondary antibodies were applied at 37 °C for 30 min. Chromogenic development was carried out using a diaminobenzidine (DAB) peroxidase substrate kit(Zhong Shan Golden Bridge Biotech, Beijing, China; Cat. #ZLI-9018), with the reaction duration optimized through microscopic monitoring. Finally, sections were counterstained with hematoxylin and mounted for scanning. Whole-slide scanning was performed to acquire high-resolution digital images by Pannoramic MIDI II Digital Scanner (3DHISTECH Ltd., Budapest, Hungary), and quantitative immunohistochemical analysis was conducted using ImageJ software. The following primary antibodies were used for immunohistochemistry: LY96(Cloud-Clone Corp., USA; Cat. #PAH705Hu01), PARP1(Cloud-Clone Corp., USA; Cat. #PAA279Mu01), ZBP1 (Cloud-Clone Corp., USA; Cat. #PAB552Hu01), CASP3 (Cloud-Clone Corp., USA; Cat. #PAA626Ra01).
Cell viability assessment by CCK-8 assay
Cells in the logarithmic growth phase were harvested and seeded in 96-well plates at a density of 3 × 10⁴ cells per well. A concentration gradient of the PARP1 inhibitor Talazoparib (Ambeed, Illinois, USA; Cat. #1207456-01-6) was added to respective wells, followed by continued incubation in a cell culture incubator. After 72 h, 10 µL of CCK-8 solution (Goonie Biotech, Guangzhou, China) was added to each well and incubated at 37 °C for 4 h. Absorbance was subsequently measured at 450 nm using an EON microplate reader (BioTek Instruments, Winooski, Vermont, USA).
Wright-Giemsa staining
Centrifuged cell pellets were resuspended in PBS to achieve a density of 1 × 10⁶ cells/mL. Using a clean glass slide, 100 µL of cell suspension was spread at a 45° angle to create monolayer smears, which were air-dried for 10–15 min. The dried smears were stained with modified Wright-Giemsa for 8 min at room temperature, then rinsed at a 45° angle under running water for 30 s. After complete air-drying for 2 h, slides were scanned at 40× magnification using the Pannoramic MIDI II Digital Scanner (3DHISTECH Ltd., Budapest, Hungary).
Statistical analysis
Statistical analyses were performed with R version 4.4.1 and its associated packages. Intergroup differences were assessed by employing non-parametric tests, and correlations were evaluated using Spearman’s method. A p-value < 0.05 was considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Results
Identification of differential PRGs and functional enrichment analysis in patients with MM
RNA sequencing data from 170 newly diagnosed patients and 6 healthy donor bone marrow samples in the GEO dataset GSE39754 were analyzed, revealing 2,821 DEGs (Fig. 1A), including 1,588 upregulated and 1,263 downregulated genes. To explore the biological functions and pathways associated with these DEGs, functional and pathway enrichment analysis was performed, revealing significant associations with innate and/or adaptive immune system regulation, cell cycle, cell death, endoplasmic reticulum stress, cytokine secretion, cytoplasmic translation, and cell adhesion (Fig. 1B–C). Among 114 PRGs, 33 were differentially expressed (Fig. 1D), of which 17 were upregulated and 16 were downregulated (Fig. 1E). A PPI network highlighted strong interactions among these genes’ encoded proteins (Fig. 1F–G).
Identification of differential PRGs in MM. A Heatmap and volcano plot (Red: upregulated genes in the MM group; Blue: downregulated genes; Gray: genes with no significant change; gene names with |logFC| ≥ 3 and P < 0.05 are labeled in the figure) of DEGs between MM and healthy bone marrow samples.) B Enriched GO terms of DEGs. BP (Biological Process), CC (Cellular Component), MF (Molecular Function); y-axis: enriched pathway names; x-axis: gene counts; colour gradient: P value. C Enriched KEGG pathways of DEGs20,21,22. y-axis: enriched pathway names; x-axis: GeneRatio (proportion of enriched genes per pathway); dot size: gene counts. D Venn diagram of 114 PRGs and 2,821 DEGs. E Thirty-three differentially expressed PRGs. F PPI network (interaction confidence > 0.7). Nodes represent proteins; edges indicate protein-protein interactions. Darker-colored central nodes exhibit stronger connectivity. G Bar chart of node numbers for these genes. Protein names (y-axis); number of interacting partners (x-axis).
Establishment of a risk prognostic model based on PRGs
A PANoptosis-related prognostic model was developed using the MMRF training cohort. Initially, the aforementioned 33 PANoptosis-related differentially expressed genes were subjected to LASSO regression, leading to the identification of 10 key genes (Fig. 2A). To ensure the robustness of the model, the same 33 genes were further incorporated into a RF model to construct decision trees, and the importance of each gene is displayed in Fig. 2B. Following this, genes exhibiting an importance score exceeding 0.01 in the RF analysis were cross-referenced with the foregoing genes identified with the LASSO regression approach. The intersection of both analytical approaches identified eight key genes, followed by univariate Cox regression analysis, which revealed seven core genes: PARP1, ZBP1, CASP3, CDK1, FAS, LY96, and TNFSF10 (Fig. 2C). Their chromosomal locations are shown in Fig. 2D. The Cox proportional hazards model was initially fitted with the coxph function, followed by stepwise regression to optimize the model. Risk scores were calculated based on the optimal model with the predict function. MM patients were stratified into two groups according to the median risk score, and their survival status was visualized in a scatter plot (Fig. 2E). As anticipated, Kaplan-Meier survival analysis demonstrated significantly shorter overall survival in the high-risk group (Fig. 2F). ROC curve analysis yielded AUC values of 0.712, 0.705, and 0.689 for 1-year, 3-year, and 5-year overall survival, respectively (Fig. 2G). To comprehensively investigate the relationship between the risk score and biological functions in MM patients, we performed GSEA. As illustrated in Fig. 2H, pathways such as the cell cycle, DNA replication, p53 signaling pathway, and alanine, aspartate, and glutamate metabolism were significantly upregulated in the high-risk group. Conversely, pathways including cell adhesion molecules, cytokine-cytokine receptor interaction, neutrophil extracellular trap formation, protein processing in the endoplasmic reticulum, and Toll-like receptor signaling pathway were markedly downregulated (Fig. 2I). Expectedly, the GSEA results indicate that pathways closely linked to cancer progression are highly active in the high-risk group, whereas those involved in anti-tumor immunity and tumor metastasis suppression are notably dysfunctional.
Development of a prognostic model based on PRGs in the training cohort. A Variable selection using LASSO regression analysis (x-axis: log(λ); left y-axis: model error rate; right y-axis: coefficients).B Variable screening using the random forest model. C Forest plot of univariate Cox analysis results. D Chromosomal locations of the seven core PRGs. Autosomes (1–22) and sex chromosomes (XY) are labeled; flanking lines denote precise gene loci on chromosomes. E Distribution of risk scores and survival status between risk groups. F KM survival analysis. G ROC curves at 1, 3, and 5 years of the prognostic index. H and I GSEA analysis results showing pathways upregulated and downregulated in the high-risk group compared to the low-risk group, respectively. Only pathways with a p-value < 0.05 and |NES| > 1 are considered significant. Each panel highlights seven major pathways.
Risk scores as independent risk factors
To evaluate the prognostic value of the risk model, clinical features, including sex, age, and disease stage, were integrated. As shown in the results, univariate Cox analysis indicated that both the risk score and disease stage were significantly associated with OS (Fig. 3A). Multivariate Cox analysis further confirmed that these factors independently served as prognostic indicators, with hazard ratios consistently exceeding 1 (Fig. 3B). Afterwards, the relationship between disease stage and risk score was illustrated through a boxplot, revealing that stage III patients exhibited higher risk scores (Fig. 3C). To facilitate clinical application, a nomogram was developed to predict the survival time of patients with multiple myeloma (Fig. 3D). As depicted in Fig. 3E, F, and G, the calibration curves for 1-year, 2-year, and 3-year survival probabilities suggested that the model could accurately predict the survival time of MM individuals.
Cox regression and nomogram analysis of the prognostic model. A and B Univariate and multivariate Cox regression analyses to identify independent prognostic factors for OS. C Relationship between disease stage and risk score. D Nomogram for forecasting the mortality probability, incorporating clinical features and risk score. Clinical variables (Risk score, Age, Gender, Stage); scale values represent variable ranges. Points: individual variable scores; Total Points: aggregated score. 1/2/3-year Death indicates 1/2/3-year mortality risk. E-G Calibration curves predicted 1-, 2-, and 3-year OS. x-axis: predicted probability; y-axis: observed probability; dashed line: ideal reference (slope = 1).
Evaluation of prognostic risk models: external validation
The prognostic model was externally validated across three independent validation cohorts. The risk scores and survival status of the cohorts were illustrated in Fig. 4A, B, and C. KM curves revealed significantly higher mortality risk in the high-risk group compared to the low-risk group across all cohorts (Fig. 4D, E, and F). The AUC further verified the model’s generalizability. In the GSE4581 cohort, the 1-year, 3-year, and 5-year OS AUC values were 0.661, 0.695, and 0.660, respectively (Fig. 4G). Similarly, in the GSE57317 cohort, the 1-year, 2-year, and 3-year OS AUC values consistently exceeded 0.7, with the 3-year OS reaching 0.807 (Fig. 4H). In the GSE136337 cohort, the AUC values for 1-year, 3-year, and 5-year OS were 0.602, 0.667, and 0.634, respectively (Fig. 4I). These findings collectively underscore the robustness and predictive accuracy of the proposed model.
Assessment of TME in MM patients based on risk scores
Considering the pivotal influence of the TME on tumorigenesis, progression, and treatment response, the ESTIMATE algorithm was utilized to assess immune cell infiltration abundance across different risk groups. Significantly lower stromal and ESTIMATE scores were detected in the high-risk group compared to the low-risk group, indicating higher tumor purity (Fig. 5A). Further analysis via CIBERSORT revealed distinct infiltration patterns of 22 immune cell types between the risk groups. Notably, significant differences were identified in the abundance of B memory cells, plasma cells, CD4 + memory resting T cells, and CD4 + naive T cells (P < 0.0001; Fig. 5B and C). Specifically, the high-risk group exhibited elevated infiltration levels of five immune cell types, including B memory cells, eosinophils, M0 macrophages, monocytes, and activated mast cells (Fig. 5B). Additionally, the potential interactions among these 22 immune cell types were investigated. A significant negative correlation was observed between B memory cells and plasma cells (P < 0.001). Monocytes demonstrated positive correlations with eosinophils, activated mast cells, neutrophils, and CD8 + T cells, while showing negative correlations with plasma cells and gamma delta T cells (P < 0.001; Fig. 5D). Further analysis revealed that eight immune cell subtypes showed positive correlations with risk score (P < 0.05), especially B memory cells and CD4 + memory resting T cells (P < 0.001). Conversely, four immune cell types demonstrated negative correlations with risk score (P < 0.05), particularly plasma cells, CD4 + naive T cells, and gamma delta T cells (P < 0.001). Beyond this, the seven core genes used to construct the prognostic model displayed strong associations with immune cell infiltration levels, especially FAS and ZBP1 (Fig. 5E). Collectively, the PANoptosis risk prognostic model emerges as a promising tool in terms of clinical usage.
Analysis of the immune landscape based on risk score. A Assessment of the relationship between risk score and TME through the ESTIMATE algorithm. B Box plots of immune cell abundance between risk groups (CIBERSORT algorithm). x-axis: 22 immune cell types; y-axis: relative abundance (%); box colors: red (high-risk group), blue (low-risk group). C Stacked bar chart depicting the abundance of immune cell infiltration across individual samples. x-axis: individual MM samples; y-axis: immune cell infiltration abundance; stacked segments: proportions of immune cell subtypes. D Heatmap depicting the interrelationships among immune cell types. E Heatmap highlighting the relation of core PRGs to immune cells.
Assessment of TMB and immune checkpoint gene expression based on risk scores
During tumor progression, genetic mutations produce immunogenic neoantigens that evade self-tolerance mechanisms, thereby triggering the immune system to attack tumor cells23. Given this mechanism, exploring potential genetic abnormalities in distinct risk groups could facilitate the identification of patients with a higher probability of benefiting from immunotherapy or targeted therapy. To investigate this, we performed next-generation sequencing analysis on high-risk and low-risk MM patients. Our findings identified IGHV2-70 and IGLV3-1 as the most frequently mutated genes in this cohort, with overall mutation frequencies of 38.5% and 32.5%, respectively. Additionally, KRAS, NRAS, IGHV2-70D, and IGLL5 demonstrated mutation frequencies exceeding 10% in both groups. Among these mutations, missense mutations were predominant, followed by multi-hit mutations (Fig. 6A and B). Further analysis showed that the high-risk group exhibited a higher TMB than the low-risk group (p < 0.0001; Fig. 6C). Moreover, Spearman correlation analysis further indicated a significant positive association between risk score and TMB (p < 0.0001; Fig. 6D). Consistent with this, KM survival analysis demonstrated that patients with elevated TMB exhibited significantly worse prognoses compared to those with lower TMB (Fig. 6E).
Mutation and immune checkpoint profiles in risk groups. A-B Oncoplots of top 20 mutated genes in high- and low-risk groups. C Tumor mutation burden comparison. D Spearman correlation analysis between risk score and TMB. E KM survival analysis of TMB. F Differential expression of immune checkpoint genes (**** p < 0.0001, *** p < 0.001, ** p < 0.01, * p < 0.05). x-axis: immune checkpoint genes; y-axis: expression levels; box colors: red (high-risk group), blue (low-risk group). G Spearman correlation analysis: risk scores and immune checkpoint genes.
Notably, immunotherapy holds potential therapeutic value for the high-risk population with higher TMB, presumably improving their OS. Furthermore, Immune checkpoints play a dual role in tumor immunity, either enhancing immune-mediated cytotoxicity or driving immunosuppressive processes. Although the primary focus of research has been on inhibitory immune checkpoints, with several inhibitors already approved and exhibiting notable therapeutic benefits, the activation of stimulatory immune checkpoints offers a promising avenue to amplify anti-tumor immunity24. Moreover, the expression levels of immune checkpoint genes could function as predictive biomarkers for tumor immune responses. To explore this, we analyzed their expression across risk groups, identifying significant differences in 36 out of 51 genes (p < 0.05; Fig. 6F). Of note, TNFRSF14, TNFRSF4, CD274 (PD-1), PDCD1LG2 (PD-L2), TIGIT, CEACAM1, and LGALS9 were significantly overexpressed in the low-risk group (p < 0.0001). Conversely, SIRPA, PVR, CD28, and CD86 exhibited markedly higher expression levels in the high-risk group (p < 0.001). Subsequently, we investigated the correlation between immune checkpoint gene expression and risk scores. Of the 18 genes positively associated with risk scores, 10 exhibited significant positive correlations: CD28, CD70, CD86, CD96, CTLA4, KIR3DL2, PVR, TNFSF18, TNFSF4, and TNFSF9 (p < 0.001; Fig. 6G).
Prediction of drug sensitivity based on risk scores in MM patients
OncoPredict is an R package designed for predicting drug responses. With this tool, we investigated the sensitivity of MM patients in different risk groups to 198 commonly used chemotherapy and molecular-targeted medicines, aiming to identify potential drugs suitable for high-risk individuals. Our analysis revealed that the IC50 values of 120 drugs significantly differed between the two risk cohorts (Supplementary Table S3). However, proteasome inhibitors, which are frequently used in first-line induction therapy, exhibited no significant difference in sensitivity between risk groups. In contrast, cyclophosphamide, another commonly used agent in first-line induction therapy, demonstrated a remarkably decreased IC50 in the high-risk group (p < 0.05; Supplementary Table S3). Wee1, a serine/threonine kinase, regulates the G2/M checkpoint by phosphorylating CDK1, inducing G2-phase cell cycle arrest following DNA damage, thereby preserving genomic stability25. The ataxia telangiectasia and Rad3-related protein serine/threonine kinase (ATR) is primarily activated by sensing extensive single-strand DNA breaks (ssDNA) generated at stalled replication forks or other DNA damage sources. This ultimately induces cell cycle arrest and stabilizes stalled replication forks to enable DNA repair26.The integration of Wee1 inhibitors and ATR inhibitor VE-822 synergistically suppresses AML cell proliferation, induces DNA damage, and promotes apoptosis27. High-risk patients exhibited significantly lower IC50 values for both agents than low-risk patients (Fig. 7A and B), implying potential therapeutic advantages for this subgroup. AZD6738, another ATR inhibitor, has recently been shown to exhibit therapeutic efficacy in various solid tumors, including gastrointestinal cancers (such as gastric, pancreatic, and hepatic cancers) as well as breast and ovarian cancers28,29,30,31,32. In contrast to VE-822, AZD6738 exhibited a significantly lower IC50 value in high-risk patients (Fig. 7C), suggesting its potential as a more promising therapeutic agent. Sinularin, a biologically active lead compound derived from soft corals, induces tumor cell apoptosis in prostate cancer and glioblastoma through distinct mechanisms: by stabilizing FOXO3 protein or triggering oxidative stress, respectively33,34,35. Consistent with previous findings, our results confirmed that MM patients, particularly those in the high-risk group, exhibited heightened sensitivity to Sinularin (p < 0.0001; Fig. 7D). Additionally, JQ1, a bromodomain and extraterminal domain inhibitor, promotes apoptosis and reduces regulatory T cells in MM, reshaping the tumor microenvironment to increase the efficacy of immunotherapy36. Our analysis also supports the therapeutic potential of JQ1 in MM patients (Fig. 7E). Osimertinib, a third-generation covalent EGFR inhibitor, has been shown to induce non-EGFR-dependent apoptosis in CD34 + myeloid leukemia stem and progenitor cells37. Notably, our findings suggest its efficacy in MM, especially for high-risk patients (Fig. 7F). These results provide valuable insights into precision therapy with small-molecule compounds for risk-stratified MM patients in the PANoptosis-based prognostic model.
Expression and validation of core PRGs in multiple myeloma
To investigate the expression patterns of seven core PRGs across distinct cellular subpopulations in the bone marrow microenvironment of multiple myeloma, we processed 4,552 high-quality cells isolated from 15 multiple myeloma samples within the preprocessed TISCH dataset (GSE175385) (Supplementary Figure S1A). Using the Seurat analytical pipeline, we normalized raw counts and identified 10 cellular clusters through t-SNE nonlinear dimensionality reduction (dims = 1:20) (Supplementary Figure S1B). Based on differentially expressed marker genes determined by FindAllMarkers algorithm, ten clusters were successfully annotated as four major immune cell types: plasma cells, B lymphocytes, dendritic cells, and macrophages (Fig. 8A). Subsequently, we investigated the distribution and expression levels of seven core PRGs. The results revealed significant enrichment of PARP1, ZBP1, LY96, and CASP3 expression in plasma cells (Fig. 8B and Supplementary Figure S1C), suggesting that PANoptosis-related signaling may predominantly originate from malignant plasma cells.
Expression patterns and functional validation of core PRGs in MM. A t-SNE plot showing 10 cellular clusters identified from 4,552 high-quality bone marrow cells (15 MM samples, GSE175385), annotated as four major immune populations based on marker gene expression. Every point on the graph represents a single cell. B t-SNE plot showing expression patterns of seven core PRGs from GSE175385. Color gradient indicates expression levels. C qPCR analysis of seven core PRGs was performed in MM samples (n = 6) and healthy controls (n = 5). Expression levels were normalized to GAPDH. Statistical significance was determined using the Mann-Whitney U test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant). D Representative histopathology (H&E) and immunohistochemical staining in high-risk (n = 4) versus standard-risk (n = 5) patients. Scale bar:20 μm. E Comparative expression analysis of four core PRGs between risk groups (Mann-Whitney U test; *p < 0.05). F Cell viability of MM.1 S cells treated with Talazoparib (0–50µM, 72 h) by CCK-8 assay. ****p < 0.0001 vs. control (Mann-Whitney U test). G Morphological analysis of MM.1 S cells with or without Talazoparib treatment (12.5 µM, 72 h) by Wright-Giemsa staining. Scale bar: 20 μm.
Furthermore, qPCR was performed to compare the expression levels of the seven core PRGs in multiple myeloma patients versus healthy controls. The results demonstrated that PARP1, CASP3, and CDK1 exhibited significantly upregulated expression in the tumor cohort (p < 0.05), whereas FAS and TNFSF10 showed predominant expression in the healthy control group (p < 0.05). No statistically significant differences were observed in ZBP1 and LY96 expression profiles between the two groups (Fig. 8C).
Moreover, to validate the prognostic relevance of four plasma cell-enriched core genes (PARP1, ZBP1, LY96, CASP3) identified by single-cell RNA sequencing, we performed IHC analysis on bone marrow biopsies from mSMART 3.0-classified MM patients (4 high-risk vs. 5 standard-risk). Figure 8D displays representative H&E staining and immunohistochemical staining results from both the high-risk and standard-risk groups. Semi-quantitative analysis of protein expression levels for four core PRGs was performed using ImageJ software, with expression levels quantified as DAB-positive area percentage (%-area fraction). The results demonstrated that PARP1 expression was significantly elevated in the high-risk group compared to the standard-risk group (p < 0.05), whereas LY96 and ZBP1 showed higher expression in the standard-risk group (p < 0.05). The median expression level of CASP3 was higher in the high-risk group (5.635%) than in the standard-risk group (3.998%) (Fig. 8E), despite the lack of statistical significance. Our PANoptosis-related prognostic model established PARP1 and CASP3 as risk genes, and LY96 and ZBP1 as protective genes. The distinct immunohistochemical expression patterns of these four genes between high-risk and standard-risk groups provided experimental validation of the model’s prognostic value.
To further investigate the biological role of PARP1 in multiple myeloma, we assessed the effect of the PARP1 inhibitor Talazoparib on the proliferation of MM.1 S using the CCK-8 assay. Remarkably, even at the lowest tested concentration (3.125 µM), Talazoparib treatment for 72 h significantly suppressed the proliferative activity of MM.1 S cells (p < 0.0001; Fig. 8F). Additionally, Wright-Giemsa staining was performed on MM.1 S cells. The results showed that Talazoparib treatment induced nuclear irregularities, enlargement of both nuclei and overall cell volume, along with loss of plasma membrane integrity(Fig. 8G). These findings establish PARP1 as both a critical prognostic biomarker and a promising therapeutic target in multiple myeloma, warranting further clinical investigation.
Discussion
The survival outcomes of MM patients exhibit considerable heterogeneity, ranging from several months to decades, with the majority eventually developing relapsed or refractory disease38. Building upon previous studies that have noted the significance of PANoptosis in hematologic malignancies, including acute leukemia and lymphoma11,12,39, we hypothesized that this programmed cell death pathway might be pivotal in MM progression. To test this hypothesis, we employed bioinformatics to investigate the expression patterns of PRGs in MM, identifying genes with distinct expression within this pathway. Using machine learning algorithms (RF and LASSO) along with univariate Cox regression, we pinpointed seven core genes which formed the basis for constructing a prognostic risk model. To assess the model’s clinical relevance, we performed KM survival analysis and AUC evaluations. Further assessment via multivariate Cox regression analysis established the risk score as an independent prognostic factor. For the purpose of enhancing the model’s utility, we constructed a nomogram and calibration curve, both of which confirmed its predictive power. Additionally, the model exhibited strong prognostic accuracy across three independent validation cohorts. We further analyzed the expression patterns of core PRGs using single-cell RNA sequencing, qPCR, and immunohistochemistry, with functional validation via CCK-8 assays and Wright-Giemsa staining.
ZBP1, a nucleic acid sensor, is essential in regulating immune responses and cell death. It interacts with proteins like RIPK3, NLRP3, and caspases-1/8/6 to form the ZBP1-PANoptosome complex, which orchestrates PANoptosis to eliminate infected or cancerous cells40,41. Our Cox regression analysis identified ZBP1 as an independent protective factor in MM patients. scRNA-seq analysis further demonstrated significantly higher ZBP1 expression in plasma cells. According to the mSMART 3.0 classification criteria, we compared ZBP1 protein expression levels between high-risk and standard-risk groups of MM patients using immunohistochemistry, which confirmed significantly lower ZBP1 expression in the high-risk group compared to the standard-risk group. Therefore, targeting ZBP1 could potentially disrupt the pro-survival pathways in MM, offering a novel therapeutic strategy.
TNFSF10, also known as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), is one of the most extensively studied members of the TNF superfamily. TNFSF10 induces apoptosis through binding to its death receptors (DcR1, DcR2, and OPG), whereas its interaction with decoy receptors (DR4 and DR5) suppresses apoptotic signaling42. Importantly, silencing the TNFSF10 gene or DR5 enhances tumor susceptibility in mice and promotes the emergence of more aggressive cancer cells, thereby enhancing metastasis43,44,45,46. Consistent with these findings, Cox regression analysis demonstrated that TNFSF10 was associated with better overall survival. qPCR validation confirmed significantly lower TNFSF10 mRNA levels in MM patients compared with healthy controls(p<0.01). Meanwhile, the research on TNFSF10 receptor agonists is progressing, with key efforts focused on optimizing pharmacokinetic properties, enhancing efficacy, and exploring combination therapies to combat drug resistance. Aponermin, a TNFSF10 peptide mimetic, has demonstrated its effectiveness when paired with thalidomide and dexamethasone in treating multiple myeloma, culminating in its approval for clinical use in China in 202347.
FAS, also known as tumor necrosis factor receptor superfamily member 6 (TNFRSF6), harbors an essential death domain. Binding of its ligand initiates the formation of a death-inducing signaling complex that activates executioner caspases, triggering apoptosis. During tumor progression, FAS is frequently downregulated to evade apoptosis, a mechanism that supports its classification as a tumor suppressor48. Quantitative PCR analysis revealed significantly decreased FAS expression in MM patients compared with healthy donors(p<0.01). In contrast, Cox regression established FAS as a poor prognostic factor (HR 1.253, 95% CI 1.106–1.419, p < 0.001). This paradox has been explored in several studies. Marcus E. Peter et al. demonstrated that FAS promotes tumor growth through the JNK and Jun pathways in mouse models of ovarian and liver cancers49. Their team further showed that FAS facilitates the transformation of non-cancer stem cells into cancer stem cells, where it supports the preservation of their stemness50. In a similar vein, M. Teodorczyk et al. confirmed that FAS is involved in preserving cancer stem cell properties in metastatic pancreatic ductal adenocarcinoma (PDAC) and primary human glioblastoma51,52.
LY96, also known as MD-2, is a lymphocyte antigen expressed predominantly in B cells, macrophages, and dendritic cells. It facilitates the binding of lipopolysaccharide (LPS) to Toll-like receptor 4 (TLR4), initiating innate immune responses53,54. However, Rajamanickam’s research has shown that LY96-driven inflammatory microenvironments can foster tumor progression55. In prostate cancer tissues, for instance, elevated LY96 expression has been linked to increased invasiveness, metastasis, and a poor prognosis56. Yet, other studies have presented contradictory findings. In head and neck cancer patients treated with OK-432, for example, IFN-γ expression was positively correlated with both TLR4 and LY96 gene expression57. Building on these insights, our findings indicate that LY96 functions as a protective factor, potentially improving immunotherapy efficacy in MM. IHC analysis confirmed significantly lower LY96 protein levels in high-risk versus standard-risk patients (p < 0.05), supporting its protective role.
PARP1, the most abundant member of the poly (ADP-ribose) polymerase family, is involved in DNA repair, apoptosis, and cell proliferation58,59. It is frequently overexpressed in various malignancies including acute and chronic leukemia, lymphoma, and neuroblastoma60,61,62,63. In MM, elevated PARP1 expression has been associated with lower OS in patients undergoing autologous hematopoietic stem cell transplantation64. Corroborating these results, PARP1 mRNA levels were significantly elevated in MM patients, with Cox regression analysis revealing a strong association with adverse prognosis (HR 1.715, 95%CI 1.328–2.214, p < 0.001). Notably, immunohistochemical analysis demonstrated significantly higher PARP1 protein expression in high-risk compared with standard-risk patients (p < 0.05). PARP1 inhibitors have shown potential to enhance the efficacy of radiotherapy and chemotherapy, and ongoing clinical trials in hematologic malignancies are yielding promising results65,66,67,68. Notably, a recent study has demonstrated that the PARP1 inhibitor niraparib synergizes with bortezomib to exert antimyeloma effects by inhibiting DNA damage repair69. CCK-8 assays in MM.1 S cells combined with Wright-Giemsa staining results demonstrated that the PARP1 inhibitor Talazoparib significantly suppressed cell viability (p < 0.0001). These findings collectively validate PARP1 as a potential therapeutic target in multiple myeloma.
The cyclin-dependent kinase (CDK) family includes 20 kinases, with CDK1 being critically involved in cell cycle control and significantly associated with the oncogenesis of diverse cancers70,71,72,73. Dong Wu et al. identified CDK1 as a poor prognostic factor in MM74. Indeed, Cox regression analysis demonstrated that elevated CDK1 expression was significantly associated with poorer overall survival (HR 1.770, 95%CI 1.521–2.061, p < 0.001). Consistent with these findings, qPCR analysis revealed markedly higher CDK1 mRNA levels in multiple myeloma patients.
Apoptosis is a vital mechanism by which cytotoxic drugs eradicate cancer cells, with CASP3 serving as a central executioner protein75,76,77. Beyond promoting apoptosis, CASP3 can also activate GSDME, leading to pyroptosis and offering a novel strategy for cancer treatment78. However, CASP3 activation may influence neighboring cells, promoting the proliferation of drug-resistant cancer cells and tumor stem cells, thereby reducing therapeutic efficacy79,80,81,82. For instance, Qian Huang et al. noted that apoptotic tumor cells following radiotherapy enable surviving cells to reform tumors83. Expanding on this point, CASP3 is involved in the migration, invasion, and vasculogenic mimicry of certain cancers, such as ovarian cancer, liver cancer, and melanoma84,85,86,87. Clinically, Qian Huang et al. found that high levels of CASP3 activation in pretreatment tumor biopsy samples correlate with higher recurrence rates and shorter survival in patients with head and neck cancer as well as breast cancer83. Our study reinforces these findings, revealing a significant correlation between high CASP3 mRNA levels and poor prognosis in MM.
The TME is integral to cancer initiation, progression, and treatment response88. Similarly, we characterized the TME of MM patients based on risk scores. As the result of it, high-risk patients displayed higher tumor purity and increased infiltration of B memory cells, eosinophils, M0 macrophages, monocytes, and activated mast cells. The risk score was positively correlated with B memory cells and CD4 + memory resting T cells, whereas negatively correlated with CD4 + naive T cells. These findings suggest that PANoptosis may drive MM progression by modulating immune responses. TMB and immune checkpoint gene expression levels operate as biomarkers for cancer immunotherapy24,89. PVR and SIRPA are immune checkpoint molecules that inhibit CD8 + T cell activation and macrophage phagocytosis, respectively24,90. We found that elevated expression of these molecules was accompanied by higher TMB in high-risk individuals. Targeting PVR and SIRPA with inhibitors could promote antitumor immune responses in this group. Drug sensitivity analysis showed that cyclophosphamide was more effective in high-risk individuals. Moreover, we identified six promising drugs—Sinularin, Wee1 inhibitors, JQ1, osimertinib, and two ATR inhibitors, VE-822 and AZD6738—that could improve outcomes for this subgroup.
By integrating bulk and single-cell RNA-seq analyses, two machine learning algorithms (LASSO and Random Forest), clinical validations (qPCR/IHC), and functional assays (CCK-8/Wright-Giemsa staining), this study establishes for the first time the pivotal regulatory role of PANoptosis-related genes in multiple myeloma pathogenesis, providing a molecular basis for precision medicine in MM. Despite these findings, several methodological constraints merit careful consideration. First, our analyses relied primarily on public databases (with TMB profiling based on genomic data whereas other analyses employed transcriptomic data), necessitating validation through prospective studies. Second, the restricted sample size of the clinical validation cohort may compromise statistical robustness, as suggested by several non-significant results in qPCR and IHC assays. Furthermore, the mechanistic contributions of pivotal regulators (e.g., PARP1) require substantiation through appropriate animal models. Additionally, the current PANoptosis-related gene set (PRGs) may overlook other critical regulatory elements.
Conclusion
This study established the first PANoptosis-related prognostic model for multiple myeloma by integrating multi-omics data with machine learning algorithms. The model demonstrated robust predictive performance across three independent validation cohorts. Through systematic validation using single-cell RNA sequencing, qPCR, IHC, and functional assays, we elucidated the clinical significance of seven core PRGs, particularly highlighting PARP1 as a critical prognostic biomarker and a promising therapeutic target in multiple myeloma. Furthermore, we identified distinct immune microenvironment characteristics and drug sensitivity profiles in high-risk patients, providing a novel framework for MM molecular subtyping and targeted therapy development. Future studies should further investigate the molecular mechanisms of PANoptosis in MM progression and validate this model in prospective clinical trials.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
References
Malard, F. et al. Multiple myeloma. Nat. Rev. Dis. Primers. 10, 45. https://doi.org/10.1038/s41572-024-00529-7 (2024).
Rajkumar, S. V. Multiple myeloma: 2024 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 99, 1802–1824. https://doi.org/10.1002/ajh.27422 (2024).
Garfall, A. L. New biological therapies for multiple myeloma. Annu. Rev. Med. 75, 13–29. https://doi.org/10.1146/annurev-med-050522-033815 (2024).
Liu, Y., Li, B., Chen, X., Xiong, H. & Huang, C. The effect of Immunomodulatory drugs on bone metabolism of patients with multiple myeloma. Expert Rev. Hematol. 17, 47–54. https://doi.org/10.1080/17474086.2024.2316090 (2024).
Rees, M. J. & Kumar, S. High-risk multiple myeloma: redefining genetic, clinical, and functional high-risk disease in the era of molecular medicine and immunotherapy. Am. J. Hematol. 99, 1560–1575. https://doi.org/10.1002/ajh.27327 (2024).
Gong, L., Huang, D., Shi, Y., Liang, Z. & Bu, H. Regulated cell death in cancer: from pathogenesis to treatment. Chin. Med. J. (Engl). 136, 653–665. https://doi.org/10.1097/cm9.0000000000002239 (2023).
Ocansey, D. K. W. et al. Current evidence and therapeutic implication of PANoptosis in cancer. Theranostics 14, 640–661. https://doi.org/10.7150/thno.91814 (2024).
Gao, L., Shay, C. & Teng, Y. Cell death shapes cancer immunity: spotlighting PANoptosis. J. Exp. Clin. Cancer Res. 43, 168. https://doi.org/10.1186/s13046-024-03089-6 (2024).
Gao, J. et al. PANoptosis: bridging apoptosis, pyroptosis, and necroptosis in cancer progression and treatment. Cancer Gene Ther. 31, 970–983. https://doi.org/10.1038/s41417-024-00765-9 (2024).
Tan, Y. T. et al. WTAP weakens oxaliplatin chemosensitivity of colorectal cancer by preventing PANoptosis. Cancer Lett. 604, 217254. https://doi.org/10.1016/j.canlet.2024.217254 (2024).
Cai, Y. et al. Activation of STING by SAMHD1 deficiency promotes PANoptosis and enhances efficacy of PD-L1 Blockade in diffuse large B-cell lymphoma. Int. J. Biol. Sci. 19, 4627–4643. https://doi.org/10.7150/ijbs.85236 (2023).
Malireddi, R. K. S. et al. Inflammatory cell death, panoptosis, mediated by cytokines in diverse Cancer lineages inhibits tumor growth. Immunohorizons 5, 568–580. https://doi.org/10.4049/immunohorizons.2100059 (2021).
Huang, H. et al. A multi-dimensional approach to unravel the intricacies of lactylation related signature for prognostic and therapeutic insight in colorectal cancer. J. Transl Med. 22, 211. https://doi.org/10.1186/s12967-024-04955-9 (2024).
Wang, J. et al. NET-related gene signature for predicting AML prognosis. Sci. Rep. 14, 9115. https://doi.org/10.1038/s41598-024-59464-y (2024).
Li, D. et al. A distinct lipid metabolism signature of acute myeloid leukemia with prognostic value. Front. Oncol. 12, 876981. https://doi.org/10.3389/fonc.2022.876981 (2022).
Pan, H., Pan, J., Li, P. & Gao, J. Characterization of PANoptosis patterns predicts survival and immunotherapy response in gastric cancer. Clin. Immunol. 238, 109019. https://doi.org/10.1016/j.clim.2022.109019 (2022).
Song, J., Xu, Z., Fan, Q., Sun, Y. & Lin, X. The PANoptosis-related signature indicates the prognosis and tumor immune infiltration features of gliomas. Front. Mol. Neurosci. 16, 1198713. https://doi.org/10.3389/fnmol.2023.1198713 (2023).
Zhang, Z. et al. Identification of PANoptosis-relevant subgroups to evaluate the prognosis and immune landscape of patients with liver hepatocellular carcinoma. Front. Cell. Dev. Biol. 11, 1210456. https://doi.org/10.3389/fcell.2023.1210456 (2023).
Yang, P. et al. Identification of PANoptosis-related biomarkers and analysis of prognostic values in head and neck squamous cell carcinoma. Sci. Rep. 14, 9824. https://doi.org/10.1038/s41598-024-60441-8 (2024).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–d677. https://doi.org/10.1093/nar/gkae909 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Xie, N. et al. Neoantigens: promising targets for cancer therapy. Signal. Transduct. Target. Ther. 8, 9. https://doi.org/10.1038/s41392-022-01270-x (2023).
Borgeaud, M. et al. Novel targets for immune-checkpoint Inhibition in cancer. Cancer Treat. Rev. 120, 102614. https://doi.org/10.1016/j.ctrv.2023.102614 (2023).
Do, K., Doroshow, J. H. & Kummar, S. Wee1 kinase as a target for cancer therapy. Cell. Cycle. 12, 3159–3164. https://doi.org/10.4161/cc.26062 (2013).
Ngoi, N. Y. L. et al. Targeting ATR in patients with cancer. Nat. Rev. Clin. Oncol. 21, 278–293. https://doi.org/10.1038/s41571-024-00863-5 (2024).
Qi, W. et al. Inhibition of Wee1 sensitizes AML cells to ATR inhibitor VE-822-induced DNA damage and apoptosis. Biochem. Pharmacol. 164, 273–282. https://doi.org/10.1016/j.bcp.2019.04.022 (2019).
Kim, H. J. et al. Anti-tumor activity of the ATR inhibitor AZD6738 in HER2 positive breast cancer cells. Int. J. Cancer. 140, 109–119. https://doi.org/10.1002/ijc.30373 (2017).
Min, A. et al. AZD6738, A novel oral inhibitor of ATR, induces synthetic lethality with ATM deficiency in gastric Cancer cells. Mol. Cancer Ther. 16, 566–577. https://doi.org/10.1158/1535-7163.Mct-16-0378 (2017).
Wallez, Y. et al. The ATR inhibitor AZD6738 synergizes with gemcitabine in vitro and in vivo to induce pancreatic ductal adenocarcinoma regression. Mol. Cancer Ther. 17, 1670–1682. https://doi.org/10.1158/1535-7163.Mct-18-0010 (2018).
Sheng, H. et al. ATR inhibitor AZD6738 enhances the antitumor activity of radiotherapy and immune checkpoint inhibitors by potentiating the tumor immune microenvironment in hepatocellular carcinoma. J. Immunother Cancer. 8 https://doi.org/10.1136/jitc-2019-000340 (2020).
Hur, J. et al. Synergism of AZD6738, an ATR inhibitor, in combination with belotecan, a camptothecin analogue, in Chemotherapy-Resistant ovarian Cancer. Int. J. Mol. Sci. 22 https://doi.org/10.3390/ijms22031223 (2021).
Islam, M. T. et al. Biological activities of sinularin: A literature-based review. Cell. Mol. Biol. (Noisy-le-grand). 66, 33–36 (2020).
Hsu, S. Y. et al. Sinularin induces oxidative Stress-Mediated apoptosis and mitochondrial dysfunction, and inhibits angiogenesis in glioblastoma cells. Antioxid. (Basel). 11. https://doi.org/10.3390/antiox11081433 (2022).
Meng, X. Y. et al. Sinularin stabilizes FOXO3 protein to trigger prostate cancer cell intrinsic apoptosis. Biochem. Pharmacol. 220, 116011. https://doi.org/10.1016/j.bcp.2023.116011 (2024).
Huang, Y. et al. BET inhibitor JQ1 selectively reduce Tregs by upregulating STAT3 and suppressing PD-1 expression in patients with multiple myeloma. Adv. Biol. (Weinh). 8, e2300640. https://doi.org/10.1002/adbi.202300640 (2024).
Xia, L. et al. Osimertinib covalently binds to CD34 and eliminates myeloid leukemia stem/progenitor cells. Cancer Res. 84, 479–492. https://doi.org/10.1158/0008-5472.Can-23-1632 (2024).
Engelhardt, M., Kortüm, K. M., Goldschmidt, H. & Merz, M. Functional cure and long-term survival in multiple myeloma: how to challenge the previously impossible. Haematologica 109, 2420–2435. https://doi.org/10.3324/haematol.2023.283058 (2024).
Tang, L., Zhang, W., Zhang, Y., Deng, W. & Zhao, M. Machine Learning-Based integrated analysis of PANoptosis patterns in acute myeloid leukemia reveals a signature predicting survival and immunotherapy. Int. J. Clin. Pract. 2024 (5113990). https://doi.org/10.1155/2024/5113990 (2024).
Wang, Y. & Kanneganti, T. D. From pyroptosis, apoptosis and necroptosis to panoptosis: A mechanistic compendium of programmed cell death pathways. Comput. Struct. Biotechnol. J. 19, 4641–4657. https://doi.org/10.1016/j.csbj.2021.07.038 (2021).
Sun, X. et al. Mechanisms, biology, and role in disease. Immunol. Rev. 321, 246–262. https://doi.org/10.1111/imr.13279 (2024).
Oh, Y. T. & Sun, S. Y. Regulation of Cancer Metastasis by TRAIL/Death Receptor Signaling. Biomolecules 11, 499. https://doi.org/10.3390/biom11040499 (2021).
Cretney, E. et al. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J. Immunol. 168, 1356–1361. https://doi.org/10.4049/jimmunol.168.3.1356 (2002).
Zerafa, N. et al. Cutting edge: TRAIL deficiency accelerates hematological malignancies. J. Immunol. 175, 5586–5590. https://doi.org/10.4049/jimmunol.175.9.5586 (2005).
Finnberg, N., Klein-Szanto, A. J. & El-Deiry, W. S. TRAIL-R deficiency in mice promotes susceptibility to chronic inflammation and tumorigenesis. J. Clin. Invest. 118, 111–123. https://doi.org/10.1172/jci29900 (2008).
Oh, Y. T. et al. Suppression of death receptor 5 enhances cancer cell invasion and metastasis through activation of caspase-8/TRAF2-mediated signaling. Oncotarget 6, 41324–41338. https://doi.org/10.18632/oncotarget.5847 (2015).
Dhillon, S. & Aponermin First Approval Drugs 84, 459–466 https://doi.org/10.1007/s40265-024-02004-9 (2024).
Ranjan, K. & Pathak, C. Cellular dynamics of Fas-Associated death domain in the regulation of Cancer and inflammation. Int. J. Mol. Sci. 25 https://doi.org/10.3390/ijms25063228 (2024).
Chen, L. et al. CD95 promotes tumour growth. Nature 465, 492–496. https://doi.org/10.1038/nature09075 (2010).
Ceppi, P. et al. CD95 and CD95L promote and protect cancer stem cells. Nat. Commun. 5, 5238. https://doi.org/10.1038/ncomms6238 (2014).
Teodorczyk, M. et al. CD95 promotes metastatic spread via Sck in pancreatic ductal adenocarcinoma. Cell. Death Differ. 22, 1192–1202. https://doi.org/10.1038/cdd.2014.217 (2015).
Drachsler, M. et al. CD95 maintains stem cell-like and non-classical EMT programs in primary human glioblastoma cells. Cell. Death Dis. 7, e2209. https://doi.org/10.1038/cddis.2016.102 (2016).
Ciesielska, A., Matyjek, M. & Kwiatkowska, K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell. Mol. Life Sci. 78, 1233–1261. https://doi.org/10.1007/s00018-020-03656-y (2021).
Kim, H. M. et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 130, 906–917. https://doi.org/10.1016/j.cell.2007.08.002 (2007).
Rajamanickam, V. et al. Selective targeting of the TLR4 co-receptor, MD2, prevents colon cancer growth and lung metastasis. Int. J. Biol. Sci. 16, 1288–1302. https://doi.org/10.7150/ijbs.39098 (2020).
Ferrari, M. G. et al. Myeloid differentiation factor-2/LY96, a potential predictive biomarker of metastasis and poor outcomes in prostate cancer: clinical implications as a potential therapeutic target. Oncogene 43, 484–494. https://doi.org/10.1038/s41388-023-02925-x (2024).
Okamoto, M. et al. Involvement of Toll-like receptor 4 signaling in interferon-gamma production and antitumor effect by Streptococcal agent OK-432. J. Natl. Cancer Inst. 95, 316–326. https://doi.org/10.1093/jnci/95.4.316 (2003).
Xie, N. et al. NAD(+) metabolism: pathophysiologic mechanisms and therapeutic potential. Signal. Transduct. Target. Ther. 5, 227. https://doi.org/10.1038/s41392-020-00311-7 (2020).
Karpińska, A. et al. Quantitative analysis of biochemical processes in living cells at a single-molecule level: a case of olaparib-PARP1 (DNA repair protein) interactions. Analyst 146, 7131–7143. https://doi.org/10.1039/d1an01769a (2021).
Muvarak, N. et al. c-MYC generates repair errors via increased transcription of Alternative-NHEJ factors, LIG3 and PARP1, in tyrosine Kinase-Activated leukemias. Mol. Cancer Res. 13, 699–712. https://doi.org/10.1158/1541-7786.Mcr-14-0422 (2015).
Tomoda, T. et al. Enhanced expression of poly(ADP-ribose) synthetase gene in malignant lymphoma. Am. J. Hematol. 37, 223–227. https://doi.org/10.1002/ajh.2830370402 (1991).
Newman, E. A. et al. Alternative NHEJ pathway components are therapeutic targets in High-Risk neuroblastoma. Mol. Cancer Res. 13, 470–482. https://doi.org/10.1158/1541-7786.Mcr-14-0337 (2015).
Machado, C. B. et al. PARP1 is overexpressed in hematological malignant cell lines: A framework for experimental oncology. Anticancer Res. 41, 2397–2402. https://doi.org/10.21873/anticanres.15014 (2021).
Thomas, M. et al. PARP1 and POLD2 as prognostic biomarkers for multiple myeloma in autologous stem cell transplant. Haematologica 108, 2155–2166. https://doi.org/10.3324/haematol.2022.282399 (2023).
Groelly, F. J., Fawkes, M., Dagg, R. A., Blackford, A. N. & Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer. 23, 78–94. https://doi.org/10.1038/s41568-022-00535-5 (2023).
Wu, Y., Xu, S., Cheng, S., Yang, J. & Wang, Y. Clinical application of PARP inhibitors in ovarian cancer: from molecular mechanisms to the current status. J. Ovarian Res. 16, 6. https://doi.org/10.1186/s13048-023-01094-5 (2023).
Ison, G. et al. FDA approval summary: niraparib for the maintenance treatment of patients with recurrent ovarian Cancer in response to Platinum-Based chemotherapy. Clin. Cancer Res. 24, 4066–4071. https://doi.org/10.1158/1078-0432.Ccr-18-0042 (2018).
Kontandreopoulou, C. N., Diamantopoulos, P. T., Tiblalexi, D., Giannakopoulou, N. & Viniou, N. A. PARP1 as a therapeutic target in acute myeloid leukemia and myelodysplastic syndrome. Blood Adv. 5, 4794–4805. https://doi.org/10.1182/bloodadvances.2021004638 (2021).
Zhang, H. et al. PARP1 inhibitor niraparib exerts synergistic antimyeloma effect with bortezomib through inducing DNA damage and inhibiting DNA repair. Free Radic Biol. Med. 228, 392–402. https://doi.org/10.1016/j.freeradbiomed.2024.12.052 (2025).
Cao, L. et al. Phylogenetic analysis of CDK and Cyclin proteins in premetazoan lineages. BMC Evol. Biol. 14, 10. https://doi.org/10.1186/1471-2148-14-10 (2014).
Alquézar, C. et al. Targeting Cyclin D3/CDK6 activity for treatment of parkinson’s disease. J. Neurochem. 133, 886–897. https://doi.org/10.1111/jnc.13070 (2015).
Malhotra, N., Gupta, R. & Kumar, P. Pharmacological relevance of CDK inhibitors in alzheimer’s disease. Neurochem Int. 148, 105115. https://doi.org/10.1016/j.neuint.2021.105115 (2021).
Wang, Q., Bode, A. M. & Zhang, T. Targeting CDK1 in cancer: mechanisms and implications. NPJ Precis Oncol. 7, 58. https://doi.org/10.1038/s41698-023-00407-7 (2023).
Wu, D. et al. CD138(-) multiple myeloma cells express high level of CHK1 which correlated to overall survival in MM patient. Aging (Albany NY). 12, 23067–23081. https://doi.org/10.18632/aging.104066 (2020).
Goodsell, D. S. The molecular perspective: caspases. Oncologist 5, 435–436. https://doi.org/10.1634/theoncologist.5-5-435 (2000).
Srivastava, N. & Saxena, A. K. Caspase-3 activators as anticancer agents. Curr. Protein Pept. Sci. 24, 783–804. https://doi.org/10.2174/1389203724666230227115305 (2023).
Green, D. R. Caspase activation and Inhibition. Cold Spring Harb Perspect. Biol. 14 https://doi.org/10.1101/cshperspect.a041020 (2022).
Jiang, M., Qi, L., Li, L. & Li, Y. The caspase-3/GSDME signal pathway as a switch between apoptosis and pyroptosis in cancer. Cell. Death Discov. 6, 112. https://doi.org/10.1038/s41420-020-00349-0 (2020).
Li, F. et al. Apoptotic cells activate the phoenix rising pathway to promote wound healing and tissue regeneration. Sci. Signal. 3, ra13. https://doi.org/10.1126/scisignal.2000634 (2010).
Pellettieri, J. et al. Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 338, 76–85. https://doi.org/10.1016/j.ydbio.2009.09.015 (2010).
Chera, S. et al. Apoptotic cells provide an unexpected source of Wnt3 signaling to drive hydra head regeneration. Dev. Cell. 17, 279–289. https://doi.org/10.1016/j.devcel.2009.07.014 (2009).
Galluzzi, L., Kepp, O. & Kroemer, G. Caspase-3 and prostaglandins signal for tumor regrowth in cancer therapy. Oncogene 31, 2805–2808. https://doi.org/10.1038/onc.2011.459 (2012).
Huang, Q. et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 17, 860–866. https://doi.org/10.1038/nm.2385 (2011).
Treps, L., Faure, S. & Clere, N. Vasculogenic mimicry, a complex and devious process favoring tumorigenesis - Interest in making it a therapeutic target. Pharmacol. Ther. 223, 107805. https://doi.org/10.1016/j.pharmthera.2021.107805 (2021).
Zhao, X. et al. Caspase-3-dependent activation of calcium-independent phospholipase A2 enhances cell migration in non-apoptotic ovarian cancer cells. J. Biol. Chem. 281, 29357–29368. https://doi.org/10.1074/jbc.M513105200 (2006).
Mukai, M. et al. Cross talk between apoptosis and invasion signaling in cancer cells through caspase-3 activation. Cancer Res. 65, 9121–9125. https://doi.org/10.1158/0008-5472.Can-04-4344 (2005).
Liu, Y. R. et al. Basal caspase-3 activity promotes migration, invasion, and vasculogenic mimicry formation of melanoma cells. Melanoma Res. 23, 243–253. https://doi.org/10.1097/CMR.0b013e3283625498 (2013).
El-Tanani, M. et al. Unraveling the tumor microenvironment: insights into cancer metastasis and therapeutic strategies. Cancer Lett. 591, 216894. https://doi.org/10.1016/j.canlet.2024.216894 (2024).
Yin, X. et al. Potential predictive biomarkers in antitumor immunotherapy: navigating the future of antitumor treatment and immune checkpoint inhibitor efficacy. Front. Oncol. 14, 1483454. https://doi.org/10.3389/fonc.2024.1483454 (2024).
Zhang, W. et al. Advances in Anti-Tumor treatments targeting the CD47/SIRPα Axis. Front. Immunol. 11, 18. https://doi.org/10.3389/fimmu.2020.00018 (2020).
Funding
This work was supported by the National Natural Science Foundation of China (NSFC, Grant No. 82270213).
Author information
Authors and Affiliations
Contributions
Yashu Feng, Shuoting Wang designed the study, performed the bioinformatics analysis, and drafted the manuscript. Jingwen Zhang, Chengcheng Liu assisted with data interpretation and provided critical revisions to the manuscript. Jiajun Liu and Ling Zhang supervised the project, provided funding, and contributed to the final editing and approval of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All procedures involving human participants were approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University (Approval No. RG2023-306-02) and performed in accordance with ethical standards. Written informed consent was obtained from all individual participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, Y., Wang, S., Zhang, J. et al. Comprehensive machine learning analysis of PANoptosis signatures in multiple myeloma identifies prognostic and immunotherapy biomarkers. Sci Rep 15, 22478 (2025). https://doi.org/10.1038/s41598-025-06376-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06376-0