Abstract
Glioma classification is crucial for effective diagnosis and treatment. Heat Shock Proteins (HSPs) have been associated with tumor development and progression. In this study, we established a prognostic model for glioma based on Heat Shock Proteins-associated genes (HSPGs). Using the Cancer Genome Atlas (TCGA) cohorts and the Chinese Glioma Genome Atlas (CGGA), we identified a signature of 4 HSPGs as an independent prognostic factor for glioma. The risk model demonstrated excellent performance in both training and validation sets. Additionally, we developed a nomogram incorporating clinical parameters and the HSPGs signature to enhance prognostic prediction. Immunoenrichment analysis revealed a correlation between the risk score and the immunosuppressive status of glioma. In the functional assays, HSPA5 was identified as a key participant in several critical biological processes associated with glioma. Silencing HSPA5 expression may lead to the inhibition of glioma cell proliferation, migration, invasion and resistance to apoptosis. These findings present a novel classification for glioma prognosis with enhanced accuracy and offer valuable insights into the potential use of HSPGs as prognostic indicators for gliomas.
Similar content being viewed by others
Introduction
Glioma, a primary brain tumor arising from glial cells and their precursors in the brain’s white matter, accounts for approximately 80–85% of all primary malignant brain tumors in adults, with glioblastoma representing 49% of these cases1. Accurate classification and grading of gliomas are of paramount importance for prognostic evaluation. Glioblastoma, the most aggressive form, typically leads to a overall survival of approximately 16 months following standard treatment, which includes surgical resection and concurrent chemoradiotherapy2,3. Conversely, low-grade gliomas may offer patients a relatively prolonged survival, extending over several decades.
The 2016 WHO classification of tumors of the central nervous system introduced a comprehensive approach that combines histopathological findings with molecular changes for glioma classification4. In the more recent 2021 edition, the significance of molecular changes in glioma classification has been further emphasized. Key molecular markers, such as IDH1/IDH2 (Isocitrate Dehydrogenase) mutation, 1p/19q co-deletion, CDKN2A/B homozygous deletion, TERT (telomerase Reverse Transcriptase) promoter mutation, and chromosome 7 gain/chromosome 10 loss, play a critical role in characterizing glioma subtypes5. It is crucial to continue exploring potential molecular alterations in glioma and incorporate these findings into disease classification to inform and tailor treatment strategies effectively.
Heat Shock Proteins (HSPs), also known as stress proteins, are highly conserved proteins that are upregulated in response to cellular stressors, such as heat, hypoxia, heavy metals, and cytotoxic drugs6,7. These proteins play essential roles in various biological processes, including protein synthesis, folding, assembly, and degradation8. The HSP family can be categorized into 6 groups: HSP27, HSP40, HSP60, HSP70, HSP90, and large HSPs9,10. Emerging evidence indicates that some HSPs exhibit persistent expression and are implicated in numerous cellular functions.
In the context of cancer, HSPs have garnered significant attention due to their involvement in tumor development and progression. Studies have demonstrated a relationship between Heat Shock Proteins-associated genes (HSPGs) expression and the occurrence, proliferation, invasion, and metastasis of various tumor. For instance, HSP40 and HSP70 have been found to synergistically regulate p53-mediated apoptosis, and they can directly bind to P53, thereby acting as tumor suppressors11,12,13.
In gliomas, HSPs have been extensively studied and have been linked to the biological behavior of tumors. Some studies have found small HSPs specifically associated with the malignancy of astrocytoma14. Notably, HSP27 is prominently overexpressed in oligodendrogliomas and has shown promise as a potential biological marker15. Another HSPs family member, HSP47, also known as collagen 2, exhibits high specificity for certain tissues and cells. HSP47 is notably absent in normal cerebral blood vessels but highly expressed in glioma cells, making it a glioma-associated antigen involved in glioma angiogenesis16. Furthermore, HSP90, functioning as a crucial molecular chaperone involved in DNA damage repair, contributes to glioblastoma resistance to both radiotherapy and chemotherapy17. However, the development of Onalespib, an HSP90 inhibitor, has shown encouraging outcomes, enhancing the efficacy of chemoradiotherapy in glioblastoma both in vitro and in vivo9,18.
In this study, our objective is to develop a prognostic signature associated with HSPGs to predict the prognosis of gliomas. To achieve this, we utilized gene expression data and clinical information sourced from the TGGA database for signature selection, while an independent dataset from the CCGA database was employed for validation purposes. Our rigorous analysis established a 4-HSPGs signature, which has been identified as a robust and independent risk factor. This HSPGs-associated signature exhibits a remarkable ability to effectively stratify gliomas into two distinct groups, each with different outcomes. Notably, this signature provides prognostic insights that complement traditional grading systems, offering a more nuanced approach to stratifying glioma patients and highlighting its potential as a valuable tool to enhance clinical prognostic assessments.
Materials and methods
Acquisition of HSPGs
The gene set was searched on the GeneCards (https://www.genecards.org/)website with “Heat Shock Protein” as the keyword, and the genes that could encode proteins were screened. Unnamed pseudogenes and genes not directly related to HSP, such as heat shock factor binding protein and histone, were excluded. Heat shock protein related gene sets were obtained.
Collection of datasets
In this study, the training cohort was initially obtained from the TCGA database (n = 702, https://portal.gdc.cancer.gov/), which includes RNA sequencing data and the corresponding clinical features, while the validation cohort was sourced from the CGGA database (n = 325, http://www.cgga.org.cn/)19, which also comprises RNA sequencing data and the corresponding clinical features. All gene expression files were normalized using a log2 transformation. After excluding samples with missing values (NA), the final datasets consisted of 562 cases from TCGA and 273 cases from CGGA for subsequent analyses. Univariate Cox proportional hazards regression was applied to the gene sets to identify heparan sulfate proteoglycans (HSPGs) associated with survival outcomes. Thereafter, LASSO (Least Absolute Shrinkage and Selection Operator) regression analysis was performed to further screen for genes significantly associated with survival. All methods were performed in accordance with the relevant guidelines and regulations.
Establishment of a prognostic gene signature
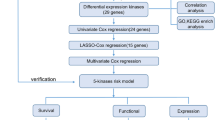
The multivariate COX regression model was used to analyze the significantly related genes obtained from the LASSO analysis, and the COX regression coefficients of the genes were used to calculate the risk score of the genes. The data in the training set were divided into low-risk and high-risk groups according to the scoring formula, and a heat map of significant genetic risk score was drawn. The same method was used to verify in the validation set.
A multivariate COX regression model was used to verify whether the risk score was an independent prognostic factor for survival. The time-ROC (Receiver Operating Characteristic) curve was drawn to evaluate the specificity and sensitivity of the risk score in predicting the survival of 1 year, 1.5 years and 2 years, respectively. The difference between groups was evaluated by log-rank test, and P value less than 0.05 was considered to be significant.
Construction and validation of the nomogram
The rms package was used to establish a prognostic nomogram to evaluate the 1-year, 1.5-year, and 2-year survival rates of patients. The nomogram included all independent predictive parameters. The C-index and calibration curve were used to calculate the discrimination and correction between the predicted value of nomogram and the true survival rate.
Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA) was conducted to determine the related pathways and molecular mechanisms of the high-risk group and low-risk group in TCGA cohorts (https://www.gsea-msigdb.org/gsea/index.jsp). Gene sets with a p value of<0.05 and false discovery rate (FDR) of <0.25 after 1000 permutations were consider significantly enriched20.
Analysis of immune cell proportions
To assess the composition of immune cell types in the RNA-seq data, we employed the CIBERSORT methodology, as described by Newman et al. in 201521. Gene expression profiles were uploaded and the CIBERSORT software was executed online (accessible at https://cibersort.stanford.edu/runcibersort.php), with the LM22 gene signature and 1000 permutations selected as parameters. Consequently, we derived the composition of 22 distinct cell types for each sample based on their gene expression profiles22.
Validation of protein expression levels of heat shock proteins-associated genes signature via the human protein atlas
To further verify the protein expression levels of the above four genes in glioma, we searched the immunohistochemical data in The Human Protain Atlas database (HPA, https://www.proteinatlas.org/), which can provide proteomics-based IHC results for multiple proteins in glioma. The serial numbers of antibodies used for immunohistochemical detection were HPA038845, HPA038846, CAB005221, HPA010642 and HPA036289.
Cell culture and transfection
U118, U251, and U87 glioma cell lines were obtained from the Chinese Academy of Sciences (Shanghai, China) with STR authentication. The cells were cultured in DMEM medium (Lonza, catalog number CC3170) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. The siRNAs targeting HSPA5 (si- HSPA5#1: 5′-UGGGUAGUUGAUUCCUAAAAACUCGCA-3′; si- HSPA5#2: 5′-UAUAAUCUCUGAGUAAUCUUCCUUUUC-3′) were synthesized by ORIGENE. Scrambled siRNA was synthesized as a negative control. Transfections were performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol.
Western blotting
Western blotting was conducted as previously described. Primary antibodies used included HSPA5 (Proteintech, catalog number 10725-1-AP) and β-actin (Santa Cruz, catalog number sc-47778). Protein bands were detected using a chemiluminescence kit (Amersham Pharmacia Biotech, Piscataway, NJ). All experiments were repeated three times to ensure reproducibility.
Functional assay
CCK8 Assay: Cells were seeded in a 96-well plate at a density of 2000 cells/well, with three replicates per group. Cell proliferation was assessed at 24, 48, and 72 h post-seeding using the CCK8 reagent (Solarbio, catalog number CA1210). For each time point, the culture medium was replaced with medium containing the CCK8 reagent and incubated for 1 h at 37 °C in a 5% CO2 incubator. The optical density (OD) at 450 nm was measured using a microplate reader. Transwell Assay: Cells were cultured in serum-free medium for 2 h before being seeded into the upper chamber of a 24-well Transwell insert at a density of 2 × 10^4 cells/well. Each group had two replicates. The lower chamber was filled with medium containing 10% fetal bovine serum (FBS). After 72 h, the lower surface of the Transwell inserts and the wells were stained using Giemsa stain (Beyotime, catalog number C0133), and the cells on the lower surface were counted. Colony Formation Assay: Cells were seeded in a 6-well plate at a density of 1000 cells/well, with three replicates per group. After 144 h, the colonies were stained with Giemsa stain, and the number of colonies per well was counted. Apoptosis analysis: After 72 h of culture, cells were harvested and stained using the Full Gold apoptosis detection kit (catalog number FA101) according to the manufacturer’s instructions. Apoptosis was then analyzed using flow cytometry.
Immunohistochemical analysis
Immunohistochemical staining for HSPA5 (GRP78) and PD-L2 was conducted on formalin-fixed, paraffin-embedded tissues, with 4-µm-thick sections prepared from each paraffin block, deparaffinized in xylene, rehydrated through a graded ethanol series, and washed in double-distilled water. Antigen retrieval was performed via heat-induced epitope retrieval by steaming slides in 10 mM sodium citrate buffer (pH 6.0) at 100 °C for 15 min. Rabbit polyclonal anti-PD-L2 (18251–1-AP, Proteintech; 1:200 dilution) and anti-GRP78 (11587–1-AP, Proteintech; 1:200 dilution) were used to detect protein expression, with stained slides independently assessed by two blinded neuropathologists. Staining was scored for intensity (0 = negative, 1 = weak, 2 = moderate, 3 = strong) and positive cell percentage (1 = 0–25%, 2 = 26–50%, 3 = 51–75%, 4 = 76–100%), with the final composite score determined by multiplying intensity and percentage scores (range: 0–12).
Statistical analysis
Statistical computations and graphical representations were executed using the R statistical software (version 4.2.0), available at http://www.r-project.org. To assess the prognostic significance of HSPs score, we employed Kaplan–Meier (KM) analysis and Cox proportional hazard model analysis with the “survival” and “survminer” packages in R. In this investigation, we conducted Pearson correlation analysis to identify genes coexpressed with HSPs score. In order to further evaluate the relationship between HSPGs and glioma prognosis, KM analysis was performed on each gene in CGGA and TCGA databases, and the expression of each gene was divided into low expression and high expression according to the median expression of HSPGs in glioma. Specifically, a positive correlation was defined as a correlation coefficient exceeding 0.6 with a p-value less than 0.05, while a negative correlation was characterized by a correlation coefficient below − 0.6 with a p-value less than 0.05. For two-group and multiple-group comparisons, the Wilcoxon test and one-way ANOVA test were utilized, respectively. In all statistical analyses, a significance level of p < 0.05 was considered indicative of statistical significance.
Results
The procedure of this study and the construction of HSPs score
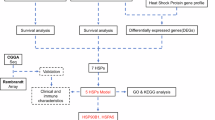
The flowchart illustrating the process of this study is depicted in Fig. 1. We searched all HSPGs in the Gene Cards database (n = 77), and then applied univariate COX regression analysis in the TCGA database to identify the HSPGs significantly associated with the survival of glioma patients (OS, p < 0.05), retaining 42 genes for further analysis. Then, Lasso regression analysis was carried out on these 42 genes with 10-fold cross-validation to select the optimal penalty parameter lambda by minimizing the mean squared error (MSE), obtaining the final screened 4 key genes. The HSPGs score was constructed using the screened genes and their multivariate COX regression coefficients. The score formula is (-0.86129822 * DNAJB12 expression level) + (-0.31879448 * DNAJC12 expression level) + (0.04657945 * DNAJC5B expression level) + (0.16766430 * HSPA5 expression level)(Fig. 2 A, B).
Construction of HSPs score and multifactorial COX survival analysis of HSPs Score. (A) Lasso coefficient profiles of included prognostic HSPs genes. (B) Lasso regression with ten-fold cross-validation identified 4 prognostic genes using the lambda.1se. (C) Multifactorial regression analysis results of HSPs score in the TGGA dataset. (D) Multifactorial regression analysis results of HSPs score in the CCGA dataset.
The predictive role of HSPs score in the survival prognosis of glioma patients
We calculated HSPs characteristic scores for each glioma patient in the TCGA (n = 562) and CGGA (n = 273) cohorts. Multivariate COX regression analysis was performed to evaluate the association between the HSPs score and clinical parameters (gender, age, tumor grade, IDH mutation status, 1p/19q co-deletion status) and survival outcomes. The results show that the HSPs characteristic score in both the TCGA cohort (Fig. 2C) and the CGGA (Fig. 22D) cohort are an independent prognostic factor reflecting the survival of glioma patients.
Patients were stratified into high-risk and low-risk groups based on the median risk score (TCGA: 281 high-risk vs. 281 low-risk; CGGA: 137 high-risk vs. 136 low-risk), and scatter plots were used to show the relationship between risk score and survival status. In both the TGGA and CCGA datasets, the survival of the high-risk group was shorter than that of the low-risk group (Fig. 3A, C). Heat maps further revealed that HSPA5 and DNAJC5B were upregulated in the high-risk group, while DNAJB12 and DNAJC12 were downregulated, consistent with their positive/negative coefficients in the risk model (Fig. 3B, D).
Risk Score Characteristics and Heatmap of HSPGs Score. The distributions of the risk score, survival time, and patient status in TGGA cohorts (A) and CGGA cohorts (C). The dotted lines represent the optimal cut-off value distinguishing between the low- and high-risk groups. Heatmap displaying gene-expression profiles of the HSPGs score in TCGA cohorts (B) and the CGGA cohort (D).
KM curve results showed the relationship between risk score and patient survival. We found significant differences in survival between high-risk and low-risk groups in the TCGA and CGGA data sets (p< 0.05)(Fig. 4A, C). Time-dependent ROC analysis showed excellent predictive performance of the risk score. In the TCGA cohort, the area under the curve (AUC) was 0.863 for 1-year survival, 0.922 for 1.5-year survival, and 0.905 for 2-year survival (Fig. 4B). In the CGGA cohort, the AUC values were 0.736, 0.782, and 0.818 for the same time points, respectively (Fig. 4D). These results highlight the risk score’s robust ability to discriminate survival outcomes across different follow-up periods. Moreover, we established a nomogram for prognostic-decision support. Our nomogram could well predict the 1 year, 1.5-year and 2-year survival probabilities for glioma patients by incorporating the HSPs score and prognostic factors including patients age, tumor WHO Grade, IDH mutation status and 1p-19q co-deletion status. Consequently, the C-index of the nomogram is 0.8751341 in TCGA cohort and 0.7667895 in CGGA cohort ( Figure S1).
The relationship between HSPs score and clinical characteristics, tumor features and tumor microenvironment in gliomas
We investigated the distribution trend of risk score in glioma patients (Fig. 5A and S2A). We found that with the increase of risk score, the WHO grade of tumor increased, the pathological type of tumor gradually turned bad, the proportion of IDH wild-type patients gradually increased, the proportion of 1p/19q non-codeletion increased, and the proportion of O-6-Methylguanine-DNA Methyltransferase (MGMT) promoter unmethylation increased. These characteristics in glioma are often associated with a worse prognosis. Boxplot shows the relationship between risk score and different tumor characteristics (Fig. 5B–F and S2B–F). It can be seen that risk score in the WHO high grade group (WHO IV) was significantly higher than in the WHO low grade group (WHOII-III) (p< 0.05) (Fig. 5B and S2B). Risk score in glioblastomas group was significantly higher than in other pathologic types (p< 0.05) (Fig. 5C and S2C). The risk score of IDH wild combined with 1p/19q non-codeletion group was significantly higher than that of IDH mutant group and 1p19q co-deletion group (Fig. 5D, E and S2D, E). We also found that in the TCGA low-grade glioma (LGG) cohort and the CGGA glioblastoma (GBM) cohort, risk scores of tumors with MGMT promoter non-methylation were significantly higher than those with MGMT promoter methylation, whereas no statistical significance was observed in the TCGA GBM cohort and CGGA LGG cohort (Fig. 5F and S2F). Prior evidence has indicated that MGMT-methylated gliomas demonstrate enhanced responsiveness to temozolomide (TMZ) chemotherapy and prolonged survival outcomes compared with MGMT-unmethylated tumors23,24,25,26. Collectively, these findings indicate that patients with higher risk scores may exhibit reduced sensitivity to temozolomide (TMZ) treatment. In the low-grade gliomas (LGGs) group, the risk score for recurrent tumors was significantly higher than that for primary tumors (Figure S2G), suggesting that some genes in the HSPs family may play a role in tumor recurrence in low-grade gliomas.
Landscape of clinical and molecular characteristics associated with HSP score in gliomas. TCGA cohorts are arranged in an increasing order of HSPs score(A). HSPs score expression pattern in glioma. WHO II–IV(B); Histology(C); 2016 WHO classification(D); IDH mutation status in LGGs and glioblastomas(E); MGMT promoter methylation status in LGGs and glioblastomas(F).
Since the HSPs characteristic score had significant differences in the survival period of tumor patients, tumor pathological types, tumor molecular pathological characteristics and other aspects, we investigated the genes that are highly correlated with this risk score and analyzed their biological functions. First of all, we got the genes associated with a significant risk score (Pearson | R | < 0.6 and p> 0.05) screened out genes with significant positive and negative correlation with risk score. We then conducted GO biological process prediction and GSEA analysis for these co-expressed genes, with results showing that the biological functions of genes positively correlated with risk score were mainly concentrated in G2M, E2F, MYC, metabolic and immune response related pathways (IFN-γ response pathway included) (Fig. 6). And the genes that were negatively associated with risk score were mainly concentrated in the regulation of synaptic signaling and membrane potential (Fig. 6A blue entries). In conclusion, we believe that HSPs score can reflect tumor proliferation, metabolic status and immune response.
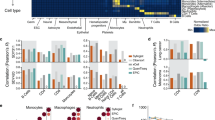
Relationship Between HSPs Score and the Tumor Microenvironment in Glioma Tissues. The results of stromal score (A), immune score (B), and tumor purity (C) evaluated by the ESTIMATE algorithm. The relationship between immune cell infiltration in gliomas and HSPs score (D,E). The correlation between HSPs score and the proportions of specific immune cells (F), the association with immune-related biological functions (G), and the relationship with expression levels of immune checkpoint genes (H). Representative immunohistochemical staining of HSPA5 and PD-L2 in four glioma patients (scale bar = 200 μm) (I). Correlation between HSPA5 and PD-L2 protein expression in glioma tissues. Scatter plot showing Pearson correlation analysis between HSPA5 IHC scores and PD-L2 IHC scores (J).
The tumor microenvironment (TME) exerts a significant influence on cancer progression, comprising various components such as tumor cells, immune cells, and stromal cells. In this study, we employed the Estimation of Stromal and Immune cells in Malignant Tumor tissues using Expression data (ESTIMATE) algorithm to assess the infiltration of immune and stromal cells. This assessment involved the computation of immune score and stromal score. Our findings revealed a positive correlation between HSPs score and stromal score (Fig. 7A), as well as immune score (Fig. 7B) in gliomas. Conversely, there was a negative correlation observed between HSPs score and tumor purity (Fig. 7C).
In recent years, immunotherapy has gradually become an important part of tumor therapy. We also found that HSPs score were correlated with immune response. Therefore, we analyzed the relationship between HSPs characteristic score and immune activity in glioma. We first used CIBERSORT software to evaluate the relative abundance of 22 immune cells in the CGGA data set, which mainly included lymphocytes, macrophages, monocytes and other immune cells. Boxplot shows the difference in immune cell content between the high-risk and low-risk groups (Fig. 7D, E). Using the Wilcox-test, we found a significant difference in the proportion of immune cells between the two groups of samples. At the same time, we also assessed the characteristics of HSPs score and immune cells, and statistical Pearson inspection | R | > 0.2 cell type, it was found that the characteristic score of HSPs were positively correlated with the immunomodulatory cells such as M0 and Treg in the samples or the cells with low immune killing ability, while negatively correlated with the pro-inflammatory and anti-tumor type of immune cells (NK cell active) in the samples. Suggesting that the HSPs score reflects the immune state within the tumor (Fig. 7F). Similarly, in the GO functional note, we analyzed immune reaction-related pathways associated with HSPs score. Among the top10 immune response pathways positively correlated with HSPs score, several pathways were associated with immune regulation (Fig. 7G), suggesting that the anti-oncologic properties of immune cells in samples with high HSPs score may be inhibited. As a protective molecule in the human immune system, immune checkpoint can prevent T cells from over-activating and inducing inflammatory damage. Tumor cells can take advantage of this property of the human immune system to inhibit immune response by over-expressing immune checkpoint molecules (Fig. 7H), so as to escape human immune surveillance and killing. Therefore, we analyzed the relationship between HSPs score and immune checkpoint protein, and the results showed that the HSPs score were positively correlated with immune checkpoint, indicating the existence of immunosuppression in gliomas with high scores. To further validate the correlation between HSPGs and immune checkpoint molecules at the protein level, we performed immunohistochemical analysis of HSPA5 and PD-L2 (most highly correlated with HSPs) in 20 glioma tissues. Representative staining results from four patients (Fig. 7I) and Pearson correlation analysis revealed a significant positive association between HSPA5 and PD-L2 expression (Pearson r = 0.6256, p = 0.032; Fig. 7J). This finding demonstrates that HSPA5, as a core component of the HSPGs signature, may promote immunosuppression in gliomas by upregulating PD-L2, thereby highlighting a potential mechanistic link between HSPGs and immune evasion.
To further validate the association between HSPs score and prognosis in glioma patients, we performed survival analysis on each of the 4 HSPGs separately in both the training set and validation set. The Kaplan-Meier curve demonstrated a significant correlation between expression levels of HSPA5, DNAJC5B, DNAJC12, and DNAJB12 with glioma prognosis (P < 0.05) (Fig. 8A). Higher expression of HSPA5 and DNAJC5B genes was associated with shorter survival time in glioma patients, while the expression of DNAJC12 and DNAJB12 genes showed opposite relationship with glioma survival time. Protein expression levels of these 4 genes are depicted in Fig. 8B. According to data from the Human Protein Atlas (HPA) database, certain genes such as DNAJB12 exhibited only low to medium expression levels in gliomas. Figure 8 C demonstrates the expression levels of HSPA5, DNAJC12, and DNAJB12 in low-grade gliomas and high-grade gliomas in the HPA database. Of note, the highest number of samples (n = 32) was obtained because HSPA5 expression was measured using three different antibodies. Specifically, only two high-grade gliomas showed significantly high expression levels of HSPA5, while both low-grade and high-grade gliomas showed persistently low expression levels of DNAJC12. In addition, it should be noted that some gliomas did not detect any expression of HSPA5, DNAJC12, or DNAJB12.
HSPA5 promotes glioma cell migration, invasion, and resistance to apoptosis
We conducted a series of in vitro functional assays to investigate the role of HSPA5 in influencing the malignant phenotype of glioma. First, we selected glioma cell lines U118, U251, and U87, known for their high malignancy. We designed and synthesized siRNA-1 specifically targeting HSPA5. The siRNA were transfected into U118, U251, and U87 cells, and Western blotting was performed to assess the knockdown efficiency. Western blotting analysis indicated that siRNA-1 significantly downregulated HSPA5 expression in U251 cells (p < 0.05); meanwhile, U118 and U87 cells exhibited a downward trend in HSPA5 levels (though statistically non-significant; Fig. 9A). Using the CCK-8 assay, we observed that knocking down HSPA5 significantly inhibited glioma cell viability (Fig. 9B). Colony formation assays showed a marked inhibition in colony formation in U118, U251, and U87 cells upon HSPA5 knockdown (Fig. 9C). To investigate the role of HSPA5 in cell migration and invasion, we conducted transwell assays. The results demonstrated that HSPA5 suppression significantly decreased glioma cell migration and invasion (Fig. 9D). Additionally, we analyzed cell apoptosis using flow cytometry and found that knocking down HSPA5 significantly increased cell apoptosis (Fig. 9E). These findings suggest that HSPA5 plays a crucial role in promoting glioma oncogenic activities.
Western blot was used to determine the expression levels of HSPA5 in U118, U87, and U251 cells transfected with HSPA5-targeting siRNA (A). CCK8 assay was used to evaluate the cell growth capacity of U118, U87, and U251 cells transfected with HSPA5 siRNA over a period of 3 days (B). HSPA5 gene knockout inhibited colony formation in U118, U87, and U251 cells; the colony formation images were captured using a camera (C). Transwell migration and invasion assays demonstrated that HSPA5 knockout inhibited the migration and invasion of U118, U87, and U251 cells; the scale is 200 μm (D). Flow cytometry showed that the proportion of early apoptotic cells increased in U118, U87, and U251 cells transfected with HSPA5-targeting siRNA (E).
Discussion
This study identified 4 HSPGs significantly correlated with glioma prognosis, all of which encode HSP-related proteins. Among them, previous research has revealed a positive role of DNAJB12 in maintaining endoplasmic reticulum calcium homeostasis, effectively inhibiting damage to the endoplasmic reticulum caused by misfolded protein synthesis. DNAJC12 is believed to be involved in the cellular response to oxidative stress, heat stress, and other environmental stressors27. This indicates that DNAJC12 could be a crucial component of cells’ ability to cope with various stressful conditions. Moreover, DNAJC12 might also play a role in protein folding and quality control. Members of the DnaJ family typically work in cooperation with the molecular chaperone Hsp70 to assist in proper protein folding and prevent the aggregation of misfolded proteins28. DNAJC5B is a gene responsible for encoding cysteine string protein beta (CSPβ). CSPs, specifically cysteine-string proteins (CSPs), have been proposed to act as genuine catalysts for proper protein folding, notably in the later stages of insulin secretion and in neuroexocytosis29. The first characterized isoform within the CSP family, CSPα, plays a central role in regulating calcium channel activity during mammalian neurosecretion, likely through its interactions with G proteins30,31. Two new predicted isoforms, CSPβ and CSPγ, were identified from human and mouse testis cDNA libraries, broadening our knowledge of this protein family32. Recent studies have unveiled a link between CSP and the neurodegenerative disease known as adult-onset neuronal ceroid lipofuscinosis (ANCL). CSP primarily prevents neurodegeneration by preserving the conformation of SNAP-25, facilitating its integration into the membrane-fusing SNARE complex33. HSPA5, also known as Heat Shock 70 kDa Protein 5, is the gene name for the protein commonly referred to as Glucose-Regulated Protein 78 (GRP78) or Binding Immunoglobulin Protein (BiP). This protein is a member of the HSP70 family and is located primarily in the endoplasmic reticulum (ER) of eukaryotic cells34. GRP78 predominantly resides within the endoplasmic reticulum’s lumen, a cellular compartment responsible for the synthesis and proper folding of proteins35. Its critical role lies in maintaining protein equilibrium by aiding in the assembly and appropriate folding of freshly generated proteins36. The involvement of GRP78 in the development and advancement of various cancer types has been documented37. It is also found that GRP78/BiP as an innovative strategy to enhance the sensitivity of malignant gliomas to chemotherapy36. Its heightened expression is linked to several distinctive cancer characteristics, including heightened cell survival, increased cell proliferation, the promotion of angiogenesis (the formation of new blood vessels to support tumor growth), and resistance to chemotherapy and radiation therapy38.
The low-risk and high-risk groups differentiated based on HSPGs showed good sensitivity and specificity in predicting survival risks at 1 year, 1.5 years, and 2 years, with an AUC value of 0.844. Additionally, as the risk score based on HSPGs increased, there was a corresponding increase in WHO grading, and the proportion of IDH wild-type and 1p19q co-deletion also gradually increased. In a previous study, Cheng W et al. found that HSPB11, a small HSP expressed exclusively in tumor cells, was an independent prognostic factor for high-grade gliomas. However, this study did not consider the significant impact of molecular markers, such as IDH mutation status, on overall survival39. Existing research indicates that DNAJC10 is associated with the malignancy of gliomas, and its expression increases with higher pathological grades40.
HSPs form a large protein family, involving numerous genes, signaling pathways, and downstream proteins. Currently, there is relatively limited research on HSPs’ involvement in tumor formation, progression, and aggressiveness. The relevant mechanisms warrant more in-depth fundamental research in the future, requiring additional support in understanding the role of HSPs in cancer.
The methylation status of the MGMT promoter is crucial for the treatment response to alkylating agents such as TMZ. MGMT is a DNA repair enzyme, and when its promoter region is methylated, its transcriptional activity is suppressed, leading to downregulation of MGMT protein expression. This further implies that DNA damage, such as O(6)-methylguanine, cannot be efficiently repaired, resulting in more severe damage to tumor cells by alkylating agents. Consequently, glioblastoma patients with methylated MGMT promoter are more sensitive to alkylating agent therapy39,41. This study found that in glioblastomas, tumors with non-methylated MGMT promoter showed significantly higher tumor HSPs score compared to tumors with methylated MGMT promoter. These findings are consistent with the previous research by Cheng W and colleagues, who also found that low expression of HSPB11 indicates a higher sensitivity to chemotherapy. They speculated that this might be related to the role of HSPB11 in regulating the cell cycle and genomic integrity39.
The immune microenvironment of glioblastoma is characterized by a relative lack of immune cells, making immunotherapy challenging for this type of tumor. This study found that genes positively correlated with the HSP risk score in glioblastomas are mainly involved in the G2M, E2F, MYC, metabolism, and immune response pathways(such as IFN-γ response pathway). The HSP-related gene score in glioblastomas may reflect the tumor’s proliferation, metabolism, and immune status. Previous research has demonstrated that DNAJC10 is associated with T cell activation, T cell complexes, and T cell signaling in glioblastomas, indicating a close relationship between DNAJC10 expression level and the immune status of glioblastomas40. A previous study designed a multi-valent autologous anti-tumor vaccine targeting HSP, which showed potential in improving the survival of glioblastoma patients in a phase II clinical trial2. Therefore, inhibiting HSPs function or related expression in glioblastomas could potentially enhance the tumor’s immune response. Furthermore, we found that HSPA5 significantly promotes the proliferation, migration, invasion, and resistance to apoptosis of glioma cells in vitro. Notably, Wang et al. demonstrated that PD-L2 is upregulated in GBM and IDH wild-type gliomas, acting as a poor prognostic biomarker for glioma patients and potentially driving immune evasion42. Separately, studies have shown that HSPA5 is significantly overexpressed in gliomas43,44. In this study, immunohistochemical analysis confirmed a significant positive correlation between HSPA5 and PD-L2 expression in glioblastoma tissues. These findings suggest that HSPA5 may contribute to immune suppression by regulating PD-L2 expression and trafficking, thereby expanding our understanding of immune microenvironment regulation in gliomas. Further studies are required to elucidate the molecular mechanisms underlying the interaction between HSPA5 and PD-L2.
In conclusion, we have established a signature composed of 4 HSPGs. This score serves as an independent prognostic factor for overall survival in glioblastoma patients. It demonstrates relatively good stability and shows higher prognostic efficiency compared to the traditional WHO classification. This method is more readily applicable in a clinical setting and exhibits higher predictive accuracy compared to conventional approaches. However, it is important to acknowledge that this study has limitations as it is retrospective, and further validation of the signature’s efficacy is required in future studies. Overall, our research provides a novel HSP-based classification criterion applicable to glioblastomas, offering a new perspective for the classification and prognosis of this brain tumor. Additionally, our experiments suggest that HSPA5 may be a potential therapeutic target for glioma.
Data availability
For this article, the RNA sequencing data and clinical features of the validation set were obtained from the CGGA database (http://www.cgga.org.cn/), and the data of the training set were obtained from the TCGA database (https://portal.gdc.cancer.gov/). All the HSPGs were obtained for the Gene Cards website (https://www.genecards.org/).
References
Schaff, L. R. & Mellinghoff, I. K. Glioblastoma and other primary brain malignancies in adults: A review. Jama 329, 574–587. https://doi.org/10.1001/jama.2023.0023 (2023).
Bloch, O. et al. Autologous heat shock protein peptide vaccination for newly diagnosed glioblastoma: impact of peripheral PD-L1 expression on response to therapy. Clin. Cancer Res. 23, 3575–3584. https://doi.org/10.1158/1078-0432.Ccr-16-1369 (2017).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant Temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996. https://doi.org/10.1056/NEJMoa043330 (2005).
Louis, D. N. et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 131, 803–820. https://doi.org/10.1007/s00401-016-1545-1 (2016).
Whitfield, B. T. & Huse, J. T. Classification of adult-type diffuse gliomas: impact of the world health organization 2021 update. Brain Pathol. 32, e13062. https://doi.org/10.1111/bpa.13062 (2022).
Hagymasi, A. T., Dempsey, J. P. & Srivastava, P. K. Heat-Shock proteins. Curr. Protoc. 2, e592. https://doi.org/10.1002/cpz1.592 (2022).
Morimoto, R. I. Cells in stress: transcriptional activation of heat shock genes. Science 259, 1409–1410. https://doi.org/10.1126/science.8451637 (1993).
Parsell, D. A. & Lindquist, S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27, 437–496. https://doi.org/10.1146/annurev.ge.27.120193.002253 (1993).
Wu, J. et al. Heat shock proteins and Cancer. Trends Pharmacol. Sci. 38, 226–256. https://doi.org/10.1016/j.tips.2016.11.009 (2017).
Ciocca, D. R. & Calderwood, S. K. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell. Stress Chaperones. 10, 86–103. https://doi.org/10.1379/csc-99r.1 (2005).
Lu, W. J. et al. Mortalin-p53 interaction in cancer cells is stress dependent and constitutes a selective target for cancer therapy. Cell. Death Differ. 18, 1046–1056. https://doi.org/10.1038/cdd.2010.177 (2011).
Silva, G. et al. Trans-chalcone increases p53 activity via DNAJB1/HSP40 induction and CRM1 Inhibition. PLoS One. 13, e0202263. https://doi.org/10.1371/journal.pone.0202263 (2018).
Feng, X., Bonni, S. & Riabowol, K. HSP70 induction by ING proteins sensitizes cells to tumor necrosis factor alpha receptor-mediated apoptosis. Mol. Cell. Biol. 26, 9244–9255. https://doi.org/10.1128/MCB.01538-06 (2006).
Babi, A. et al. Targeting heat shock proteins in malignant brain tumors: from basic research to clinical trials. Cancers (Basel). 14. https://doi.org/10.3390/cancers14215435 (2022).
Castro, G. N. et al. Hsp27 (HSPB1): a possible surrogate molecular marker for loss of heterozygosity (LOH) of chromosome 1p in oligodendrogliomas but not in Astrocytomas. Cell. Stress Chaperones. 17, 779–790. https://doi.org/10.1007/s12192-012-0350-6 (2012).
Hosono, J., Morikawa, S., Ezaki, T., Kawamata, T. & Okada, Y. Pericytes promote abnormal tumor angiogenesis in a rat RG2 glioma model. Brain Tumor Pathol. 34, 120–129. https://doi.org/10.1007/s10014-017-0291-y (2017).
Önay Uçar, E., Şengelen, A. & Mertoğlu Kamalı, E. Hsp27, Hsp60, Hsp70, or Hsp90 depletion enhances the antitumor effects of Resveratrol via oxidative and ER stress response in human glioblastoma cells. Biochem. Pharmacol. 208, 115409. https://doi.org/10.1016/j.bcp.2022.115409 (2023).
Chan, K. C. et al. A novel Hsp90 inhibitor AT13387 induces senescence in EBV-positive nasopharyngeal carcinoma cells and suppresses tumor formation. Mol. Cancer. 12, 128. https://doi.org/10.1186/1476-4598-12-128 (2013).
Zhao, Z. et al. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci. DATA. 4 https://doi.org/10.1038/sdata.2017.24 (2017).
Wu, T. et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. (Cambridge (Mass)). 2, 100141. https://doi.org/10.1016/j.xinn.2021.100141 (2021).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12, 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. Circlize implements and enhances circular visualization. Bioinf. (Oxford England). 30, 2811–2812. https://doi.org/10.1093/bioinformatics/btu393 (2014).
Esteller, M. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med. 343, 1350–1354. https://doi.org/10.1056/NEJM200011093431901 (2000).
Stupp, R. et al. Effects of radiotherapy with concomitant and adjuvant Temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. LANCET Oncol. 10, 459–466. https://doi.org/10.1016/S1470-2045(09)70025-7 (2009).
Dunn, J. et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given Temozolomide and radiotherapy. Br. J. Cancer. 101, 124–131. https://doi.org/10.1038/sj.bjc.6605127 (2009).
Hegi, M. E. et al. MGMT gene Silencing and benefit from Temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003. https://doi.org/10.1056/NEJMoa043331 (2005).
Kampinga, H. H. et al. Guidelines for the nomenclature of the human heat shock proteins. Cell. Stress Chaperones. 14, 105–111. https://doi.org/10.1007/s12192-008-0068-7 (2009).
Cunnea, P. M. et al. ERdj5, an Endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 278, 1059–1066. https://doi.org/10.1074/jbc.M206995200 (2003).
Boal, F., Le Pevelen, S., Cziepluch, C., Scotti, P. & Lang, J. Cysteine-string protein isoform beta (Cspbeta) is targeted to the trans-Golgi network as a non-palmitoylated CSP in clonal beta-cells. Biochim. Biophys. Acta. 1773, 109–119. https://doi.org/10.1016/j.bbamcr.2006.08.054 (2007).
Magga, J. M., Jarvis, S. E., Arnot, M. I., Zamponi, G. W. & Braun, J. E. Cysteine string protein regulates G protein modulation of N-type calcium channels. Neuron 28, 195–204. https://doi.org/10.1016/s0896-6273(00)00096-9 (2000).
Natochin, M. et al. Characterization of the G alpha(s) regulator cysteine string protein. J. Biol. Chem. 280, 30236–30241. https://doi.org/10.1074/jbc.M500722200 (2005).
Gorleku, O. A. & Chamberlain, L. H. Palmitoylation and testis-enriched expression of the cysteine-string protein beta isoform. Biochemistry 49, 5308–5313. https://doi.org/10.1021/bi100550h (2010).
Evans, G. J., Morgan, A. & Burgoyne, R. D. Tying everything together: the multiple roles of cysteine string protein (CSP) in regulated exocytosis. Traffic 4, 653–659. https://doi.org/10.1034/j.1600-0854.2003.00127.x (2003).
Akinyemi, A. O. et al. Unveiling the dark side of glucose-regulated protein 78 (GRP78) in cancers and other human pathology: a systematic review. Mol. Med. 29, 112. https://doi.org/10.1186/s10020-023-00706-6 (2023).
Lee, A. S. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem. Sci. 26, 504–510. https://doi.org/10.1016/s0968-0004(01)01908-9 (2001).
Pyrko, P., Schonthal, A. H., Hofman, F. M., Chen, T. C. & Lee, A. S. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 67, 9809–9816. https://doi.org/10.1158/0008-5472.CAN-07-0625 (2007).
Dong, D. et al. Vascular targeting and antiangiogenesis agents induce drug resistance effector GRP78 within the tumor microenvironment. Cancer Res. 65, 5785–5791. https://doi.org/10.1158/0008-5472.CAN-05-0754 (2005).
Sedighzadeh, S. S., Khoshbin, A. P., Razi, S. & Keshavarz-Fathi, M. Rezaei, N. A narrative review of tumor-associated macrophages in lung cancer: regulation of macrophage polarization and therapeutic implications. Transl Lung Cancer Res. 10, 1889–1916. https://doi.org/10.21037/tlcr-20-1241 (2021).
Cheng, W. et al. Association between small heat shock protein B11 and the prognostic value of MGMT promoter methylation in patients with high-grade glioma. J. Neurosurg. 125, 7–16. https://doi.org/10.3171/2015.5.Jns142437 (2016).
Liu, F. et al. DNAJC10 correlates with tumor immune characteristics and predicts the prognosis of glioma patients. Biosci. Rep. 42 https://doi.org/10.1042/bsr20212378 (2022).
Chemotherapy for high-grade glioma. The Cochrane database of systematic reviews, Cd003913. (2002). https://doi.org/10.1002/14651858.Cd003913
Wang, Z. et al. PD-L2 expression is correlated with the molecular and clinical features of glioma, and acts as an unfavorable prognostic factor. Oncoimmunology 8 https://doi.org/10.1080/2162402X.2018.1541535 (2019).
Li, T. et al. New progresses on cell surface protein HSPA5/BiP/GRP78 in cancers and COVID-19. Front. Immunol. 14 https://doi.org/10.3389/fimmu.2023.1166680 (2023).
Pyrko, P., Schoenthal, A. H., Hofman, F. M., Chen, T. C. & Lee, A. S. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 67, 9809–9816. https://doi.org/10.1158/0008-5472.CAN-07-0625 (2007).
Funding
This work was supported by the National Natural Science Foundation of China (82151310), the National Key R&D Program of China (2022YFC2405301), Beijing Natural Science Foundation (L246018), Beijing Municipal Health Commission (BRWEP2024W032040205), Beijing Municipal Administration of Hospitals Incubating Program (PX2024019) and the Postdoctoral Fellowship Program of CPSF (GZC20231749).
Author information
Authors and Affiliations
Contributions
D.B.Z and P.Y designed this study. Z.Y.S and H.B.W analyzed the data. Z.Y.S and P.Y wrote the manuscript. M.H.D revised the manuscript and performed experimental analysis. D.B.Z mainly provided many comments and suggestions, and edited the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, Z., Wan, H., Du, M. et al. Heat shock proteins related signature characterizes immune status and predicts prognosis of gliomas. Sci Rep 15, 28652 (2025). https://doi.org/10.1038/s41598-025-12509-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-12509-2