Abstract
The cranial base synchondrosis (CBS) is a critical growth center in the craniofacial region, and its abnormal development can lead to various craniofacial deformities. In a hypoxic microenvironment, hypoxia-inducible factor-1α (HIF-1α) is a crucial regulatory factor for cellular adaptation to low oxygen conditions. However, the role of HIF-1α in the CBS and its mechanisms regulating the function of chondrocytes remain unclear. This study aims to investigate the expression characteristics of HIF-1α in the CBS and its potential mechanisms in regulating mouse spheno-occipital synchondrosis (SOS) chondrocytes (SOSCs). Histological and immunohistochemical staining were utilized to observe the growth pattern of the SOS and the expression characteristics of HIF-1α in the SOS of 1–8 week-old mice. Chemical hypoxia simulation and siRNA technology modulated HIF-1α expression, and potential signaling pathways were detected through transcriptome sequencing. Results indicate that HIF-1α is expressed in all layers of the mouse SOS and is closely associated with cell proliferation and differentiation. In vitro studies demonstrate that enhancing HIF-1α expression enhances cell proliferation and matrix synthesis capacity, improves cell apoptosis, and enhances the expression of chondrogenic markers SOX9 and Collagen II while diminishing osteogenic marker RUNX2 expression. We found that with the upregulation of HIF-1α expression, the PI3K/Akt signaling pathway is activated. In conclusion, our study revealed that HIF-1α regulates the proliferation and differentiation of SOSCs by activating the PI3K/Akt signaling pathway.
Similar content being viewed by others
The cranial base plays a crucial role in the development of craniofacial morphology and facial skeleton. Longitudinal growth and development of the cranial base are closely related to craniofacial morphology, with abnormalities in anterior cranial base development often accompanied by midface hypoplasia, while the length and inclination of the posterior cranial base affect the position of the mandible1. In recent years, there has been increasing attention and recognition of the integrative role of the cranial base in craniofacial growth and the impact of cranial base growth on craniofacial deformities. Therefore, exploring the growth and development characteristics as well as regulatory mechanisms of the cranial base is of paramount importance for understanding craniofacial skeletal development and craniofacial deformities.
Persistent cartilaginous segments existing between ossification centers in the cranial base represent various cartilaginous junctions, among which the cranial base synchondrosis (CBS) is a crucial growth center driving longitudinal growth of the cranial base2,3. According to anatomical location, three types of cartilaginous junctions exist in the cranial base from anterior to posterior: spheno-ethmoidal synchondrosis (SES), intersphenoid synchondrosis (ISS), and spheno-occipital synchondrosis (SOS). There is considerable variation in the ossification timing of these three cranial base cartilage junctions, the ISS ossifies at 2–3 years of age, whereas the SOS ossifies much later between 16 and 18 years, coinciding with the cessation of peak growth in humans4,5. SOS has received almost exclusive attention in clinical studies. The CBS originates from endochondral ossification, a fundamental process in hard tissue biology where cartilaginous primordia are gradually replaced by mature bone. Unlike the unidirectional growth of long bones, the cranial base cartilage junction consists of bipolar growth plates, with a resting zone in the center flanked by proliferative and hypertrophic zones on either side, thus exhibiting bidirectional growth. However, the cranial synchondrosis is different from the long bone growth plate in the following aspects. First, given the orientation of chondrocytes, the synchondrosis produces growth in opposing directions. Second, the synchondrosis is not overlaid by an articular synovial layer, which is typical of the growth plate in developing long bones. Third, the secondary ossification center is absent in the cranial synchondrosis. Fourthly, although both long bones and the cranial base develop through endochondral ossification, key signaling pathways regulating bone elongation exhibit functional divergence at cranial base synchondroses versus long bone growth plates6,7. Investigating the growth and developmental mechanisms of the cranial base cartilage junction is of significant importance and value for understanding craniofacial skeletal development anomalies and their clinical treatment strategies.
Hypoxia-inducible factor-1α (HIF-1α) as a transcription factor stabilized and enhanced in low-oxygen environments, is essential for the survival of chondrocytes8,9. It forms a heterodimer comprising α and β subunits, with the α subunit being the primary functional regulator of HIF-1, particularly under hypoxic conditions10,11. Due to the lack of a vascular system, the cranial base cartilage junction resides in a hypoxic microenvironment, prompting cells to activate different molecular mechanisms to adapt to their surroundings. Previous studies have indicated that HIF-1α metabolism can regulate collagen synthesis and modification in chondrocytes12. In summary, chondrocytes and cartilage cells exist in a physiologically hypoxic environment, and the regulation of chondrocyte survival and metabolism under hypoxia has been a focus of research.
In this study, we aim to investigate the expression characteristics of HIF-1α in the mouse spheno-occipital synchondrosis (SOS) in vivo and explore the effects of HIF-1α enhancement or silencing on the biological properties of SOSCs in vitro, along with potential pathways, to provide a theoretical basis for further understanding the growth regulatory mechanisms of the cranial base cartilage junction.
Materials and methords
Animals
All the animal experiments were approved by the Ethics Committee of the Animal Laboratory of Kunming Medical University (no. Kmmu20231512). The Laboratory Animal Centre at Kunming Medical University provided 1, 2, 4, 6, and 8-week-old, healthy female C57BL/6J mice. The mice were maintained in a barrier environment throughout the experiments and fed conventional food and water. The testing was conducted in line with the ARRIVE recommendations, and all methods were carried out in accordance with relevant guidelines and regulations.
Micro‑CT analysis
Tissue blocks from the skull base of 1, 2, 4, 6, and 8-week-old C57BL/6J mice were collected and fixed with 4% paraformaldehyde for 24 h. Subsequently, micro-CT scans were performed with the scanning conditions set to 90 kV,60 µA tube flow. The acquired images were reconstructed and analyzed using Avatar 1.50 software (Pingsheng Medical, China).
Histomorphological examination
Tissue blocks from the skull base of 1, 2, 4, 6, and 8 weeks C57BL/6J mice were collected and fixed with 4% paraformaldehyde for 24 h. The fixed specimens were rinsed with running water for 24 h. The decalcification solution was used to decalcify the specimens for 1–4 weeks until a fine needle could penetrate the tissue specimens without resistance indicating that the decalcification was completed. The specimens were rinsed with running water overnight and subjected to gradient alcohol dehydration, and paraffin sections were prepared and subsequently stained with histological morphology. Samples were cut into 5 mm sections using a microtome. Sections(Samples) were stained with hematoxylin-eosin stain(H&E), Alcian Blue & Nuclear Fast Red staining, and Safranin O/Fast green staining according to the manufacturer’s instructions. Chondrocyte and matrix structures were visualized using Hematoxylin-Eosin stain staining, and cartilage matrix proteoglycans were visualized using Alcian Blue & Nuclear Fast Red staining, and Safranin O &Fast green staining.
Immunohistochemistry
The skull base tissue blocks were routinely paraffin-embedded, and immunohistochemical staining was performed on the sections. The sections were hydrated by double dewaxing with xylene and graded ethanol and washed three times with PBS. The sections were thermally repaired at high temperature with EDTA anti-repair solution, cooled to room temperature, and then blocked and sealed with 3% endogenous hydrogen peroxide blocking agent and 10% normal goat serum sealing solution, respectively. The sections were washed three times in PBS, and incubated overnight at 4 °C with primary antibodies specific for HIF-1α (Abcam, USA, 1:500); Next day, the sections were incubated with the HRP-coupled secondary antibody at 37 °C for 20 min. The reaction was visualized by incubating the sections with a DAB kit (Fuzhou Maixin). After air-drying, the slices were sealed with neutral resin and photographed under the microscope. The number and area of positive cells were analyzed by Image-Pro Plus 6.0 (National Institutes of Health, Bethesda, MD).
Isolation and characterization of SOSCs
This study was approved by the Research Ethics Committee of the Hospital of Stomatology, Kunming Medical University. Primary spheno-occipital synchondrosis chondrocytes were isolated from the spheno-occipital synchondrosis of 1-day-old C57BL/6 mice(n = 6). These mice were euthanized by over-injection of anesthetics (0.3% pentobarbital sodium, Sigma); Briefly, the spheno-occipital synchondrosis tissue block of the skull base was excised under aseptic conditions under direct visualization of the intracranial surface of cranial base, the cartilage block was rinsed three times in PBS, chopped to approximately 1 mm3 in size, and then collected into centrifugal tubes filled with PBS. The cells were digested with 0.25% trypsin at 37 °C for 15 min and 0.1% type II collagenase at 37 °C for 2 h. The cells were routinely cultured at 37 °C in culture flasks supplemented with 10% FBS(Gibco) and 0.1% DMEM/F12 (Gibco) medium with penicillin/streptomycin (Gibco), and the medium was changed every 3 days, and photos were taken to record the growth status, and cells of the 2nd-4th generations were used for subsequent experiments.
Generation 3 chondrocytes were collected for characterization. Morphological staining of chondrocytes was performed, cells were cultured in 24-well plates (1*104 cells per well), cells were fixed with 4% paraformaldehyde when the density of cells grew to 80%, followed by staining with, Alcian Blue, Toluidine Blue, and Safranin O according to the manufacturer’s instructions, and the staining was observed under an inverted microscope. Immunofluorescence staining was used to detect cell surface markers, cells were inoculated in confocal dishes at a density of 1*104 cells/well, fixed with 4% paraformaldehyde when the cells grew to 80%, washed with PBS and stained with collagen II according to the antibody instruction for dilution ratios: Collagen II (Santa Cruz, 1:200), and then stained with fluorescent goat anti-mouse di-pit (Abbkine, 1:250) for color development. Cell nuclei were stained with a DAPI solution. Subsequently, the cells were observed under an inverted microscope and photographed for image-taking. The growth curve of SOSCs was observed using CCK-8 assay. The cells were routinely digested to adjust the concentration and then inoculated in 96-well plates with a cell density of 3*103 cells/well. After overnight starvation, each well was incubated with 10 µL of CCK-8 solution with 90 µL of the serum-free medium at the same time points of the following 1–7 days for 1.5 h and then up-examined, and the OD value at 450 nm was measured on the enzyme counter in each well, and the growth curves were plotted after the calculation of the mean number.
RNA-sequencing and bioinformatics analysis
The differential gene expression analysis was performed using DEGseq under the condition of P-value threshold of < 0.05 and |log2(fold change)|> 1. Further analysis using the Gene Ontology (GO) database and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database was conducted using gplots package in R software 3.5.0.
The 3rd generation chondrocytes (1 *105 cells/well) were inoculated in 6-well plates, grown to 80% confluence, and the medium was changed according to different groups (Control: normal medium; CoCL2 group: a medium group containing 100 µM/LCoCL2; siRNA group: a medium group of siHIF-1α + 100 µM cobalt chloride), and cultured for 24 h. Total RNA was extracted from the cells in each group using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. After 24 h of incubation, total RNA was extracted from the cells of each group using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions.RNA sequencing and bioinformatics analysis were entrusted to NOVO ZYUAN. Sequencing results were further analyzed using R programming language and software, to identify differentially expressed genes (DEGs) and perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
Drug treatments and CCK-8 assay
When the SOSCs had reached 80% confluence, the culture medium was supplemented with different concentrations of cobalt chloride (0, 50, 10, 150, and 200 µM; Sigma‑Aldrich; Merck KGaA, Darmstadt, Germany) for the indicated times. Collection of cell samples for subsequent experiments.
To evaluate the effect of cobalt chloride on the viability of chondrocytes, the assay was performed with CCK-8 (Meilunbio, China) according to the manufacturer’s protocol. Briefly, cells were routinely digested and counted, adjusted the cell concentration, and then planted into 96-well plates(3,000 cells/well). After the cells were attached to the wall, the medium was replaced with DMEM/F-12 without FBS to starve the cells overnight. Then each well was replaced with DMEM/F12 medium containing different concentrations of cobalt chloride the next day. The cells were incubated in a 90 µL serum-free DMEM/F12 and 10 µL CCK-8 solution at 37℃ under protected light for 2 h. The OD value at 450 nm of each well was determined by an enzyme marker.
Immunofluorescence
The second-generation cells were inoculated in confocal dishes (1*105 cells/well) for routine culture, and when the cells grew to 80% confluence, the cells were replaced with different media (A: Control (normal medium); B: experimental group (DMEM/F12 medium with 100 µM/LCoCL2)) and cultured for 24 h. The cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X‑100 for 15 min, and blocked at room temperature with 0.1% BSA for 1 h. The cells were incubated with primary antibodies HIF-1α (Abcam,1:200), SOX9 (Abcam,1:200), and RUNX2 (CST,1:200) overnight at 4 °C; The cells were rinsed three times with PBS and incubated with the corresponding secondary antibodies (Abkine,1:250). the cells were stained with DAPI solution for 5 min at 25 °C. Images were captured under light-protected conditions with an autofluorescence microscope. Fluorescence intensity was measured using Image J 1.51 software( National Institutes of Health, USA).
Cell cycle, apoptosis
To evaluate the impact of cobalt chloride (CoCL2) pretreatment on cell proliferation viability and apoptosis rate, cell cycle and apoptosis experiments were performed. Well-cultured SOSCs were subjected to routine cell digestion and counting. After adjusting the cell density, cells were seeded at a density of 1 × 10^5 cells per well in a six-well plate and treated with either regular DMEM/F12 medium or DMEM/F12 medium containing 100µM/L CoCL2. The cells were then conventionally cultured at 37 °C, 5% CO2 for 24 h. Subsequently, the cell cycle staining solution was prepared following the instructions. The working solution was formulated by combining staining buffer, propidium iodide (PI) staining solution (20X), and RNase A (50X) in a ratio of 100:5:2. Next, 500µL of the working solution was added to gently and thoroughly resuspend the cells, followed by incubation at 37 °C in the dark for 30 min. Apoptosis detection was performed using the Annexin V-FITC/PI double-staining apoptosis detection kit (Keygen). Cells were collected by digesting with trypsin without EDTA, and a single-cell suspension was prepared by adding 500µL of Binding Buffer and gently blowing to resuspend the cells. After mixing with 5µL Annexin V-FITC, 5µL Propidium Iodide was added and thoroughly mixed. The reaction was carried out at room temperature in the dark for 10 min. Finally, cell detection was performed using a flow cytometer (Agilent NovoCyte Flow Cytometer, Agilent Technologies).
Western blot analysis
The extraction of total protein from SOSCs was performed using RIPA lysis buffer (Solarbio, China), supplemented with 1%PMSF protease inhibitor (Solarbio, China). The protein concentration was measured using a BCA protein assay kit (Beyotime, China). Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a 0.45 μm polyvinylidene difluoride (PVDF) membrane (Millipore, USA). After blocking with rapid blocking solution for 30 min, the membranes were incubated overnight at 4 °C with primary antibodies against HIF-1α (Abcam, 1:5000), SOX9 (Abcam, 1:5000), Collagen II (ProteinTech, 1:1000), VEGF (Santa Cruz, 1:500), RUNX2 (Cell Signaling Technology, 1:1000), β-actin (Affinity, 1:10000), PI3K (HuaBio, 1:1000), p-Akt (HuaBio, 1:5000), and Akt (HuaBio, 1:5000). Subsequently, the membranes were incubated with corresponding secondary antibodies (ProteinTech, 1:10000). After washing with TBST, proteins on the membranes were detected using an electrochemiluminescence (ECL) detection kit. The images were captured by Image Lab (Bio-Rad) and analyzed using Image J 1.51 software( National Institutes of Health, USA).
Total RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA extraction was performed using the Total RNA Extraction Kit (TaKaRa MiniBEST Universal RNA Extraction Kit (Takara, Osaka, Japan) in strict accordance with the manufacturer’s instructions. Subsequently, cDNA synthesis was performed using the PrimeScript™ RT Master Mix (Perfect Real Time) (Takara). RT-qPCR analysis was performed on the system using a QuantStudio™ Real-Time.
PCR System (Thermo Fisher Scientific, USA) with the TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara)under the following conditions: Initial denaturation for 30 s at 95 °C; followed by 40 cycles of 95 °C for 5 s; melting at 60 °C for 34 s; and cooling at 4 °C for 30 s. The primer sequences used during amplification can be found in Table 1. Expression levels were determined based on threshold cycling (Ct) and relative expression levels were calculated using the 2-ΔΔCT method, with β-actin serving as the reference gene.
Statistical analysis
The data are presented as mean ± standard deviation (SD). Independent Student’s t-test and one-way analysis of variance (ANOVA) were employed for data following a normal distribution in this study. Statistical significance was set at p < 0.05. SPSS 20.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 8.3 (GraphPad Software Inc., La Jolla, CA, USA) were used for statistical analysis. All experiments were independently repeated at least three times.
Results
HIF-1α exhibited sustained expression within the spheno-occipital synchondrosis of mice aged 1–8 weeks and was closely associated with cell proliferation and differentiation
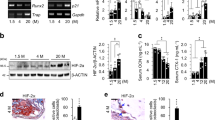
Micro-CT and histological staining including Hematoxylin and Eosin (HE), Safranin O-Fast green, and Alcian Blue-Nuclear Fast Red demonstrated that the spheno-occipital synchondrosis in mice consisted of mirrored bipolar growth plates, with a resting zone centrally, and proliferative and hypertrophic zones on either side. Clear cellular boundaries were observed between the layers of the synchondrosis in the 1 and 2-week-old groups, while boundaries between the proliferative and hypertrophic zones became indistinct in the 1, 6, and 8-week-old groups. With growth, the width of the spheno-occipital synchondrosis gradually decreased, eventually merging with the mineralized bone at both ends(Fig. 1A). Immunohistochemical staining revealed varying degrees of HIF-1α expression in the resting, proliferative, and hypertrophic zones of the spheno-occipital synchondrosis. Comparison of HIF-1α expression in different growth stages showed that positive brown staining for HIF-1α was predominantly observed in the cell nuclei. Among the 1–8 week-old groups, the expression intensity of HIF-1α was significantly higher in the 2-week-old group compared to the other groups (P < 0.05) (Fig. 1B, C). The expression levels of HIF-1α in different layers of the spheno-occipital synchondrosis also followed a certain pattern: in the resting zone, there was no significant difference in HIF-1α expression intensity between the 1 and 2-week-old groups, with a gradual decrease observed in the 4, 6, and 8-week-old groups; in the proliferative zone, no significant change in HIF-1α expression intensity was observed between the 1, 2, and 4-week-old groups, with an increase in expression observed in the 6-week-old group and a decrease in the 8-week-old group; in the hypertrophic zone, HIF-1α expression intensity gradually decreased with age, with no significant differences observed in the 4, 6, and 8-week-old groups (Fig. 1D). These findings suggest that HIF-1α may be involved in the early development of the mouse spheno-occipital synchondrosis and correlates with cell proliferation and differentiation.
The growth pattern of the sphenoid-occipital synchondrosis in 1–8 week-old mice and the expression characteristics of HIF-1α within it. (A) Micro-CT imaging, hematoxylin-eosin (H&E) staining, Safranin O & Fast green staining, and Alcian Blue & Nuclear Fast Red staining reveal the tissue morphological characteristics of the spheno-occipital synchondrosis in 1-8-week-old mice. Scale bar:5 mm、200 μm; (B) IHC staining of HIF-1α in the sphenoid-occipital synchondrosis of 1–8 week-old mice. Scale bar: 200 μm; (C,D) The quantitative expression of positive optical densities in IHC staining using the Image-Pro Plus software. n = 3, *p < 0.05; **p < 0.01; ***p < 0.001;#p < 0.0001;ns: no significance.
Enhancing the expression of HIF-1α promotes proliferation and extracellular matrix synthesis in SOSCs, improving apoptosis
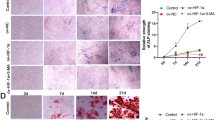
The present study utilized modified tissue block culture to cultivate SOSCs, which exhibited typical chondrocyte morphology (Fig. 2A). CCK-8 assays demonstrated robust proliferative activity of the cells (Fig. 2J). Toluidine blue staining, Safranin O staining, and Alcian Blue staining all exhibited positive results, while immunofluorescence revealed positive expression of type II collagen, indicating a rich extracellular matrix (Fig. 2B-I). Cobalt chloride intervention was employed to enhance HIF-1α expression in SOSCs, and CCK-8 assays demonstrated a significant enhancement in cell proliferation compared to the control group following treatment with 100µM cobalt chloride (P < 0.05) (Fig. 3A). A concentration of 100µM cobalt chloride was selected for subsequent experiments. Cell cycle analysis indicated an increased proportion of cells in the S phase and G2 phase of the cell cycle after enhancing HIF-1α expression in SOSCs, suggesting enhanced proliferative activity (Fig. 3B). Apoptosis assays revealed an increase in the proportion of normal cells and a decrease in the apoptosis rate following enhancement of HIF-1α expression, suggesting an improvement in apoptosis of SOSCs (Fig. 3C). Alcian Blue staining performed on chondrocyte micromasses cultured for 2 days revealed that intervention with 100µM CoCL2 to enhance HIF-1α expression can promote extracellular matrix synthesis (Fig. 3D-E).
Isolation, culture, and identification of SOSCs; (A) Schematic diagram of isolation and culture of mouse sphenooccipital synchondrosis cells; (B–D) First-generation SOSCs exhibit typical triangular or quadrilateral shapes; (E) Late-stage growth of SOSCs shows cobblestone-like arrangement; (F–H) Alcian blue staining, Safranin O staining, and toluidine blue staining demonstrate abundant proteoglycans in SOSCs; (I) Immunofluorescence staining of cells shows positive expression of type II collagen in the extracellular matrix of SOSCs; (J) CCK-8 assay detects the proliferation activity of SOSCs.
Enhanced expression of HIF-1α on the proliferation, apoptosis, and extracellular matrix of SOSCs; (A,B) CCK-8 and cell cycle experiments assess the impact of enhanced HIF-1α expression on the proliferation of SOSCs; (C) Apoptosis assay evaluates the effect of enhanced HIF-1α expression on the apoptosis of SOSCs; (D,E) Alcian blue staining examines the influence of enhanced HIF-1α expression on the extracellular matrix of SOSCs. Scale bar:1 cm. n = 3, ***P<0.001.
Enhanced expression of HIF-1α promotes maintenance of chondrogenic phenotype in SOSCs
Western blot experiments showed that treatment of SOSCs with CoCL2 in a time- and concentration-dependent manner resulted in a stable upregulation of HIF-1α protein expression levels, with subsequent changes in the expression of the downstream target gene VEGF. Additionally, varying concentrations of CoCL2 led to differential upregulation of chondrogenic markers such as SOX9 and Collagen II protein expression levels, while decreasing the expression level of the osteogenic marker RUNX2 (P < 0.05) (Fig. 4A–D). Immunofluorescence experiments demonstrated that intervention with 100µM CoCL2 enhanced HIF-1α expression in SOSCs, concomitantly upregulating SOX9 expression while downregulating RUNX2 expression (P < 0.05) (Fig. 4E,F).
Enhancing the expression of HIF-1α impacts the expression of proliferation and differentiation-related factors in SOSCs. (A,B) Expression changes of chondrogenic differentiation-related factors in SOSCs after 24 h of treatment with different concentrations of cobalt chloride, the original protein blot is shown in Supplementary Fig. 4A; (C,D) Expression changes of chondrogenic differentiation-related factors in SOSCs after treatment with 100µM cobalt chloride at different time points, the original protein blot is shown in Supplementary Fig. 4C; (E,F) Effects of enhanced HIF-1α expression on the expression changes of SOX9 and RUNX2. (The blue fluorescence corresponds to the nuclei of SOSCs stained with DAPI, while the red fluorescence indicates the target protein; ) n = 3, *P < 0.05 **P < 0.01, ***P < 0.001, #P < 0.0001,ns: no significance.
Silencing the expression of HIF-1α reduces the proliferation and extracellular matrix synthesis capabilities of SOSCs. HIF-1α regulates the proliferation and differentiation of SOSCs
The results of siRNA transfection experiments showed that the transfection efficiency was highest at a final concentration of 50nM. RT-qPCR and Western blot experiments demonstrated that siRNA3 had the highest transfection efficiency (P < 0.05) (Fig. 5A, B,C).CCK-8 assays revealed that inhibition of HIF-1α expression in SOSCs affected their proliferative activity (Fig. 5E). Alcian Blue staining results indicated that inhibition of HIF-1α expression affected extracellular matrix synthesis capability (Fig. 5D). Western blot and RT-qPCR experiments revealed that enhancing HIF-1α expression upregulated the expression levels of proliferation and differentiation-related factors SOX9, Collagen II, and VEGF proteins and genes in SOSCs while decreasing the expression levels of RUNX2 protein and gene (P < 0.05). Conversely, inhibition of HIF-1α expression downregulated the expression levels of SOX9, Collagen II, and VEGF proteins and genes in SOSCs, while enhancing the expression levels of RUNX2 protein and gene (P < 0.05) (Fig. 5F–H).
The effect of enhancing or inhibiting HIF-1α expression on SOSCs proliferation and differentiation; (A–C) Western blot and RT-qPCR to assess the transfection efficiency of siRNA, the original protein blot is shown in Supplementary Fig. 5A; (D) Alcian blue staining to examine the impact of inhibiting HIF-1α expression on the extracellular matrix of SOSCs; (E) CCK-8 assay to evaluate the effect of HIF-1α expression on SOSCs proliferation activity; (F,G) Western blot to investigate the effect of enhancing and inhibiting HIF-1α expression on the expression of chondrogenic differentiation-related proteins, the original protein blot is shown in Supplementary Fig. 5F. (H) RT-qPCR to determine the effect of enhancing and inhibiting HIF-1α expression on the expression of chondrogenic differentiation-related genes; n = 3, *P < 0.05;**P < 0.01; ***P < 0.001; #P < 0.0001; ns: no significance.
HIF-1α mediates the PI3K/Akt signaling pathway to regulate proliferation and matrix synthesis activities of mouse sphenoid-occipital synchondrosis cells
Following treatment according to group requirements and sample collection, RNA-seq, transcriptome sequencing, and bioinformatics analysis revealed a total of 8253 significantly differentially expressed genes between the enhanced and silenced groups, comprising 4142 upregulated genes and 4111 downregulated genes (Fig. 6A, B). KEGG analysis indicated that differentially expressed genes were mainly enriched in the PI3K/Akt signaling pathway (Fig. 6C)13,14,15. Further validation through Western blot experiments revealed that in the enhanced group, the protein expression levels of HIF-1α, PI3K, and p-Akt were upregulated, whereas they were downregulated in the silenced group, with no change observed in the expression level of total Akt protein (Fig. 6D,E). After the inhibition of AKT expression using AKT inhibitor, the results of western blot experiments showed that with the inhibition of p-AKT expression, the expression of PI3K in the MK group was weakened compared with that in the control group, indicating that the PI3K/AKT pathway was successfully blocked in the experiments. Compared with the control group, the protein expression levels of Collagen II and SOX9, which are factors related to the proliferation and differentiation of chondrocytes, were weakened in the MK group, whereas with the addition of CoCL2, with the enhancement of the protein expression level of HIF-1α, the expression levels of Collagen II and SOX9 were also enhanced; for the expression of VEGF, after blocking the PI3K/AKT pathway, the expression level of VEGF was reduced. pathway, no change in the protein expression level of VEGF was observed, whereas in the CoCL2-only group, it was significantly higher compared with the control and MK groups; changes in RUNX2, an osteogenesis-related assay, were reduced in the MK group compared with the control group, and the trend of reduction was more pronounced in the MK + CoCL2 group (Fig. 6F, G). To further analyze the changes in cell proliferation viability after blocking the PI3K/AKT pathway, EDU experiments were performed, and the results showed that the proliferation viability of cells was significantly reduced after blocking the PI3K/AKT pathway (Fig. 6H, I). Overall, the proliferative vigor of SOSCs was significantly reduced when the PI3K/AKT pathway was blocked, and the addition of CoCL2 to enhance the expression of HIF-1α slightly reversed this situation.
Impact of enhancing or inhibiting HIF-1α expression on the PI3K/Akt signaling pathway; (A–C) Transcriptomic sequencing data; A: Heatmap illustrating gene expression changes in SOSCs (Stem-like osteosarcoma cells) following treatment with cobalt chloride and siRNA. B: Gene Ontology (GO) classification of genes with differential expression in SOSCs treated with cobalt chloride and siRNA. C: Kyoto Encyclopedia of Genes and Genomes (KEGG) classification of genes with distinct expression patterns in SOSCs treated with cobalt chloride and siRNA. (D,E) Western blot analyses assessing the expression changes of PI3K and Akt in SOSCs after enhancement or inhibition of HIF-1α expression, along with quantitative analysis of protein expression levels, the original protein blot is shown in Supplementary Fig. 6D. (F,G) Western blot analysis was performed to examine the expression changes of proliferation- and differentiation-related proteins in SOSCs following inhibition of AKT expression, and protein expression levels were quantitatively analyzed, the original protein blot is shown in Supplementary Fig. 5F. (H,I) EDU assay was conducted to assess the proliferation activity of SOSCs after inhibition of AKT expression. n = 3, *p < 0.05; **p < 0.01; ***p < 0.001; #p < 0.0001; ns: no significance.
Discussion
Extensive research has elucidated the mechanistic role of HIF-1α in bone and cartilage development. However, most investigations have focused predominantly on long bone growth plates and mandibular condylar cartilage, where conditional knockout of HIF-1α induces proliferative and metabolic disturbances in chondrocytes. These disruptions lead to skeletal dysplasia and subsequent developmental complications. In contrast, the function of HIF-1α in murine cranial base synchondroses remains poorly understood, with no relevant phenotypic manifestations reported to date16,17,18.
In this study, we found that HIF-1α is expressed in all layers of the murine spheno-occipital synchondrosis from 1 to 8 weeks of age, with the highest levels localized in the resting zone at 2 weeks of age. By modulating the expression of HIF-1α in vitro in murine sphenoid-occipital synchondrosis chondrocytes through enhancement and silencing techniques, we demonstrated that HIF-1α can promote the proliferation and matrix synthesis capacity of these cells. Furthermore, transcriptomic sequencing and bioinformatics analysis revealed that HIF-1α can regulate the proliferation and matrix synthesis activities of murine sphenoid-occipital synchondrosis cells by activating the PI3K/Akt signaling pathway.
Chondrocyte ossification within the cranial base is a complex and finely regulated process involving the coordination of multiple cytokines19. Studies have indicated that HIF-1α regulates collagen synthesis and modification during chondrocyte differentiation, playing roles in chondrocyte survival, energy metabolism, cartilage formation, and endochondral ossification12,20,21. This study observed the growth characteristics of the murine spheno-occipital synchondrosis (SOS) at different developmental stages from 1 to 8 weeks through histomorphological staining, followed by exploration of the expression pattern of HIF-1α within. It was found that HIF-1α is expressed to varying degrees in all layers of the murine SOS from 1 to 8 weeks of age, with the highest levels localized in the resting zone at 2 weeks of age. In the 1-2-week-old groups, HIF-1α expression intensity was significantly pronounced, gradually decreasing in all layers with increasing mouse age, except for the proliferative layer at 6 weeks. Analysis of HIF-1α expression patterns in murine SOS indicates its multifaceted roles in cranial base cartilage growth. Not only does it facilitate the induction of undifferentiated mesenchymal cells into resting zone cells, but its expression is also evident in hypertrophic zone cells, suggesting its regulatory role in chondrocyte maturation and differentiation. This underscores the importance of HIF-1α expression in maintaining the phenotype and normal growth and development of the murine SOS during active stages of proliferation and differentiation.
The proliferation and differentiation of chondrocytes are influenced by various intrinsic and extrinsic factors. The cranial base cartilaginous joints develop through endochondral ossification, wherein chondrocytes undergo proliferation, hypertrophy, and differentiation, followed by apoptosis, and eventual replacement by mature bone. Throughout this intricate process, HIF-1α orchestrates a series of responses to ensure the smooth progression of chondrocyte differentiation, including regulation of SOX9, Collagen II, VEGF, and RUNX2 expression22,23. SOX9 primarily functions in the early proliferation and differentiation of chondrocytes, being a crucial factor necessary for cartilage formation. It promotes chondrocyte survival and can activate relevant regulatory factors24. HIF-1α can enhance proliferation, differentiation, and synthetic metabolic capacity of chondrocytes in vivo by upregulating SOX9 expression, thereby maintaining cartilage phenotype25,26. Collagen II, an essential extracellular matrix secreted by chondrocytes, exhibits specific expression in chondrocytes, with its expression increasing with increased matrix synthesis metabolism and vice versa. Literature suggests that Collagen II expression during chondrogenesis is directly regulated by SOX9 protein27,28, with its expression in chondrocytes positively correlated with SOX9, reflecting the differentiation status of chondrocytes29. In contrast to the initiation of chondrogenesis by SOX9, RUNX2 is typically considered a major driver in the late stages of chondrocyte ossification30. RUNX2, a member of the essential RUNX gene family for organogenesis and survival, is primarily expressed in pre-hypertrophic chondrocytes and maintained until terminal hypertrophic differentiation31. Studies report that RUNX2-/- mice lose the ability to form osteoblasts and bone tissue, with inhibition of chondrocyte maturation32. Additionally, it has been indicated that expression of HIF-1α and RUNX2 increases simultaneously in hypertrophic chondrocytes, where RUNX2 protein transcriptionally induces HIF-1α expression, accelerating ossification activity33. As chondrocytes mature and blood vessels invade, hypertrophic chondrocytes express VEGF, promoting endochondral ossification. VEGF, a downstream gene of HIF-1α, is the most critical factor in angiogenesis, widely recognized for its role in treating inflammatory diseases (including osteoarthritis) and malignancies34. Studies by J. YU et al. reported35that mechanical pressure can simultaneously enhance HIF-1α and VEGF expression in juvenile rat condylar chondrocytes. Literature suggests that in ischemic osteonecrosis of the femoral head, part of the upregulation of VEGF in chondrocytes is mediated by HIF-1α. Our study, through in vitro modulation of HIF-1α expression, found that cobalt chloride enhances HIF-1α expression, promoting the proliferation and matrix synthesis capacity of murine spheno-occipital synchondrosis cells. Cobalt chloride is commonly used in vitro to simulate cellular hypoxic microenvironments36, with its mechanism widely recognized as Co2+ substituting for Fe2+ in prolyl hydroxylases (PHDs), preventing degradation of hypoxia-inducible factors under normoxic conditions37. Although the extent to which cobalt chloride can mimic hypoxia-induced hypoxia models in terms of cellular metabolism, gene transcription, etc., is not fully understood, it is a reliable model capable of stabilizing and enhancing HIF-1α expression in cells.
The maintenance of cartilage homeostasis is a dynamic equilibrium process, wherein the growth and differentiation of chondrocytes predominantly bifurcate into chondrogenic and osteogenic pathways. In the enhancement group, chondrogenic differentiation of synchondrosis cells gains a favorable advantage over osteogenic activity, whereas in the silencing group, cells primarily undergo osteogenic activity. Worth discussing is the inconsistent expression of osteogenic markers VEGF and RUNX2 following modulation of HIF-1α expression. Considering possible reasons, there exists a dynamic balance between the expression of RUNX2 and SOX9 during cartilage formation and development. SOX9, by reducing β-catenin signaling and RUNX2 expression, maintains chondrocyte proliferation, delays hypertrophic differentiation, and subsequently prevents chondrocyte osteogenic differentiation38. When SOX9 expression in chondrocytes is inhibited, chondrocyte apoptosis occurs, accompanied by increased osteogenic differentiation, whereas enhanced SOX9 expression restricts RUNX2-induced endochondral ossification activity. Additionally, a large body of literature has identified VEGF as a downstream target gene of HIF-1α, with its expression changes largely consistent with those of HIF-1α. This aligns with the expression of VEGF by hypertrophic layer chondrocytes with chondrocyte maturation and vascular invasion, promoting endochondral ossification and other activities. In summary, we find that the role of HIF-1α in murine spheno-occipital synchondrosis cells primarily manifests in regulating cell proliferation and differentiation, promoting extracellular matrix synthesis, and facilitating chondrocyte survival and maintenance of the cartilage phenotype.
Previous studies have found that HIF-1α regulates the normal synthesis, degradation metabolism, and glycolysis of chondrocytes to promote normal development of cartilage tissue, with multiple signaling pathways and cellular molecules involved. Y Tang et al.18discovered that HIF-1α promotes mandibular condylar cartilage growth through AMPK signaling mediated osteoclast induction. Keita Okada et al.39reported that HIF-1α can maintain mouse articular cartilage phenotype by inhibiting the NF-κB signaling pathway. Studies have confirmed that HIF-1α and Notch intracellular domain (Notch ICD) can directly interact to regulate chondrocyte proliferation, and differentiation, promote the transition from pre-hypertrophic chondrocytes to hypertrophic chondrocytes, maintain chondrocyte phenotype, and regulate energy metabolism40. This study, through transcriptomic sequencing and bioinformatics analysis, found that differentially expressed genes between the enhancement and silencing groups are mainly enriched in the PI3K/Akt pathway. HIF-1α can regulate the proliferation and matrix synthesis activities of murine spheno-occipital synchondrosis cells by activating the PI3K/Akt signaling pathway. The PI3K/Akt signaling pathway is a classical protein kinase signaling pathway, which plays extremely important biological functions in cell growth, survival, proliferation, apoptosis, angiogenesis, autophagy, and other processes41. Studies have found that in regulating chondrocyte proliferation and differentiation, fluoride can regulate chondrocyte proliferation and autophagy through the PI3K/Akt/mTOR signaling pathway, and blocking activation of the PI3K/Akt pathway can inhibit chondrocyte hypertrophy42. Qifei Wang et al.43found that the PI3K/Akt pathway promotes cell proliferation through anaerobic glycolysis. Jia Gu Huang et al.44found that estrogen promotes chondrocyte proliferation in a rat osteoarthritis model through the PI3K/Akt pathway. Yan-Lin Chen et al.45 found that the knockdown of PTEN promotes the expression of extracellular matrix and proteoglycans in chondrocytes by increasing Akt phosphorylation under oxidative stress. Previous studies also indicate that HIF-1α is regulated by the PI3K/Akt signaling pathway46. Under hypoxic conditions, the PI3K/HIF pathway plays an important role in cardiac protection and neuroprotection. HIF-1α expression significantly increases in various ischemic organs and tissues, including the myocardium, nervous system, and retina47. p-Akt and HIF-1α protein levels show increased responses to hypoxia in human mesenchymal stem cells. Moreover, the peak expression of p-Akt precedes that of HIF-1α. To further verify the specific role of the PI3K/Akt signaling pathway in the spheno-occipital synchondrosis (SOSCs), we inhibited PI3K/Akt signaling using the Akt inhibitor MK2206. The experimental results demonstrated that blocking PI3K/Akt signaling significantly reduced the proliferative activity of SOSCs. However, the addition of CoCL2 to enhance HIF-1α expression partially reversed the decrease in cell proliferation induced by PI3K/Akt pathway blockade. Therefore, we speculate that HIF-1α can regulate the proliferation, differentiation, and matrix synthesis activities in the murine spheno-occipital synchondrosis partially through the PI3K/Akt signaling pathway. Nevertheless, under hypoxic conditions, the regulatory mechanisms of HIF-1α on chondrocytes exhibit multifactorial complexity. Furthermore, given the unique microenvironment of the cranial base, the role of HIF-1α within it shares both similarities and differences with its role in chondrocytes located in other parts of the body. This warrants further investigation.
Through transcriptome sequencing and bioinformatics analysis, a large number of differentially expressed genes were obtained from the sequencing results. We screened the top-ranked genes, mainly including Gipr, A2m, Rab39, Smtnl2, Krt19, Trpm2, Sncb, Ascl2, Jakmip3, Egln3, etc. By constructing Venn diagrams of differentially expressed genes with genes in the PI3K pathway and Hypoxia pathway from the GSEA database, a new target gene SLC2A1 was screened. Previous studies have reported a close relationship between SLC2A1 and HIF-1α in articular cartilage, and these data indicate the direction for subsequent research.
Subsequently, we performed Gene Ontology (GO) enrichment analysis to classify significantly differentially expressed genes according to Molecular Function (MF), Cellular Component (CC), and Biological Process (BP). MF analysis showed that these differentially expressed genes were involved in regulating biological processes such as transferase activity (transferring phosphorus-containing groups) and ubiquitin-like protein transferase activity. BP analysis revealed their participation in cellular protein modification process, protein modification process, DNA replication, and phosphate-containing compound metabolic process. CC analysis indicated that these genes were involved in regulating membrane-bounded organelle, nucleus, endomembrane system, etc.
KEGG pathway enrichment analysis further showed that differentially expressed genes were mainly enriched in PI3K/Akt, Human papillomavirus infection, Focal adhesion, and other pathways. In this study, we only investigated one related signaling pathway, which is clearly insufficient. For other signaling pathways and significantly changed differentially expressed genes, we will continue to explore them in future research.
Conclusion
Finally, through observation and modulation of HIF-1α expression in mouse sphenoid-occipital synchondrosis, we found that HIF-1α can regulate the proliferation and matrix synthesis activities of mouse sphenoid-occipital synchondrosis chondrocytes by activating the PI3K/Akt signaling pathway. The research findings provide a theoretical basis for further elucidating the regulatory mechanisms of endochondral ossification within cranial base cartilaginous joints. However, this section of the study also has certain limitations. Considering that the growth and development of cranial base cartilaginous joints and their endochondral ossification involve multiple pathways and coordinated actions of signaling factors, there exists a complex interplay among these pathways. Therefore, whether the regulatory function of HIF-1α on synchondrosis cells and its role in synchondrosis development intersect with other pathways and cellular factors requires further investigation in future studies.
Data availability
The datasets generated and/or analysed during the current study are available in the https://dataview.ncbi.nlm.nih.gov/object/PRJNA1221781?reviewer=jj3b71pv6o164l1c04u984b0ud, PRJNA1221781.
References
O’Rahilly, R. Guide to the staging of human embryos. Anat. Anz. 130(5), 556–559 (1972).
Mankarious, L. A. & Goudy, S. L. Craniofacial and upper airway development. Paediatr. Respir Rev. 11(4), 193–198 (2010).
Noden, D. M. Interactions and fates of avian craniofacial mesenchyme. Development. 103(Suppl), 121–140 (1988).
Noden, D. M. The embryonic origins of avian cephalic and cervical muscles and associated connective tissues. Am. J. Anat. 168(3), 257–276 (1983).
Le Douarin, N. M., Creuzet, S., Couly, G. & Dupin, E. Neural crest cell plasticity and its limits. Development 131(19), 4637–4650 (2004).
Thorogood, P. The developmental specification of the vertebrate skull. Development. 103(Suppl), 141–1 53 (1988).
Funato, N. New insights into cranial synchondrosis development: a mini review. Front. Cell Dev. Biol. 8, 706 (2020).
Kronenberg, H. M. Developmental regulation of the growth plate. Nature 423(6937), 332–336 (2003).
Hojo, H., Ohba, S., Yano, F. & Chung, U. I. Coordination of chondrogenesis and osteogenesis by hypertrophic chondrocytes in endochondral bone development. J. Bone Min. Metab. 28(5), 489–502 (2010).
Long, F. & Ornitz, D. M. Development of the endochondral skeleton. Cold Spring Harb Perspect. Biol. 5(1), a008334 (2013).
Hall, B. K. & Miyake, T. The membranous skeleton: the role of cell condensations in vertebrate skeletogenesis. Anat. Embryol. (Berl). 186(2), 107–124 (1992).
Stegen, S. et al. HIF-1α metabolically controls collagen synthesis and modification in chondrocytes. Nature 565(7740), 511–515 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53(D1), D672–d7 (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. Protein Sci. 28(11), 1947–1951 (2019).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30 (2000).
Schipani, E. Hypoxia and HIF-1alpha in chondrogenesis. Ann. N Y Acad. Sci. 1068, 66–73 (2006).
Hu, S. et al. Stabilization of HIF-1α alleviates osteoarthritis via enhancing mitophagy. Cell. Death Dis. 11(6), 481 (2020).
Tang, Y. et al. HIF-1α mediates osteoclast-induced mandibular condyle growth via AMPK signaling. J. Dent. Res. 99(12), 1377–1386 (2020).
Mackie, E. J., Ahmed, Y. A., Tatarczuch, L., Chen, K. S. & Mirams, M. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int. J. Biochem. Cell. Biol. 40(1), 46–62 (2008).
Lee, J. W., Bae, S. H., Jeong, J. W., Kim, S. H. & Kim, K. W. Hypoxia-inducible factor (HIF-1)alpha: its protein stability and biological functions. Exp. Mol. Med. 36(1), 1–12 (2004).
Montemurro, C. et al. IAPP toxicity activates HIF1α/PFKFB3 signaling delaying β-cell loss at the expense of β-cell function. Nat. Commun. 10(1), 2679 (2019).
Schipani, E. et al. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 15(21), 2865–2876 (2001).
Lee, H. J. et al. BICD1 mediates HIF1α nuclear translocation in mesenchymal stem cells during hypoxia adaptation. Cell. Death Differ. 26(9), 1716–1734 (2019).
Wuelling, M. & Vortkamp, A. Chondrocyte proliferation and differentiation. Endocr. Dev. 21, 1–11 (2011).
Tsuchida, S. et al. HIF-1α-induced HSP70 regulates anabolic responses in articular chondrocytes under hypoxic conditions. J. Orthop. Res. 32(8), 975–980 (2014).
Wang, P. et al. Icariin increases chondrocyte vitality by promoting hypoxia-inducible factor-1α expression and anaerobic glycolysis. Knee 27(1), 18–25 (2020).
Bell, D. M. et al. SOX9 directly regulates the type-II collagen gene. Nat. Genet. 16(2), 174–178 (1997).
Ng, L. J. et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev. Biol. 183(1), 108–121 (1997).
Zhang, C. et al. Hypoxia-inducible factor-1 is a positive regulator of Sox9 activity in femoral head osteonecrosis. Bone 48(3), 507–513 (2011).
Akiyama, H., Chaboissier, M. C., Martin, J. F., Schedl, A. & de Crombrugghe, B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16(21), 2813–2828 (2002).
Komori, T. Regulation of proliferation, differentiation and functions of osteoblasts by Runx2. Int. J. Mol. Sci. 20(7), (2019).
Komori, T. et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89(5), 755–764 (1997).
Kong, P. et al. HIF-1α repairs degenerative chondrocyte glycolytic metabolism by the transcriptional regulation of Runx2. Eur. Rev. Med. Pharmacol. Sci. 25(3), 1206–1214 (2021).
Mu, G. et al. Calmodulin 2 facilitates angiogenesis and metastasis of gastric cancer via STAT3/HIF-1A/VEGF-A mediated macrophage polarization. Front. Oncol. 11, 727306 (2021).
Yu, J., Liang, F., Huang, H., Pirttiniemi, P. & Yu, D. Effects of loading on chondrocyte hypoxia, HIF-1α and VEGF in the mandibular condylar cartilage of young rats. Orthod. Craniofac. Res. 21(1), 41–47 (2018).
Yang, C. et al. N-Acetylcysteine protects against Cobalt chloride-induced endothelial dysfunction by enhancing glucose-6-phosphate dehydrogenase activity. FEBS Open. Bio. 12(8), 1475–1488 (2022).
Yuan, Y., Hilliard, G., Ferguson, T. & Millhorn, D. E. Cobalt inhibits the interaction between hypoxia-inducible factor-alpha and von Hippel-Lindau protein by direct binding to hypoxia-inducible factor-alpha. J. Biol. Chem. 278(18), 15911–15916 (2003).
Dy, P. et al. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev. Cell. 22(3), 597–609 (2012).
Okada, K. et al. Hypoxia-inducible factor-1 alpha maintains mouse articular cartilage through suppression of NF-κB signaling. Sci. Rep. 10(1), 5425 (2020).
Liu, Z. et al. A dual role for NOTCH signaling in joint cartilage maintenance and osteoarthritis. Sci. Signal. 8(386), ra71 (2015).
De Santis, M. C., Gulluni, F., Campa, C. C., Martini, M. & Hirsch, E. Targeting PI3K signaling in cancer: challenges and advances. Biochim. Biophys. Acta Rev. Cancer. 1871(2), 361–366 (2019).
Ma, L., Zhang, R., Li, D., Qiao, T. & Guo, X. Fluoride regulates chondrocyte proliferation and autophagy via PI3K/AKT/mTOR signaling pathway. Chem. Biol. Interact. 349, 109659 (2021).
Wang, Q. et al. PI3K/AKT pathway promotes keloid fibroblasts proliferation by enhancing glycolysis under hypoxia. Wound Repair. Regen. 31(2), 139–155 (2023).
Huang, J. G. et al. 17β-Estradiol promotes cell proliferation in rat osteoarthritis model chondrocytes via PI3K/Akt pathway. Cell. Mol. Biol. Lett. 16(4), 564–575 (2011).
Chen, Y. L. et al. Maslinic acid prevents IL-1β-induced inflammatory response in osteoarthritis via PI3K/AKT/NF-κB pathways. J. Cell. Physiol. 236(3), 1939–1949 (2021).
Xiao, Y. et al. PDGF promotes the Warburg effect in pulmonary arterial smooth muscle cells via activation of the PI3K/AKT/mTOR/HIF-1α signaling pathway. Cell. Physiol. Biochem. 42(4), 1603–1613 (2017).
Yao, H. C. et al. Intravenous high mobility group box 1 upregulates the expression of HIF-1α in the myocardium via a protein kinase B-dependent pathway in rats following acute myocardial ischemia. Mol. Med. Rep. 13(2), 1211–1219 (2016).
Funding
This work was financially supported by the Yunnan Provincial Department of Science and Technology Basic Research Program(202201AY070001—173). It is also supported by the National Natural Science Foundation of China (Grant No. 82360200), and the First-Class Discipline Team Program of Kunming Medical University (Grant No. 2024XKTDTS08).
Author information
Authors and Affiliations
Contributions
Cun Liang: Data curation (equal); formal analysis (equal); funding acquisition (equal); methodology (equal); project administration (equal); validation (equal); writing – original draft (equal). Shuhui Yin: Data curation (equal); methodology (equal); validation (equal). Zhenjin Yang: Data curation (equal); methodology (equal); validation (equal). Yingchao Wang: Software (equal); validation (equal). Jiangtian Hu: Conceptualization (equal); funding acquisition (equal); project administration (equal); supervision(lead); writing– review and editing (equal). Guojie Gao: Conceptualization (lead); funding acquisition (equal); validation (equal); writing – review and editing (equal).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liang, C., Yin, S., Yang, Z. et al. HIF-1α regulates the proliferation and differentiation of mouse cranial base sphenoid-occipital synchondrosis chondrocytes via PI3K/Akt signaling. Sci Rep 15, 31240 (2025). https://doi.org/10.1038/s41598-025-11055-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-11055-1