Abstract
The migration of gonadal distal tip cells (DTCs) in Caenorhabditis elegans serves as an excellent model for studying the migration of epithelial tubes during organogenesis. Mutations in the mig-17/ADAMTS gene cause misdirected DTC migration during gonad formation, resulting in deformed gonad arms. An amino acid substitution in RPL-20, the ortholog of mammalian RPL18a/eL20, a component of the 60 S ribosomal large subunit, exhibited a slow-growth phenotype and strongly suppressed the mig-17 gonadal defects. Slow-growing mutations clk-1 and clk-2 also suppressed mig-17. Intestine-specific overexpression of mutant RPL-20 protein resulted in a slow-growth phenotype and suppressed the mig-17 gonadal defects, but these effects were much weaker when wild-type RPL-20 was overexpressed, suggesting that the mutant RPL-20 protein acquired a novel function. Analysis of ribosome profiles revealed reduced biogenesis of the 60 S subunit, leading to a reduction of 80 S ribosomes in the rpl-20 mutant. These results suggest that DTC migration defects in mig-17/ADAMTS mutants can be partly suppressed by growth retardation caused by the rpl-20 mutation. While defective ribosome biogenesis may contribute to the observed growth retardation, further investigation is needed to clarify the molecular basis of this phenomenon.
Similar content being viewed by others
Introduction
The ADAMTS (a disintegrin and metalloprotease with thrombospondin motifs) family of secreted zinc metalloproteases plays important roles in morphogenetic processes during animal development. ADAMTS proteases often degrade extracellular matrix proteins in these processes1. For example, ADAMTS-9 and − 20 cleave the proteoglycan versican and are required for digit formation and closure of the palate in mice2,3. ADAMTS-1 is also a versicanase, and the ADAMTS-1 null mice exhibit growth retardation with malformation of adipose, ureteral and adrenal tissues4,5,6. However, the precise roles of ADAMTS proteases in development still remain elusive.
An ADAMTS protease, MIG-17, in C. elegans, is involved in the directional regulation of gonadal leader cells, the distal tip cells (DTCs), during the formation of the U-shaped gonad arms in larval development. In the mig-17 mutants, the misdirected migration of DTCs leads to the deformation of the gonad arms7. Through genetic suppressor analyses, several gain-of-function amino acid substitutional mutations were identified in extracellular matrix protein genes, including fbl-1/fibulin-1, emb-9/collagen IV a1 chain, and let-2/collagen IV a2 chain8,9,10,11. Genetic and molecular analyses of these suppressor mutations revealed the regulatory pathway by which MIG-17 recruits fibulin-1 to the basement membrane and activates fibulin-1 and collagen IV to modulate downstream events affecting the directional migration of DTCs10,11. Since MIG-17 is a matrix metalloprotease, it is reasonable that genetic suppressors were found in genes encoding extracellular matrix proteins.
In the present study, we identified a novel suppressor mutation, tk73, in the ribosomal protein gene rpl-20, which corresponds to mammalian RPL18a/eL20, one of the components of the 60 S large ribosomal subunit12. The suppressor rpl-20(tk73) mutation was an amino acid substitution resulting in growth retardation and acted as a dominant allele to suppress the mig-17 DTC migration abnormality. The amount of 60 S large ribosomal subunit was markedly reduced in the rpl-20(tk73) mutants. Genetic analyses suggest that growth retardation induced by rpl-20(tk73) is causative for mig-17 suppression.
While ribosomal protein mutations are generally thought to affect translation efficiency, defects in ribosome biosynthesis caused by mutations in single ribosomal protein genes lead to a tissue-specific phenotypic abnormality called ribosomopathy in humans13. Our findings should help to understand the molecular mechanism underlying ribosomopathies.
Results
An amino acid substitution in the ribosomal protein gene rpl-20 suppresses the abnormal distal tip cell migration of mig-17 mutants
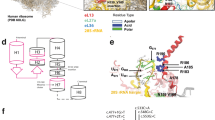
In C. elegans, the distal tip cells (DTCs) located at the anterior and posterior ends of the gonad primordium act as leader cells to elongate and form the U-shaped gonad arms. The ADAMTS family metalloprotease MIG-17 is required for the directional migration of DTCs7. The mig-17 mutants show deformed gonad formation due to meandering and straying of DTCs14. Through forward genetics screening, we identified a mutation rpl-20(tk73) that could alleviate the gonadal defect of mig-17 mutants (Fig. 1A-C). RPL-20 is an ortholog of the mammalian ribosomal protein RPL18a/eL20, a component of the 60 S large ribosomal subunit12. rpl-20(tk73) was an amino acid substitution of evolutionarily conserved glycine 82 (glycine 79 in mammals) into arginine (Fig. 1D). It suppressed the mig-17(k174) null allele strongly as a homozygote and weakly as a heterozygote, while alone showed no gonadal defects (Fig. 1B). The deletion allele rpl-20(ok2256), which totally deletes the coding region, exhibited a homozygous early larval lethal phenotype and rpl-20(ok2256)/+ failed to suppress mig-17(tk174) (Fig. 1B). rpl-20(tk73) also suppressed missense mig-17 alleles k135 and k169, as well as mutations in mig-18 that encodes a cofactor for the MIG-17 metalloprotease15although the suppression for mig-18 appeared to be weaker than for mig-17 (Fig. 1E). However, it failed to suppress the gonadal defects caused by RNAi knockdown of gon-1, another ADAMTS metalloprotease that functions in gonadogenesis16 (Fig. 1F). It is possible that partial suppression could not be detected, as the phenotype of gon-1 RNAi is much more pronounced compared to that of mig-17 mutants. Similarly, rpl-20(tk73) could not suppress the mig-17-like meandering DTC migration defects caused by mutations in sqv-5 (chondroitin synthase) and mig-22 (chondroitin polymerizing factor)17 (Fig. 1G). These results suggest that rpl-20(tk73) specifically suppresses the gonadal defects caused by mutations in mig-17 and its cofactor mig-18.
Suppression of mig-17 and mig-18 DTC migration defects by rlp-20(tk73). (A) Gonad morphology (arrows) of wild type, mig-17(k174), and mig-17(k174); rpl-20(tk73) young-adult hermaphrodites. Posterior gonads are shown. Anterior to the left, dorsal to the top. Bar, 20 μm. (B) Quantitative analysis of mig-17 gonadal defects combined with rpl-20(tk73). N = 60 for each strain. P-values for Fisher’s exact test against mig-17(k174) for mig-17(k174) carrying strains are indicated: ***, P < 0.001; NS, not significant. (C) Transgenic rescue experiments of mig-17. mig-17 animals introduced with transgenic arrays containing PCR-amplified fragments of the rpl-20(tk73) gene were suppressed for their gonadal defects. #1 and #2 are independently established transgenic lines. N = 60 for each strain. P-values for Fisher’s exact test against mig-17(k174) are indicated: ***, P < 0.001. (D) Amino acid sequence homology between RPL-20 and human RPL18a. Identical amino acids are shown in black boxes. G82 mutated in rpl-20(tk73) is depicted with an arrow. (E-G) rpl-20(tk73) suppresses the DTC migration defects of mig-17 and mig-18 alleles (E), but not gon-1(RNAi) (F) or sqv-5(k175) and mig-22(k141) mutants (G). sqv-5(k175) and mig-22(k141) were marked with unc-13(e1091) and unc-32(e189), respectively. N = 60 for each strain. P-values for Fisher’s exact test against mig-17, mig-18, rpl-20(tk73) control RNAi, sqv-5, and mig-22 for the respective double mutants are indicated: ***, P < 0.001; **, P < 0.01; NS, not significant.
It is reported that the guidance molecule semaphorin regulates epidermal morphogenesis by stimulating general translation through inhibition of the target of rapamycin complex 2 (TORC2) pathway, which in turn activates TORC1 repressing the phosphorylation of eukaryotic initiation factor 2α (eIF2α)18,19. We suspected that MIG-17 may also affect translation to regulate DTC migration. This possibility seemed consistent with the fact that a mutation in a ribosomal protein RPL-20 suppresses the mig-17 defects. Semaphorin signaling promotes eIF2α dephosphorylation by repressing the activities of GCN-1 and the PEK-1 protein kinase18. Semaphorin also represses RICT-1 (TORC2 component)19. We generated gcn-1; mig-17, pek-1; mig-17 and rict-1; mig-17 double mutants and found that they failed to suppress the mig-17 gonadal defects (Supplementary Fig. S1), suggesting that MIG-17 does not control DTC migration through the stimulation of general translation.
Growth rate deceleration suppresses the gonadal abnormality of mig-17 mutants
During the analysis of the rpl-20(tk73) mutants, we observed that they grew slower compared to the wild type or mig-17 mutant animals. rpl-20(tk73); mig-17(k174) double mutants also grew slowly. We analyzed their growth rates based on the stages of vulval development (Fig. 2A, B). At 43 h after hatching, while wild-type or mig-17(k174) animals had reached the late L4 or adult stage, those carrying rpl-20(tk73) were still at mid-late L4 or early L4 stages. By 48 h after hatching, most wild-type or mig-17(k174) animals had reached the adult stage, whereas those with rpl-20(tk73) remained primarily in the L4 stage.
Growth rate analysis of mig-17, rpl-20, mig-17; rpl-20, clk-1, and clk-2 mutants. (A-C) Percentages of larval stages were determined at 43 h (A) and 48 h (B) and 67 and 72 h (C) after hatching, as described in Methods. Color codes represent larval stage 2 (L2), larval stage 3 (L3), early larval stage 4 (EL4), mid-larval stage 4 (ML4), mid-late larval stage 4 (MLL4), late larval stage 4 (LL4), and adult stage (A), respectively. (D, E) Suppression of mig-17(k174) (D) and mig-17 and mig-18 alleles (E) by clk-1 and clk-2. N = 60 for each strain. P-values for Fisher’s exact test against mig-17 and unc-42 mig-17 for the respective double mutants are indicated: ***, P < 0.001; **, P < 0.01; *, P < 0.05.
To investigate whether the growth retardation caused mig-17 suppression, we created double mutants between mig-17 or mig-18 alleles and slow-growing mutants clk-1(qm30) or clk-2(qm37)20,21. We found that these clk mutations partially suppressed mig-17 and mig-18 gonadal defects (Fig. 2D, E). clk-1 encodes a mitochondrial hydroxylase that is necessary for ubiquinone biosynthesis, while clk-2 encodes a component of DNA damage response and telomere metabolism22,23 as well as the nonsense-mediated mRNA decay pathway24. Thus, these genes have no functional relevance to rpl-20, suggesting that the slow growing phenotype of these mutants is partly contribute to the suppression. Since clk-2(qm37) exhibited a growth delay similar to rpl-20(tk73) and clk-1(qm30) animals grew much slower than rpl-20(tk73) (Fig. 2A-C), it is possible that an additional mechanism other than growth retardation acts in mig-17 suppression by rpl-20(tk73).
Intestine-specific expression of mutant RPL-20 efficiently suppresses mig-17
We generated a translational fusion construct, rpl-20p::mCherry::rpl-20(WT). This construct was partially functional as it rescued the early larval lethal phenotype of the rpl-20(ok2256) deletion allele when introduced as an extrachromosomal array containing multicopy rpl-20p::mCherry::rpl-20(WT) transgenes, although the adult animals with the array were sterile. The mCherry expression was detected ubiquitously except in the germline, likely due to gene silencing of the multicopy array25. The expression appeared most intense in the intestine (Fig. 3A). The same expression pattern was observed in rpl-20p::mCherry::rpl-20(tk73) (Fig. 3A) and it partially rescued rpl-20(ok2256) as observed in rpl-20p::mCherry::rpl-20(WT).
Tissue-specific expression of RPL-20. (A) Expression of mCherry-RPL-20(WT) and mCherry-RPL-20(tk73). Upper panels are Nomarski and confocal merged images for young adult animals and lower panels represent close-up images of the gonadal tip regions. Arrows and arrowheads indicate intestine and the distal tip cells, respectively. Bars, 50 μm. (B, C) Rescue experiments of the mig-17 gonadal defect by expressing rpl-20(tk73) (B) or rpl-20(WT) (C) genes under various tissue-specific promoters. N = 60 for each strain. P-values for Fisher’s exact test against mig-17(k174) for mig-17(k174) carrying strains are indicated: ***, P < 0.001; **, P < 0.01; NS, not significant. #1 and #2 are independently established transgenic lines. (D, E) Growth rate analysis of mig-17 animals carrying various plasmid constructs. Percentages of larval stages were determined at 43 h (D) and 48 h (E) after hatching, as described in Methods. #1 and #2 are independently established transgenic lines. Color codes represent larval stages, early L4 (EL4), mid-L4 (ML4), mid-late L4 (MLL4), late L4 (LL4), and adult (A), respectively. N ≥ 60 for each strain.
To determine the tissues in which expression of the mutant RPL-20(tk73) protein is important for suppressing mig-17, we expressed the mutant rpl-20(tk73) gene under tissue-specific promoters. We found that DTC-specific expression (mig-24p::rpl-20(tk73))26 pharyngeal muscle-specific expression (myo-2::rpl-20(tk73))27 and body wall muscle-specific expression (myo-3::rpl-20(tk73))27 weakly suppressed mig-17, whereas intestine-specific expression (elt-2p::rpl-20(tk73))28 strongly suppressed the gonadal defects of mig-17 mutants (Fig. 3B). The suppressor activity of elt-2p::rpl-20(WT) was much weaker than that of elt-2p::rpl-20(tk73) (Fig. 3C). Next, we analyzed these transgenic lines for their growth rates. Although all these lines exhibited growth retardation (Fig. 3D, E), elt-2p::rpl-20(tk73) was the slowest, and its growth rate was equivalent to that of the rpl-20(tk73) mutant (Fig. 3D, E). The effects of mig-24p::rpl-20(tk73) or myo-3::rpl-20(tk73) were weaker than those of elt-2p::rpl-20(tk73), but they also showed a marked decrease in growth rate compared to the wild type (Fig. 3D, E). The growth rate for elt-2p::rpl-20(WT) was slightly slower than that of the wild-type animals (Fig. 3D, E). The non-cell-autonomous dominant effect of rpl-20(tk73) implies its gain-of-function or dominant-negative effect. These results support the idea that the growth rate retardation caused by the rpl-20(tk73) allele is causative for mig-17 suppression and that RPL-20(G82R) expressed in the intestine plays an important role in the suppression.
rpl-20(tk73) does not affect levels of expression of FBL-1C
We previously reported that amino-acid substitutions in the basement membrane protein FBL-1C can suppress the gonadal defect of mig-17 mutants. From the genetic analysis, we demonstrated that MIG-17 activity is necessary for efficient recruitment and accumulation of FBL-1C to the basement membrane, and that this activity is essential for the directional migration of gonadal DTCs10. FBL-1C is a basement membrane protein that is expressed and secreted from the intestine8. As rpl-20(tk73) expression in the intestine had the strongest effect on mig-17 suppression, it is possible that FBL-1C is involved in rpl-20(tk73) suppression of mig-17. We compared the levels of FBL-1C-Venus expression between wild type and rpl-20(tk73) backgrounds using anti-GFP immunoblotting and observed that the amount of FBL-1C-Venus protein was slightly lower in rpl-20(tk73) compared to wild type, indicating that the suppressor activity of rpl-20(tk73) does not result from the upregulation of FBL-1C expression (Supplementary Fig. S2). Using a transgenic line in which the mNeonGreen reporter is placed at the N-terminus of fbl-1 coding region by the CRISPR/Cas9 method29 we examined the basement membrane localization of the FBL-1C isoform. The fbl-1 gene produces various splicing isoforms having the same N-terminus and either one of the two types of C-terminal domains, generating two groups of splicing variants classified as FBL-1C or FBL-1D8,30. Although it is known that FBL-1C, but not FBL-1D, localizes to the gonadal basement membrane8 the mNeonGreen fluorescence was not detectable in this location, probably because of its accumulation below the detection level. Thus, we examined the levels of mNeonGreen-FBL-1 accumulation in the pharynx. In the pharynx, FBL-1D accumulates along the flexible tracks at the anterior tip of the pharynx, whereas FBL-1C does so in the pharyngeal basement membrane30. We quantified the intensity of mNeonGreen fluorescence in the pharyngeal basement membrane, which is presumed to predominantly represent the level of FBL-1C, using confocal images. We found that the fluorescence intensity did not differ significantly among mig-17(k174); rpl-20(tk73), mig-17(k174) and wild type (Supplementary Fig. S2). We also examined the accumulation levels of FBL-1C-Venus in the gonadal basement membrane but did not detect a significant difference between mig-17(k174); rpl-20(tk73) and mig-17(k174) (Supplementary Fig. S2). Thus, the levels of expression as well as basement membrane accumulation of FBL-1C appear not to be affected by the rpl-20(tk73) mutation.
Reduction of 60 S ribosomal subunit in rpl-20(tk73) mutants
Western blot analysis was performed on ribosomes extracted from animals carrying rpl-20p::mCherry::rpl-20(WT) and rpl-20p::mCherry::rpl-20(tk73) arrays using anti-mCherry. The results showed that both the wild-type and mutant RPL-20 proteins were incorporated into the ribosome (Fig. 4A). We analyzed ribosome profiles by sucrose density gradient centrifugation using extracts from wild-type and rpl-20(tk73) animals. It was observed that there was a concomitant reduction of the 60 S subunit and the 80 S ribosomes in rpl-20(tk73), indicating that the RPL-20(tk73) mutant protein partially interferes with the biogenesis of the 60 S subunit, resulting in a decrease in the amount of 80 S mature ribosomes (Fig. 4B).
Analysis of ribosomes. (A) Western blot analysis of ribosomes for wild type animals expressing mCherry-RPL-20(tk73) or mCherry-RPL-20(WT). Ribosomes were precipitated by dual sucrose cushion centrifugation and the samples of supernatants and precipitates were analyzed. Sup 1 and 2 and PPT 1 and 2 represent supernatant and precipitate samples from first and second centrifugation, respectively. Arrow indicates the bands corresponding to mCherry-RPL-20. The double bands may be generated due to post-translational modification of RPL-2044. Original blot is presented in Supplementary Figure S3. (B) Polysome profiles of wild type and rpl-20(tk73) animals. The amounts of 60 S subunit and 80 S ribosome in rpl-20(tk73) were reduced relative to the wild type.
Discussion
In this study, we identified a novel missense mutation in the ribosomal subunit-encoding gene rpl-20 that strongly suppresses the gonadal DTC migration defect of mig-17(tk174) null mutants. The suppression was specific for mig-17 and mig-18 alleles, but not for other mutations having gonadal DTC migration defects. The rpl-20 mutants grew slower than wild-type animals. The slow-growing mutants, clk-1 or clk-2, also suppressed mig-17.
It is surprising that a mutation in a ribosome subunit strongly suppresses the gonadal defect of mutations in mig-17, which encodes a matrix metalloprotease that acts in remodeling the basement membrane. The ribosome is a cellular factory comprising multiple ribosomal RNAs and proteins and functions in the translational production of proteins from mRNAs. Although mutations in ribosomal proteins can affect translational efficacy in general, defects in ribosome biogenesis caused by mutations in single ribosomal protein genes result in tissue-specific phenotypic abnormalities. In humans, mutations in nineteen out of eighty-one ribosomal protein genes, not including RPL18a/eL20 thus far, are known to be causative for dominant genetic disorders called ribosomopathies. Ribosomopathies are mainly characterized by erythroid hypoplasia in the bone marrow (Diamond-Blackfan anemia; DBA), but they also associate with a series of phenotypic spectra, including craniofacial, limb, cardiac, and genitourinary malformations, as well as growth retardation and increased risks of cancers. DBA is caused by inhibition of the hematopoietic system through nucleolar stress-dependent stabilization of p53, which leads to cell cycle arrest and apoptosis13,31. DBA patients with mutations in RPL9 exhibit a reduction of the 60 S subunit coupled with a reduction of the 80 S ribosomes, which is reminiscent of our observation in rpl-20(tk73)32. In mice, Belly spot and tail (Bst) is a semi-dominant, homozygous lethal mutation in RPL24, and Bst/+ mice have decreased pigmentation, a kinked tail, a reduced number of retinal ganglion cells, and an extra preaxial digit33. Tail short (Ts) is a dominant, homozygous lethal mutation in RPL38, and Ts/+ animals exhibit a short and kinked tail that is associated with skeletal patterning defects. Vertebrae extending along the anterior-posterior body axis exhibit homeotic transformations. Interestingly, a specific subset of Hox genes (Hoxa4, a9, a11, b3, c8, and d10) specifying the vertebral patterning are markedly decreased in polysome association in Ts/+ embryos, suggesting that RPL38 has a specialized function in translational control of these mRNAs34.
As we observed in rpl-20(tk73) mutants, growth retardation is a common feature of ribosomopathies. We found that rpl-20(tk73) is a gain-of-function mutation that suppresses the mig-17 gonadal defects. Although the overexpression of rpl-20(tk73) in the intestine strongly suppressed mig-17, the suppression was marginal when we used rpl-20(WT). Thus, it is likely that rpl-20(tk73) could gain a novel function rather than simply an elevated wild-type activity. In mig-17 mutants, the DTCs often detach from the body wall, their natural substratum, and instead attach to the intestine or gonad and migrate over these tissues7. This suggests that the activity of MIG-17 is important for the adhesion of DTCs to the body wall, which may lead to appropriate responses to guidance molecules such as UNC-6/netrin and Wnt35,36. When mig-17(k174) mutants were cultured at 16 °C, the growth rate was delayed by about 1.5 times, and the DTC phenotype was weakened (DTC migration defect: 37% anterior and 72% posterior at 20 °C, N = 60; 18% anterior and 53% posterior at 16 °C, N = 60). Since k174 is a null mutation, the mig-17 gene function cannot be restored even at low temperatures. The penetrance of mig-17(k174) mutants with an abnormal DTC migration phenotype is around 70 to 80%. Therefore, a second pathway may regulates DTC migration, besides the MIG-17 pathway. One possibility is that at 16 °C, the duration for adhesion to the body wall and response to guidance molecules is prolonged, which may allow more time for the second pathway to function in the absence of MIG-17. It might be possible that a similar situation occurs in the rpl-20(tk73) mutants even at 20 °C.
The rpl-20(tk73) mutation suppressed mig-17 more strongly than clk-1 and clk-2 mutations, despite clk-2 showing a similar degree of growth retardation and clk-1 exhibiting even stronger growth retardation than rpl-20(tk73). These results suggest that a mechanism other than growth retardation may be involved in rpl-20(tk73)-dependent suppression of mig-17. Although we considered the possibility that the rpl-20 mutation might affect the expression or basement membrane localization of FBL-1 C, our results did not provide evidence to support this hypothesis. Since FBL-1 C recruits NID-1/nidogen to the gonadal basement membrane to control DTC migration10 it is possible that misregulation of NID-1 expression in rpl-20 mutants may contribute to the suppression. In addition to FBL-1 C, we previously reported that EMB-9 and LET-2, corresponding to the α1 and α2 chains of type IV collagen, respectively, are also downstream targets of MIG-17-dependent control of DTC migration10,11. Although alterations in the levels of these collagens in the gonadal basement membrane cannot be detected in mig-17 mutants, genetic evidence suggests an increase in collagen accumulation11. Thus, it is possible that rpl-20(tk73) downregulates expression or gonadal accumulation of type IV collagen molecules to suppress mig-17. As reported in mammals, it is also possible that stress responses involving p53 stabilization is triggered by the ribosomal mutation rpl-20(tk73). Further molecular analysis will be needed to understand the precise mechanism downstream of rpl-20(tk73).
Methods
Strains and genetic analysis
Culture, handling, and ethyl methanesulfonate (EMS) mutagenesis of C. elegans were conducted as described37. The following mutations were used in this work: mig-17(k135, k169, k174), mig-18(k140, gm321), fbl-1(tk45, qy62[mNG + loxP::fbl-1]), rpl-20(ok2256), unc-13(e1091), unc-25(e156), unc-32(e189), unc-42(e270), unc-64(e246), dpy-13(e184), unc-119(e2498), clk-1(qm30), clk-2(qm37), sqv-5(k175) and mig-22(k141), gcn-1(nc40), pek-1(ok275) and rict-1(nc41)8,14,15,17,18,19,20,22,29,37,38. The strains used in this study are listed in Supplementary Table S1.
Microscopy
Gonad migration phenotypes were scored using a Nomarski microscope (Axioplan 2; Zeiss). Analysis of gonadal phenotypes was performed at the young-adult stage as described14. Confocal laser scanning microscopy was conducted with LSM5 (Zeiss) controlled by PASCAL version 3.2 SP2 or ZEN software (Zeiss) to capture mCherry images. The fluorescence intensities of mNeonGreen were quantified as follows. For each sample, confocal images of sagittal sections of the pharynx were obtained using a TSC SP8 confocal microscope (Leica). The captured images were analyzed with Fiji software. Using the Segmented Line tool, lines with a width of 4 and a length of 0.6 were drawn over the pharyngeal basement membrane and inside the pharynx to define the background. The mean fluorescence intensity of the background was subtracted from the mean fluorescence intensity of the basement membrane to determine the fluorescence intensity of the basement membrane. The fluorescence intensities of Venus was quantified as described15.
Molecular identification of rpl-20(tk73)
The tk73 mutation was isolated as a genetic suppressor of the gonadal defect of mig-17(k174) mutants using EMS mutagenesis37. Single-nucleotide polymorphism mapping experiments39 placed tk73 between a 4581-4788 kb region on linkage group IV. Whole-genome sequence analyses comparing mig-17(k174) and tk73; mig-17(k174) genomes identified a single mutation in the coding region of the rpl-20 gene (GGA to AGA), resulting in the amino acid substitution G82R. Microinjection experiments using a PCR-amplified mutant rpl-20 gene fragment successfully rescued the gonadal defects of mig-17 mutants, confirming that rpl-20 is the gene responsible for tk73 (Fig. 1C).
Analysis of growth rate
Growth rate was analyzed as described40. Briefly, newly hatched larvae were synchronized for one hour, grown at 20 °C, and the growth rate was assessed based on the stages of vulval development at the L4 stage. The L2 and L3 stages were assessed by gonad morphology: L2, before the first turn, and L3, during the first and second turns of the DTCs.
Constructs
To produce the rpl-20p::rpl-20(tk73) plasmid, the genomic region containing 1190 bp 5’-untranslated region (UTR) to 591 bp 3’-UTR of the rpl-20 gene was PCR amplified from genomic DNA of mig-17(k174); rpl-20(tk73) and cloned into the NotI and Acc65I sites of pBluescript II KS(-). To produce the plasmids for tissue-specific expression, the 5’-UTR region was replaced with PCR amplified 5’-UTR fragments of mig-24 (1156 bp), myo-2 (1463 bp), myo-3 (2452 bp) or elt-2 (5044 bp). mig-24p::rpl-20(WT) and let-2p::rpl-20(WT) plasmid were generated by site-directed mutagenesis of mig-24p::rpl-20(tk73) and let-2p::rpl-20(tk73) plasmids, respectively. The rpl-20p::mCherry::rpl-20(WT) plasmid was constructed using pPD95.79 in which the coding region of GFP was replaced with that of mCherry. fbl-1 C cDNA::Venus plasmid was constructed form the fbl-1 C::3HA(k201, ΔD) plasmid8 by replacing the fragment from the fourth exon to 3HA into fbl-1 C cDNA.
Germline transformation
Germline transformation was carried out as described (Mello et al. 1991). Transgenic strains were made by injecting plasmids into unc-119(e2498) hermaphrodites and the generated transgenic arrays were transferred to appropriate genetic backgrounds having unc-119(e2498) by mating. mig-24p::rpl-20(tk73), myo-3p::rpl-20(tk73), elt-2p::rpl-20(tk73), mig-24p::rpl-20(WT), myo-3p::rpl-20(WT), and elt-2p::rpl-20(WT) plasmids were injected at 10 ng/µl with 30 ng/µl unc-119+ plasmid (pDP#MM016B)38 and 110 ng/µl pBluescriptII KS(–) (carrier DNA). myo-2p::rpl-20(tk73) plasmid was injected at 2 ng/µl with 38 ng/µl unc-119+ plasmid (pDP#MM016B) and 110 ng/µl pBluescriptII KS(–). fbl-1 C(-D)cDNA::Venus plasmid was injected at 5 ng/µl with 10 ng/µl unc-119+ plasmid (pDP#MM016B) and 135 ng/µl pBluescriptII KS(–).
Western blot analysis
Western blot analysis was done as described41 using anti-α-tubulin (DM1A, 1:1000; abcam), anti-GFP (3E6, 1:200; Molecular Probes) or anti-mCherry (5f8 1:1000, Funakoshi).
Ribosome assay
Preparation of worm samples and sucrose density gradient centrifugation were conducted as described42. Sucrose cushion centrifugation was conducted as described43.
Data availability
Strains and plasmids are available from the corresponding author upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article and figures.
References
Apte, S. S. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J. Biol. Chem. 284, 31493–31497. https://doi.org/10.1074/jbc.R109.052340 (2009).
Enomoto, H. et al. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development 137, 4029–4038. https://doi.org/10.1242/dev.050591 (2010).
McCulloch, D. R. et al. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev. Cell. 17, 687–698. https://doi.org/10.1016/j.devcel.2009.09.008 (2009).
Russell, D. L., Doyle, K. M., Ochsner, S. A., Sandy, J. D. & Richards, J. S. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J. Biol. Chem. 278, 42330–42339. https://doi.org/10.1074/jbc.M300519200 (2003).
Shindo, T. et al. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J. Clin. Invest. 105, 1345–1352. https://doi.org/10.1172/JCI8635 (2000).
Mittaz, L. et al. Adamts-1 is essential for the development and function of the urogenital system. Biol. Reprod. 70, 1096–1105. https://doi.org/10.1095/biolreprod.103.023911 (2004).
Nishiwaki, K., Hisamoto, N. & Matsumoto, K. A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans. Science 288, 2205–2208. https://doi.org/10.1126/science.288.5474.2205 (2000).
Kubota, Y., Kuroki, R. & Nishiwaki, K. A fibulin-1 homolog interacts with an ADAM protease that controls cell migration in C. elegans. Curr. Biol. 14, 2011–2018. https://doi.org/10.1016/j.cub.2004.10.047 (2004).
Kubota, Y., Nagata, K., Sugimoto, A. & Nishiwaki, K. Tissue architecture in the Caenorhabditis elegans gonad depends on interactions among fibulin-1, type IV collagen and the ADAMTS extracellular protease. Genetics 190, 1379–1388. https://doi.org/10.1534/genetics.111.133173 (2012).
Kubota, Y., Ohkura, K., Tamai, K. K., Nagata, K. & Nishiwaki, K. MIG-17/ADAMTS controls cell migration by recruiting Nidogen to the basement membrane in C. elegans. Proc. Natl. Acad. Sci. U S A. 105, 20804–20809. https://doi.org/10.1073/pnas.0804055106 (2008).
Imanishi, A. et al. Genetic interactions among ADAMTS metalloproteases and basement membrane molecules in cell migration in Caenorhabditis elegans. PLoS One. 15, e0240571. https://doi.org/10.1371/journal.pone.0240571 (2020).
Ban, N. et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 24, 165–169. https://doi.org/10.1016/j.sbi.2014.01.002 (2014).
Kang, J. et al. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal. Transduct. Target. Ther. 6, 323. https://doi.org/10.1038/s41392-021-00728-8 (2021).
Nishiwaki, K. Mutations affecting symmetrical migration of distal tip cells in Caenorhabditis elegans. Genetics 152, 985–997 (1999).
Kim, H. S. et al. The novel secreted factor MIG-18 acts with MIG-17/ADAMTS to control cell migration in Caenorhabditis elegans. Genetics 196, 471–479. https://doi.org/10.1534/genetics.113.157685 (2014).
Hesselson, D., Newman, C., Kim, K. W. & Kimble, J. GON-1 and Fibulin have antagonistic roles in control of organ shape. Curr. Biol. 14, 2005–2010. https://doi.org/10.1016/j.cub.2004.11.006 (2004).
Suzuki, N., Toyoda, H., Sano, M. & Nishiwaki, K. Chondroitin acts in the guidance of gonadal distal tip cells in C. elegans. Dev. Biol. 300, 635–646. https://doi.org/10.1016/j.ydbio.2006.08.037 (2006).
Nukazuka, A., Fujisawa, H., Inada, T., Oda, Y. & Takagi, S. Semaphorin controls epidermal morphogenesis by stimulating mRNA translation via eIF2alpha in Caenorhabditis elegans. Genes Dev. 22, 1025–1036. https://doi.org/10.1101/gad.1644008 (2008).
Nukazuka, A. et al. A shift of the TOR adaptor from rictor towards raptor by semaphorin in C. elegans. Nat. Commun. 2, 484. https://doi.org/10.1038/ncomms1495 (2011).
Wong, A., Boutis, P. & Hekimi, S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics 139, 1247–1259. https://doi.org/10.1093/genetics/139.3.1247 (1995).
Lakowski, B. & Hekimi, S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272, 1010–1013. https://doi.org/10.1126/science.272.5264.1010 (1996).
Benard, C. et al. The C. elegans maternal-effect gene clk-2 is essential for embryonic development, encodes a protein homologous to yeast Tel2p and affects telomere length. Development 128, 4045–4055. https://doi.org/10.1242/dev.128.20.4045 (2001).
Ahmed, S., Alpi, A., Hengartner, M. O. & Gartner, A. C. Elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 11, 1934–1944. https://doi.org/10.1016/s0960-9822(01)00604-2 (2001).
Guo, Y., Tocchini, C. & Ciosk, R. CLK-2/TEL2 is a conserved component of the nonsense-mediated mRNA decay pathway. PLoS One. 16, e0244505. https://doi.org/10.1371/journal.pone.0244505 (2021).
Robert, V. J., Sijen, T., van Wolfswinkel, J. & Plasterk, R. H. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19, 782–787. https://doi.org/10.1101/gad.332305 (2005).
Tamai, K. K. & Nishiwaki, K. bHLH transcription factors regulate organ morphogenesis via activation of an ADAMTS protease in C. elegans. Dev. Biol. 308, 562–571. https://doi.org/10.1016/j.ydbio.2007.05.024 (2007).
Okkema, P. G., Harrison, S. W., Plunger, V., Aryana, A. & Fire, A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135, 385–404. https://doi.org/10.1093/genetics/135.2.385 (1993).
Fukushige, T., Hawkins, M. G. & McGhee, J. D. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198, 286–302 (1998).
Keeley, D. P. et al. Comprehensive Endogenous Tagging of Basement Membrane Components Reveals Dynamic Movement within the Matrix Scaffolding. Dev Cell 54, 60–74 e67, (2020). https://doi.org/10.1016/j.devcel.2020.05.022
Muriel, J. M., Dong, C., Hutter, H. & Vogel, B. E. Fibulin-1 C and Fibulin-1D splice variants have distinct functions and assemble in a hemicentin-dependent manner. Development 132, 4223–4234. https://doi.org/10.1242/dev.02007 (2005).
Teng, T., Thomas, G. & Mercer, C. A. Growth control and ribosomopathies. Curr. Opin. Genet. Dev. 23, 63–71. https://doi.org/10.1016/j.gde.2013.02.001 (2013).
Lezzerini, M. et al. Ribosomal protein gene RPL9 variants can differentially impair ribosome function and cellular metabolism. Nucleic Acids Res. 48, 770–787. https://doi.org/10.1093/nar/gkz1042 (2020).
Oliver, E. R., Saunders, T. L., Tarle, S. A. & Glaser, T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse minute. Development 131, 3907–3920. https://doi.org/10.1242/dev.01268 (2004).
Kondrashov, N. et al. Ribosome-mediated specificity in hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397. https://doi.org/10.1016/j.cell.2011.03.028 (2011).
Levy-Strumpf, N. & Culotti, J. G. Netrins and Wnts function redundantly to regulate antero-posterior and dorso-ventral guidance in C. elegans. PLoS Genet. 10, e1004381. https://doi.org/10.1371/journal.pgen.1004381 (2014).
Levy-Strumpf, N., Krizus, M., Zheng, H., Brown, L. & Culotti, J. G. The Wnt frizzled receptor MOM-5 regulates the UNC-5 Netrin receptor through small GTPase-Dependent signaling to determine the Polarity of migrating cells. PLoS Genet. 11, e1005446. https://doi.org/10.1371/journal.pgen.1005446 (2015).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Maduro, M. & Pilgrim, D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141, 977–988 (1995).
Wicks, S. R., Yeh, R. T., Gish, W. R., Waterston, R. H. & Plasterk, R. H. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28, 160–164. https://doi.org/10.1038/88878 (2001).
MacNeil, L. T., Watson, E., Arda, H. E., Zhu, L. J. & Walhout, A. J. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell 153, 240–252. https://doi.org/10.1016/j.cell.2013.02.049 (2013).
Ihara, S. & Nishiwaki, K. Prodomain-dependent tissue targeting of an ADAMTS protease controls cell migration in Caenorhabditis elegans. EMBO J. 26, 2607–2620. https://doi.org/10.1038/sj.emboj.7601718 (2007).
Saijou, E., Fujiwara, T., Suzaki, T., Inoue, K. & Sakamoto, H. RBD-1, a nucleolar RNA-binding protein, is essential for Caenorhabditis elegans early development through 18S ribosomal RNA processing. Nucleic Acids Res. 32, 1028–1036. https://doi.org/10.1093/nar/gkh264 (2004).
Fujiwara, T., Ito, K. & Nakamura, Y. Functional mapping of ribosome-contact sites in the ribosome recycling factor: a structural view from a tRNA mimic. RNA 7, 64–70. https://doi.org/10.1017/s1355838201001704 (2001).
Simsek, D. & Barna, M. An emerging role for the ribosome as a nexus for post-translational modifications. Curr. Opin. Cell. Biol. 45, 92–101. https://doi.org/10.1016/j.ceb.2017.02.010 (2017).
Acknowledgements
We thank Nami Okahashi, Noriko Nakagawa and Asami Sumitani for technical assistance. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
Funding
This work was supported by a Grant-in-Aid for Challenging Research by Ministry of Education, Culture, Sports, Science and Technology to KN (26650086).
Author information
Authors and Affiliations
Contributions
H.-S.K. and K.N. designed research; H.-S.K., K.M., S.K., R.Y., Y.S., S.I., T.F., H.O., C.Y., K.K., F.M., M.T., Y.K. and T.T. performed research and analyzed data; H.-S.K. and K.N. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, HS., Mitsuzumi, K., Kondo, S. et al. Ribosomal protein mutation suppresses gonadal leader cell migration defects in mig-17/ADAMTS mutants in Caenorhabditis elegans. Sci Rep 15, 26435 (2025). https://doi.org/10.1038/s41598-025-10316-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10316-3