Abstract
The Coronavirus disease 2019 (COVID-19) has rapidly become the worst pandemic since the 1918 influenza pandemic. Studies have shown that severe COVID-19 patients have immune dysfunction. To characterize the dysregulated immune response to SARS-CoV-2 infection, we performed a comprehensive analysis of scRNA-seq and scV(D)J-seq in peripheral blood mononuclear cells from mild, moderate, and severe patients. We observed that as the severity of the disease increased, several CD8 + T cell subsets and Treg cells continued to decrease, while CD4 + T subsets, natural killer cells and plasma cells continued to increase. Several aberrantly expressed biomarkers associated with SARS-CoV-2 severity were identified. For example, RPS26 was down-regulated, while the ZFP36, IL-32, and IgM genes were up-regulated with increasing disease severity. Functional analysis showed multiple immune-related pathways, such as interleukin-2 and interleukin-10 production pathways, were dysregulated. As the disease severity increased, intercellular interactions fluctuated. Particularly, naive CD8 + T cells regulated memory and activated CD8 + T cells, and the weakening in Treg cells’ regulation of other immune cells was especially obvious. The expression of the MIF signaling pathway, mediated by CD74 + CXCR4, was higher throughout SARS-CoV-2 infection and the intensity of immune cell-cell interactions mediated by TGF-β was enhanced from mild to severe. Subsequently, scV(D)J-seq analysis showed a decreasing trend in the number of clonotypes, repertoire diversity and clonotypes overlap of monoclonal B cell receptor (BCR) and T cell receptor (TCR) as the SARS-CoV-2 progresses. The CDR3 sequence length in COVID-19-specific clonotypes showed a bias towards being longer as the severity of COVID-19 increases. Our findings may provide new clues for understanding COVID-19 immunopathogenesis and help identify optimal biomarkers for new therapeutic strategies.
Similar content being viewed by others
Introduction
In December 2019, pneumonia caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was reported in Wuhan, Hubei Province, China. Until today, according to the incomplete statistics of the World Health Organization (WHO) the number of global cases caused by Novel Coronavirus infection has reached 4.6 million and the number of deaths has reached 300,000 people1.
The clinical manifestations of COVID-19 are very diverse, ranging from mild to severe. Mild COVID-19 is described as COVID-19 without pneumonia or with a mild course of disease2.In moderate cases, infected patients develop lung injury characterized by bilateral infiltrates, as well as dyspnea and hypoxia, requiring hospitalization and supplemental oxygen. Finally, some patients develop severe or fatal COVID-19, which also includes respiratory failure due to acute respiratory distress syndrome (ARDS), as well as shock and/or hypercytokinemia leading to multi-organ dysfunction3. In particular, lymphopenia and inflammatory cytokine storm are typical abnormalities found in COVID-19 patients and may be related to the severity of the disease, highlighting the potent capacity of SARS-CoV-2 to suppress the adaptive immune response4. All these events lead to invasive mechanical ventilation and ultimately to patient death. Fortunately, not all individuals infected with SARS-CoV-2 develop severe respiratory disease; as most infections remain as essentially mild5. The reasons for the evolution of these different clinical manifestations remain largely unknown, so there is an urgent need to better understand COVID-19, not only to control the spread of the pandemic, but also to identify accurate biomarkers that may help to identify as soon as possible those patients who are at risk of developing the most severe and lethal forms of COVID-19, so that the most adequate preventive and therapeutic measures can be taken6.
Advances in biotechnology have facilitated biomedical research. Single-cell RNA sequencing (scRNA-seq) technology, which enables high-throughput sequencing at the single-cell level, has been widely used in a variety of disease studies7. In particular, single-cell RNA sequencing technology played an important role in the global pandemic of the novel coronavirus, revealing multiple immune features of the novel coronavirus at the cellular and molecular levels8. Peripheral blood mononuclear cells (PBMCs) are key components of the immune system that play a crucial role in defense against viral infections and contain many immune cells (e.g., leukocytes) and immune molecules (e.g., specific plasma proteins). Studies based on PBMCs are more conducive to discovering the proportion changes of immune cells and immune molecules related to the disease severity in the COVID-19 patients9. According to previous studies on SARS-CoV and MERS-CoV, dysregulation of the host immune response and production of inflammatory cytokines during SARS-CoV and MERS-CoV infections have been found to correlate with disease severity and poor prognosis10. Several pro-inflammatory cytokines and chemokines (e.g., CCL-2, CCL-3, RANTES, IL-2, and IL-8) were abnormally elevated during MERS-CoV infection of human peripheral blood monocyte-derived macrophages11. Interestingly, recent studies have reported elevated plasma IGKV2D-29, IGLV1-3, and TNFα levels in severe COVID-19 cases as compared to mild cases, suggesting that the release of inflammatory cytokines is critical for COVID-19 progression12. Additionally, the analysis of changes in the proportion of immune cells in PBMC showed that lymphocytopenia was common in SARS-CoV-2-infected patients, probably due to lymphocyte-induced apoptosis13.
The T cell receptor (TCR) and B cell receptor (BCR), which are present on the surface of T and B cells, respectively, are key molecules in the adaptive immune response14. The TCR/BCR-mediated immune response is one of the major defense mechanisms of the adaptive immune system against viruses. Both strands of the TCR (α and β) consist of a variable (V) region, a junction (J) region, and a constant (C) region. The diversity (D) region connects the V and J regions to form a complete β chain. Thus, the TCR recombination process generates highly diverse complementary-determining regions (CDRs) located in the TCR α and β strands, forming a functional and highly diverse TCR library15.The CDR sequences determine the specificity of TCR binding to pMHC, where the third CDR (CDR3) is highly variable and contributes to direct peptide recognition. Thus, revealing the size and diversity of the TCR repertoire may aid in understand the T cell-mediated immune response against SARS-CoV-2 infection16. Previous studies have reported TCR clonal bias in COVID-19 patients. For example, Minerva A. et al. revealed memory T-cell dynamics in patients with mild COVID-19 infection by T cell receptor (TCR) sequencing17.Snyder T. et al. developed a diagnostic method for SARS-CoV-2 infection based on TCR sequencing of blood samples18. Luo et al. found that TCR clonotypes and BCR diversity were reduced in individuals infected with COVID-1919. Several studies have demonstrated bias in TCR and BCR after COVID-19 infection. However, the changes in TCR and BCR clonotypes and CDR3 regions in COVID-19 patients targeting different severities remain unknown.
Therefore, in order to investigate the immune response to COVID-19 and its correlation with severity, we applied scRNA-seq and scV(D)J-seq to perform a comparative analysis of peripheral blood mononuclear cells from two mild, two moderate, and five severe COVID-19 patients. Our study provides an unbiased and comprehensive view of the immune response of PBMCs across mild, moderate and severe COVID-19 patients, illustrates the changing nature of the cellular response, and reveals key elements of antiviral immunity and pathogenesis in moderate and severe patients. The transcriptomic data, combined with detailed TCR- and BCR-derived lineage information, may serve as a rich resource for insights into peripheral lymphocytes from COVID-19 patients and pave the way for the rational design of SARS-CoV-2-specific therapeutic strategies.
Materials and methods
Sample data collection
We obtained paired single-cell RNA sequencing (scRNA-seq) and single-cell V(D)J sequencing (scV(D)J-seq) data from 9 COVID-19 patients (2 mild, 2 moderate, and 5 severe cases) from the same individuals (Table S1). These data, sourced from http://www.microbiome-bigdata.com/project/SARS-CoV-2/, provide coverage across varying disease severities. The dataset comprised 8,163 cells from mild cases, 6,841 cells from moderate cases and 16,466 cells from severe cases. Specifically, we classified the samples into severity categories (mild/moderate/severe) according to the following criteria. Briefly, moderate cases have fever, respiratory symptoms, and pneumonia evidenced by computed tomography (CT) imaging. This study enrolled two patients with moderate infection with bilateral pneumonia, labeled M3and M4 two patients with mild infection, labeled M1and M2 Patients with severe pneumonia were diagnosed on the basis of one of the following criteria: (1) respiratory distress with respiratory rate ≥ 30 times min − 1; (2) fingertip oxygen saturation ≤ 93% at room air; (3) ratio of partial pressure of arterial oxygen to fraction of inspired oxygen (PaO2/FiO2) ≤ 300 mmHg (1 mmHg = 0.133 kPa); and (4) obvious progression of lesions in 24–48 h shown by pulmonary CT imaging > 50%. Two patients with severe pneumonia were enrolled, labeled as S1, S2, S3, and S5. These four patients showed PaO2/FiO2 ≤ 300 mmHg. The patients with critical pneumonia (S4) were diagnosed on the basis of one of the following criteria: (1) respiratory failure and an artificial airway required for invasive mechanical ventilation; (2) shock; and (3) combined failure of other organs that required intensive care unit monitoring. S4 is the patient with critical pneumonia received invasive mechanical ventilation20.
scRNA-seq expression profile analysis
The scRNA-seq data were downscaled, clustered and analyzed using Seurat21. To visualize the data, we further reduced the dimensionality of all 31,470 cells using Seurat and projected the cells into two-dimensional space using t-distributed stochastic neighborhood embedding (t-SNE)22. The steps included (1) calculating gene expression levels using the LogNormalize method of the “Normalization” function of the Seurat software; (2) performing principal component analysis (PCA) using the normalized expression levels, and selecting the top 10 principal components (PCs) for clustering and t-SNE analysis; and (3) finding clusters using the Run Harmony clustering-based method. Marker genes for each cluster were identified using the FindAllMarkers function in Seurat using default parameters23. Specifically, we identified ten major cell types using standard markers (T cells: Naive CD4 + T cell; NK cell: Cytotoxic CD4 + T cell; Memory CD4 + T cell; Memory CD8 + T cell; Treg; Naive CD8 + T cell; Activated CD8 + T cell; B cells: Plasma; Naive B cell).
Differential gene expression analysis
Differential gene expression analysis was performed using Seurat v.3 (Find Markers function) and default parameters24. We identified differentially expressed genes for each type of COVID-19 sample by comparing Treg, Activated CD8 + T cells, and Plasma cells of across COVID-19 sample different severities. Genes with an adjusted P < 0.05 and a fold change > 1.5 were considered to be significantly upregulated in patients and were used for further GO enrichment analysis.
GO and pathway enrichment analysis
GO and pathway enrichment analyses were performed using clusterProfiler (P value Cutoff = 0.01 and q value Cutoff = 0.05). For each comparison, the top ten enriched pathways containing at least five upregulated genes were retained.
Intercellular communication analysis
Intercellular communication of cell clusters was calculated and analyzed using the CellChat software package25. We analyzed immune cell communication patterns in PBMCs of COVID-19 patients with different disease severities following the standard procedure recommended by CellChat. First, CellChat objects were generated using the “create CellChat” function. Subsequently, communication probabilities were derived by using the “compute CommunProb” and “compute CommunProb Pathway” functions. The network centrality score and the contribution of each ligand-receptor pair on the signaling pathway were calculated using the “netAnalysis_computeCentrality” and “netAnalysis_contribution” functions, respectively. Finally, all visualizations describing cellular communication were generated using the embedded functions in the Cell Chat package. Heat maps and bubble diagrams were used to visually represent potential interactions between ligands and receptors, as well as cell-cell communication networks in the TGF-β signaling pathway.
scV(D)J-seq data processing and analysis
The R package scRepertoire was used to further combine the contig_annotation data from different samples into a single list object (function combineTCR)26. The combined TCR contig list file was then integrated with the corresponding Seurat object of the scRNA-seq data using the function combineExpression (cloneCall = “gene + nt”). Only the cells with both TCR and scRNA-seq data were kept for downstream clonotype analysis. The clonotype was defined according to the genes comprising the TCR and the nucleotide sequence of the CDR3 region. The frequency of the each clonotype in each patient was then calculated as clone count. To get a normalized clone count size for each clonotype, we also calculated the clone size proportion (clone count divided by number of cells per patient). The clone count and clone size proportion were added to metadata of the single-cell matrices. The number of TCR/BCR clonotypes was calculated using the quantContig function. The AbundanceContig function was used to calculate clonal abundance. Only cells with productive, paired TCR α and β chains were retained for downstream analyses27,28. CDR3 sequences were required to have length greater than or equal to 5 amino acids29. The distribution of CDR3 amino acid sequences (whole, TRA, and TRB) was calculated using the lengthContig function. The compareClonotypes function was used to compare the rate of clonotype overlap in patients with different severity levels.
Results
Single-cell transcriptome profiles of PBMCs in COVID-19 patients with different severity levels
In order to characterize the adaptive immune systems of patients with different severities of COVID-19 and understand their immune heterogeneity, we performed scRNA-seq analyses on immune cells from mild (M1, M2), moderate (M3, M4) and severe (S1-S5) cases (Fig. 1A). After quality control, a total of 31,470 cells were obtained for downstream analysis. Using the Louvain clustering algorithm and based on CellMaker 2.030, combined with the expression of canonical genes, we identified 8 different clusters representing different T-cell subpopulations and 2 different clusters representing different B cell subpopulations. Then t-distributed stochastic neighbor embedding (t-SNE) was performed to observe the cells in two-dimensional space (Fig. 1B), and the samples with different severity levels were clustered separately (Fig. 1C). To reveal changes in the composition of COVID-19-associated T-cell or B-cell subpopulations of different severity levels, we quantified the proportion of all cell types for each individual in mild, moderate and severe samples (Fig. 1D). Regulatory T cells (Treg) and Naive CD4 + T cells were identified in T cell subpopulations based on the expression of marker genes CCL4 and CD25 as well as ACTN1 and CCR7, respectively. Plasma Cells and Naive B cells were identified in B cell subsets based on the expression of marker genes CXCR4 and CD38 as well as CD20 and IgD, respectively (Fig. 1E).
Single-cell sequencing data analysis of peripheral blood mononuclear cells from COVID-19 patients. (A) Specific information of the experimental individuals. (B) t-SNE representation of PBMCs from all experimental samples (n = 9), colored by manually annotated cell types. (C) t-SNE representations of PBMCs from mild (n = 2), moderate (n = 2) and severe (n = 5) samples respectively, colored by manually annotated cell types. (D) A t-SNE plot showing the expression of typical cell marker genes used to identify the above immune cell types. Each point represents a cell and the colors in the graph are labelled according to the logarithmic expression level of each marker gene. (E) Percentage of immune cell subtypes in all experimental individuals (n = 9). Horizontal coordinates indicate individual patients, vertical coordinates indicate the proportion of immune cell subpopulations, and different colors represent individual immune cell subtypes. (F) Dynamic changes of immune cell subtypes related to the severity of the disease. The horizontal coordinate of each of these line graphs represents the disease severity, the vertical coordinate represents the proportion of immune cell subtypes, and the colors represent different disease severities. The pie chart illustrates the proportion of each immune cell subtype across different severity levels. The cell fractions represent an integrated analysis combining all samples.
Overall, we observed that as the severity increased, several T cell subsets continued to decrease, including memory CD8 + T cells, activated CD8 + T cells, naive CD8 + T cells, and Tregs. In contrast, the proportions of cytotoxic CD4 + T cells, naive CD4 + T cells, B cells, natural killer cells and plasma cells gradually increase with increasing disease severity (Fig. 1F), suggesting that the proliferation of these cells is associated with COVID-19. Notably, the increased proportion of plasma cells in patients with severe COVID-19 suggests that more severe cases may be associated with a stronger humoral immune response, similar to previous literature. Lymphopenia is a prominent feature of severe COVID-19 immune disorders31. Here, we found that the lymphopenia associated with COVID-19 included not only depletion of CD8 + T cells but also a significant loss of Treg. In addition, we noted an increased frequency of CD4 + T cells such as memory CD4 + T cells, naive CD4 + T cells, and cytotoxic CD4 + T cells (Fig. 1F).
Comparison of PBMC cell subtypes related to the severity of COVID-19
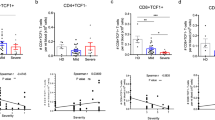
To further characterize the functional status of T and B cells related to the severity of COVID-19, we screened Treg, activated CD8+, and plasma cell subtypes that have persistent changes and important roles in COVID-19, and then performed differentially expressed gene (DEG) analysis and gene pathway enrichment analysis. We compared and analyzed the differentially expressed genes between different types of samples, respectively, and further investigated the typical pathways in multiple COVID-19 patients (Fig. 2A). As a result, we found that the ribosomal protein S26 (RPS26) was down-regulated and the ZFP36 gene was up-regulated with increasing disease severity (Fig. 2B). Functional analysis demonstrated that differential down-regulated genes were enriched in the “negative regulation of interleukin-2 production” signaling pathway in both Treg and activated CD8 + T cells in moderate COVID-19 patients compared with mild COVID-19 patients (Fig. 2C). And we observed that the differential up-regulated genes in the comparison between moderate and severe patients were significantly enriched in the “positive regulation of interleukin-10 production” and “regulation of interleukin-32 production”. In plasma cells, the signaling pathways of “positive regulation of leukocyte activation” and “regulation of adaptive immune response” were highly enriched in patients with mild disease. The signaling pathways of “regulation of viral gene replication”, “regulation of RNA splicing”, and “regulation of lymphocyte differentiation” were enriched in patients with moderate to severe disease (Fig. 2C). And we found that IgM gene was slightly upregulated in severe patients. Humoral immunity is mainly mediated by antibodies produced by plasma cells32. Among them, antibody-mediated immune response plays a crucial role in SARS-CoV-2 infection. Another study found that the intensity of the neutralizing antibody response was positively correlated with disease severity. Higher levels of antibodies, such as having high IgM levels, were present in patients with severe COVID-19. Anti-SARS-CoV-2 antibody-mediated immune response inhibits viral infection while inducing the secretion of pro-inflammatory cytokines, which may lead to cytokine storm and acute respiratory distress syndrome33. It thus exacerbates the severity of the disease.
Comparison of PBMC cell subtypes associated with the severity of COVID-19 disease. (A) Analysis of differential gene expression in Treg, activated CD8 + T cell and plasma cells related to the severity of the disease. Each dot represents a gene, where the color of the dot represents the expression of the gene and the color of the gene name represents the comparison between different severities. (B) Differential up-regulated genes in the three immune cells were selected for GO pathway enrichment, respectively. (C) Expression levels of key genes related to the severity of the disease. In all box plots, boxes show median, upper quartile and lower quartile. Dots represent cells expressing the gene and the color of the dot indicates the severity of the disease.
Comparison analysis identifies crucial signaling changes responsible for disease severity related to COVID-19
The immune response and elevated TGF-β levels led to the elevation of the level of viral-specific IgA, which may exacerbate severe COVID-19. An effective defense against SARS-CoV-2 infection requires cooperation among multiple, gradually strengthened immune cell subtypes. Bioinformatics methods developed from scRNA-seq data can predict intercellular communication through ligand-receptor interaction networks. To date, a number of scRNA-seq studies have analyzed intercellular communication in COVID-19 patients. Our study analyzed the altered intercellular communication between immune cell types in PBMC of COVID-19 patients with different severity. We first comparatively analyzed changes in the intensity of immune cell interactions in different severity levels of the disease. The results showed that the strength of immune cell interactions gradually weakened as the severity of the disease increases (Fig. 3A). Next, the study analyzed multiple ligand-receptor mediated cellular interactions (Fig. 3B and D). We found that the expression of the MIF signaling pathway, mediated by CD74 + CXCR4, was higher throughout the novel coronavirus infection, especially in Naive CD8 + T cell-regulated Activated CD8 + T cells, and that macrophage migration inhibitory factor (MIF), a pleiotropic immunomodulatory cytokine, prevented random immune cell movement. MIF has direct chemokine functions, promoting “directed” cell migration (i.e. chemotaxis) and plays an important role in inflammatory leukocyte recruitment34. At the molecular level, these activities are based on non-homologous interactions between MIF and the CXC chemokine receptors CXCR2 and CXCR4. Importantly, MIF also interacts with surface CD74, a type II transmembrane protein, inducing its phosphorylation and CD44 recruitment, ultimately leading to ERK1/2 phosphorylation. Several studies have pointed out the utility of MIF as a biomarker for different diseases with an inflammatory component35.
Immune cell interaction network of COVID-19 samples with different severity levels. (A) Immunocyte interactions network diagram, in order of mild, moderate and severe, respectively. Where the dots represent all identified immune cell subtypes, the line between two dots represents cell interactions, and the weight of the line represents the intensity of cell interactions. (B-D) Bubble plots representing the detailed pathways of cell-cell interactions between all identified immune cell subtypes in mild, moderate, and severe samples, respectively. Horizontal coordinates are the immune cell-to-cell interactions and vertical coordinates are the specific pathways of cell-to-cell interactions.
Transforming growth factor β (TGF-β) is a major pleiotropic factor that regulates the immune response. It targets antibody-like recombination to IgA, inhibits T-cell and NK-cell activity36, and supports the differentiation of regulatory T-cells37. In addition to its central role in regulating the immune response, it is also involved in tissue regeneration. Thus, it may also regulate fibrosis and thrombosis in COVID-19, as well as the reactivation of herpesviruses, including Epstein-Barr virus, which is characteristic of COVID-19 pathogenesis. Many immune cell types express TGF-β, but there is little information on the TGF-β signaling pathway. We therefore comparatively analyzed TGF-β-mediated interactions between immune cells (Fig. 4A and B), and the expression of some key genes in the TGF-β pathway in immune cell subpopulations (Fig. 4C), and statistically analyzed the expression of TGF β 1 gene in immune cells (Fig. 4D). The results showed that the intensity of immune cell-to-cell interactions mediated by TGF-β was weakened from mild to severe and TGF-β1 expression was up-regulated with increasing severity.
The TGF-β signaling pathway plays a significant role in patients with COVID-19 of different severity levels. (A) Network diagram of TGF-β signaling pathway, in order of mild and severe samples. In the hierarchical diagram, solid and hollow circles indicate sources and targets, respectively. The thicker the line, the stronger the reciprocal signals. The target in the middle of the left diagram is our selected target cell. The right figure is another set of cells other than the selected target cells treated as target cells to analyze the interactions. (B) Heat map of immune cell interactions in the TGF-β signaling pathway, in order of mild and severe samples. The vertical axis is the cell that sends the signal, the horizontal axis is the cell that receives the signal, and the color shade of the heat map represents the signal strength. The bars on the upper and right sides are cumulative of the intensity of the vertical and horizontal axes. (C) Expression of all genes in the TGF-β signaling pathway in immune cell populations, in order of mild and severe samples. (D) Statistical chart of TGF β 1 gene expression in Memory CD4 + T cells, Treg, and Naive CD4 + T cells as the severity of COVID-19 increases.
Prediction of potentially sensitive drugs for COVID-19 patients with different severity levels
In order to explore the sensitivity of COVID-19 patients with different severity levels to common non-chemotherapy drugs, we used the R package oncoPredict to analyze the half-maximal inhibitory concentration (IC50) values of COVID-19 patients with different severity levels38,39. The oncoPredict used the gene expression profile and IC50 of the cancer cell lines to drugs from CTRP and GDSC (training dataset) to build ridge regression models40. The IC50 value of each drug in each sample from COVID-19 patients with different severity levels was assessed via the “oncoPredict” package with GDSC and CTRP as the training set, representing the potential drug response in that patient. Drug sensitivity across COVID-19 patient groups with different severity levels was analyzed using Wilcoxon rank sum tests. A significance threshold of P < 0.05 was set for statistical significance. In drug sensitivity prediction, the IC50 value is an important indicator for measuring the inhibitory effect of a drug on cell growth or biological functions. A lower IC50 value indicates that the drug can effectively inhibit the proliferation or other biological functions of the target cells at a lower concentration, thus a lower IC50 value usually indicates higher drug sensitivity41. Finally, we identified 6 non-chemotherapy drugs, including Vilobelimab, Baricitinib, SB-225,002, Paxlovid, Anakinra, and Tocilizumab, that exhibited significantly lower IC50 values (indicating enhanced drug sensitivity) with worsening COVID-19 severity (Fig. 5). Additionally, drugs that showed no significant differences in IC50 values across COVID-19 severity levels were selected as the negative control group (Fig. S1). Compared with mild samples, these drugs show lower IC50 values in moderate to severe samples, indicating better therapeutic effects on moderate to severe COVID-19. For example, existing studies have confirmed that the drugs Vilobelimab, Baricitinib, Anakinra, Paxlovid and Tocilizumab are suitable for patients with severe COVID-19, and they significantly reduce the risk of death42,43. Vilobelimab attenuates excessive C5a-mediated inflammatory responses, including cytokine storm, endothelial dysfunction, and coagulopathy, while preserving the immunological activity of the membrane attack complex (C5b-9). It is indicated for adult patients with severe COVID-19 who have undergone invasive mechanical ventilation (IMV) or extracorporeal membrane oxygenation (ECMO) within 48 h44. Baricitinib exerts its therapeutic effect by inhibiting the JAK-STAT signaling pathway, thereby suppressing the release of pro-inflammatory cytokines such as IL-6 and IFN-γ. The World Health Organization (WHO) strongly recommends its use in combination with corticosteroids for patients requiring oxygen support due to critical or life-threatening illness45. Paxlovid inhibits SARS-CoV-2 viral replication through protease inhibition and is recommended for individuals at high risk of disease progression46. Anakinra, a recombinant human interleukin-1 receptor antagonist, neutralizes IL-1β-driven hyperinflammatory reactions. A prospective cohort study conducted at the University of Paris (n = 52) demonstrated a 75% reduction in the risk of mechanical ventilation requirement or mortality47. Tocilizumab, an anti-IL-6 receptor monoclonal antibody, directly targets a central mediator of the cytokine storm. Clinical data indicate that among patients with severe pneumonia and serum IL-6 levels exceeding 40 pg/mL, tocilizumab reduces the 28-day risk of death or need for mechanical ventilation by 19.4%48. Moreover, we identified SB-225,002 as a potential therapeutic candidate, demonstrating lower IC50 values in both moderate and severe COVID-19 patients. Although SB-225,002 has no data on treating COVID-19 in clinical trials, as an antiviral drug, it has a certain inhibitory effect on inflammation49. As a disease caused by a novel coronavirus, the treatment methods and drug choices for COVID-19 are not yet fully established. Drug sensitivity prediction, as an auxiliary technology for treatment decision-making, can provide personalized treatment plans based on the gene expression data of patient samples. Drug sensitivity prediction offers new ideas for the treatment of COVID-19, especially in the context of the increasing importance of individualized medicine and precision treatment. In the future, more precise drug sensitivity prediction combined with clinical data will further promote the individualization and precision of COVID-19 treatment plans.
Drug sensitivity analysis of potential non-chemotherapy drugs in COVID-19 patients across different severity levels. Drug sensitivity across COVID-19 patient groups stratified by severity level was analyzed using Wilcoxon rank sum tests, with a statistical significance threshold set at P < 0.05. A “low” IC50 value was defined as significantly lower values observed in comparative analyses among the mild-moderate, mild-severe, or moderate-severe groups, indicating enhanced drug sensitivity in the corresponding COVID-19 severity group. Each point represents the predicted IC50 value of a specific drug for a COVID-19 patient. To improve clarity and comparability, we used standardized IC50 values as the y-axis ranges across all plots.
TCR and BCR expansion in patients with COVID-19
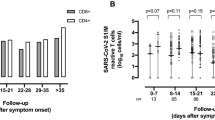
T-cell receptors (TCRs) and B-cell receptors (BCRs) are critical in immune cell-mediated virus clearance, and their diversity and specificity are fundamental to adaptive immunity. To date, a TCR/BCR bias has been observed in a variety of diseases. Activated T cells undergo a process known as “clonal expansion”, in which activated T cells proliferate rapidly and generate large numbers of T cells with the same TCR50. To explore the TCR and BCR bias in COVID-19, we performed a comparative analysis of COVID-19 patients with different severities using single-cell V(D)J sequencing. A total of 29,275 T cells with complete TCR alpha and beta chain sequences and 20,022 B cells with complete BCR heavy and light chain sequences were obtained from COVID-19 patients. The number of TCR unique clonotypes was first comparatively analyzed between different patients and between different severities (Fig. 6A-B). From the resultant graphs, we could clearly see a continuous decreasing trend in the number of TCR clonotypes as the disease progresses. And it is consistent with the resulting graph showing the number of clonotypes folding plot (Fig. 6C). Next we calculated the diversity indices including Chao index, Shannon index, ACE index and inverse Simpson index, respectively (Fig. 6D). The results showed that TCR diversity was lower in patients with severe COVID-19. This decrease in diversity suggests a more focused and restricted T-cell response in severe COVID-19 patients, which may be due to the specific targeting of SARS-CoV-2 antigen51. In contrast, patients with mild to moderate disease exhibited higher TCR diversity. We conclude that the effective clonal expansion of T cells in severe COVID-19 patients leads to decreased TCR diversity, which may contribute to their impaired ability to establish effective viral control52. Next, we analyzed the distribution patterns of TCR repertoire proportions across varying severity stages of COVID-19 to determine whether significant differences existed between these groups (Fig. 6E and F). The x-axis in the proportion of clonal clonotypes represents the clonotype index (Fig. 6F). A small clonotype index indicates an expanded clonotype, and a large index indicates a small clonotype group53. From the result graph, we can observe that in the [1:10] and [1001:10000] clone types, samples of different severity levels show significant differences (Fig. 6F). Severe COVID-19 patients have a relatively high proportion in the small clonotype index, indicating significant clonal expansion, which subsequently leads to a decrease in TCR clonal diversity in severe samples. Finally, the relative abundance of the TCR clonotypes was measured. The x-axis of Fig. 6G represents the clonotype groups, which were divided based on the expression levels of the clonotypes. The results show that, compared with the mild to moderate samples, the proportion of small clonetypes in the severe samples was relatively low, while the proportions in the large and hyperexpanded clonetypes significantly increased (Fig. 6G). We further investigated the presence of shared TCR clonotypes between COVID-19 patients of varying severity (Fig. 7A). Contrary to the expectation that shared clonotypes are prevalent, we found that the majority of TCR clones were private in the sample, suggesting significant heterogeneity in the TCR repertoire of COVID-19 patients54. This heterogeneity suggests that the TCR repertoire may reflect the severity of the disease and may serve as a valuable tool for immunodiagnosis. Indeed, perfect accuracy was achieved in classifying COVID-19 patients using the identified SARS-CoV-2-associated TCRs, demonstrating the potential of TCR analysis as a diagnostic test based on high-throughput sequencing of peripheral blood55. In addition, the distribution of TCR CDR3 sequence lengths showed significant differences in the CDR3 length distribution with different severity levels (Fig. 7B). The length range of CDR3 in mild samples is mainly concentrated in 25–30 aa, while that in severe samples is mainly concentrated in 27–33 aa. TCR CDR3 could vary in length and sequence, which enables them to recognize multiple antigens. In fact, we found that compared with mild samples, severe COVID-19 patients tended to choose longer clonal (Fig. 7C). The immune system of critically ill patients is often in a highly activated state, generating more long cloned T cells to cope with severe viral infections. However, the immune response of mild patients is relatively weak, and the distribution of clone length is also more limited56.
Comparative analysis of TCR differences related to the severity of COVID-19. (A) Analysis of the number of TCR clonotypes in individuals of different severity, the left panel shows the total number of clonotypes per sample and the right panel shows the number of clonal species. (B) Analysis of the number of TCR clonotypes in samples of three severity levels; the left panel shows the total number of clonotypes per sample and the right panel shows the number of clonal species. (C) The fold plot demonstrates the relative distribution of TCR clonotypes in individuals of different severities by abundance. (D) TCR clonotype diversity analyzed by Shannon, inverse Simpson, Chao, and ACE indices, where dots indicate different levels of severity. (E) Clonal spatial homeostasis analysis. The stacked barplot with patient ID on x-axis, relative abundance/occupied space on y-axis and color as clonotype group/clonal indices. (F) Differences in TCR clone occupied repertoire space among different severity levels of COVID-19. The x-axis represents the clonotype index, indicating in the proportion of top clonal clonotypes. (G) Differences in TCR clonal relative abundances among different severity levels of COVID-19. The horizontal axis represents the relative abundance of TCR clones, where “Small” indicates < 0.1%, “Medium” indicates 0.1%−1%, “Large” indicates 1%−10%, and “Hyperexpanded” indicates an abundance exceeding 10%. The vertical axis represents the relative abundance of the small, medium, large, and hyperexpanded clonotypes. Non-significant p-values (ns) and significant p-values (*, p < 0.05) were determined using t-test.
Analysis of the overlap rate of TCR clonal types and CDR3 length related to the severity of COVID-19. (A) Bar graph demonstrating the similarity of TCR clonotypes overlap between individuals of different severity. Colours indicate TCR clonotypes. (B) Bar graphs show CDR3 aa length distribution of TCRs in COVID-19 patients with different severity levels (TRA: α chains, TRB: β chains, aa: amino acid). (C)Comparison of average CDR3 length of COVID-19 of different severity levels.
In addition to its application in TCR, scV(D)J-seq was also used to study the BCR response profile in COVID-19 patients. The results of the BCR study were more consistent compared to the TCR57. We similarly comparatively analyzed the number of BCR clonotypes (Fig. 8), clonal diversity abundance (Fig. 8B) and clonotype overlap (Fig. 8C) in COVID-19 patients. The results showed a decreasing trend in the number of BCR clonotypes, lower BCR diversity and lower clonotype overlap, which were highly similar to those in the TCR analysis. Based on high-throughput single-cell transcriptome data and paired TCR/BCR sequence data, our study comprehensively portrayed the proportions of each cell type in each sample, clonal characteristics, and other features of immune responses in patients with different symptoms of COVID-19 infection. It provided valuable data references and resources for further understanding, controlling and attacking the novel coronavirus pneumonia epidemic58.
Comparative analysis of BCR differences related to the severity of COVID-19. (A) Analysis of the number of BCR clonotypes in samples of three severity levels; the left panel shows the total number of clonotypes per sample and the left panel shows the total number of clonotypes per sample and the right panel shows the number of clonal species. (B) BCR clonotype diversity was analyzed by Shannon, inverse Simpson, Chao, and ACE indices, where dots indicate different severity. (C) Heat map indicating the overlapping similarity of BCR clonotypes among individuals of different severity.
Discussion
Patients with severe COVID-19 present primarily with respiratory disease, but as the disease progresses, it can lead to severe lung disease and associated complications with high morbidity and mortality. COVID-19 is now largely controlled worldwide, but this fatal lung disease remains a significant threat to human health59. In this study, we analyzed scRNA-seq data and scV(D)J-seq data from PBMCs of COVID-19 patients with different disease severities to construct a global landscape of COVID-19 immunity. The study revealed changes in immune cell subtype occupancy, cytokines, and cellular interaction pathways related to the severity of COVID-19. Analysis of scV(D)J-seq data revealed TCR/BCR clonal amplification and differences in CDR3 regions across severity levels.
Our study provides an unbiased visualization of the immunological profile of COVID-19 patients. First, lymphopenia is one of the important immunological features in patients with severe COVID-1960. In our study, we clearly observed a reduced percentage of CD8 + T cells and Treg cells in severe compared to mild. It should be noted that the increased proportion of plasma cells in patients with severe COVID-19 suggests that this may be related to a stronger humoral immune response61. Moreover, there was a tendency for the CD4 + T-cell subpopulation to expand as the severity of COVID-19 increased, suggesting that they were activated in the severe samples. Therefore, we suspect that disturbed T-cell compartmentalization may be one of the immunopathogenic mechanisms of COVID-19, such as impaired antiviral responses of Treg and CD8 + T cells, and inflammatory expansion and induction of aberrant antibody responses of CD4 + T cells, which are induced to produce cytokines locally, likely leading to cytokine storm as well as peripheral blood lymphocytopenia. Interferons, interleukins, chemokines and TNF are the main factors responsible for the occurrence of cytokine storm in COVID-19 patients. In this study, we found that the RPS26 was down-regulated and the ZFP36 gene was up-regulated with increasing disease severity, RPS26 plays a key role in ribosome assembly and protein translation, and has been shown to be associated with important physiological and pathological processes, including the regulation of T cell development and immune-related diseases62. ZFP36 plays a regulatory role in the pro-inflammatory response of coronavirus-infected cells. By binding to these Adenosine Rich Element (ARE)-containing transcripts, including IL-6, IL-8, IL-17 A, IL-33, TNF-α (tumor necrosis factor-α), CXCL2, ZFP36 regulates the mRNA levels of key cytokines and other important genes related to pro-inflammatory and host innate immune responses63. ZFP36 regulates the mRNA levels of key cytokines and other important genes associated with pro-inflammatory and host innate immune responses, leading to dysregulation and excessive release of pro-inflammatory cytokines, which is one of the clinical hallmarks of immunopathogenesis in coronavirus disease64.
Secondly, we identified several functional pathways that were significantly enriched in samples of COVID-19 of different severity levels, such as “negative regulation of interleukin-2 production”, “regulation of interleukin-10 production” and “regulation of interleukin-32 production”. Dysregulation of interleukin production can exacerbate the severity of COVID-19 by leading to cytokine storms and tissue damage. Up-regulation of the IL-2 cytokine family has been shown to be a clinical outcome of the disease and correlates with disease severity. Studies have shown that increased levels of IL-2 may lead to downregulation of CD8 + T cells and a reduction in their immunological activity. Therefore, the IL-2 cytokine family could be used as a potential biomarker to monitor severity of COVID-19 and therapeutic efficacy45. The key adverse prognostic factor in the pathophysiology of SARS-CoV-2 infection is cytokine storm, a hyper inflammatory process in which cytokine release leads to an acute systemic response65. This particular state of immune response drives the disease state toward acute respiratory distress syndrome (ARDS). Although the rapid response of the innate immune system following SARS-CoV-2 infection is the first line of defense against COVID-19, an overactive immune response can produce serious complications66. During SARS-CoV-2 infection, irreversible and severe damage occurs in the lungs due to elevated plasma concentrations of circulating interleukins. IL-10 is a major pleiotropic interleukin involved in both inflammatory and anti-inflammatory response observed in humans during any infection. IL-10 is responsible for the maintenance of homeostatic balance of the immune system by exerting anti-inflammatory effects67. Similarly, interleukin-32 is a unique cytokine that regulates immune cell physiology, including cell death, survival, inflammation, and pathogen response68. However, excessive amounts of IL-10 and IL-32 are one of the causes of cytokine storms. This phenomenon causes additional damage rather than eliminating infectious agents. IL-10 and IL-32 are the key inflammatory mediators in patients with severe COVID-19. Therapeutic agents that inhibit IL-10 and IL-32 signaling mechanisms (e.g., sarilumab, tocilizumab) have been reported to be effective in COVID-1969. Finally, the strength of immune cell interactions can be obtained by constructing cellular interaction networks that diminish over the course of the disease. As the severity of the disease increased and the new coronavirus continued to invade the body, the strength of intercellular interactions first strengthened and then weakened, especially in the regulation of Memory CD8 + T cells and Activated CD8 + T cells by Naive CD8 + T cells, and this weakening was especially obvious in the regulation of other immune cells by Treg cells. Treg cells are immunosuppressive cells, maintaining the immune system’s tolerance to its own components and keeping the body in immune homeostasis. Typically, the absence or abnormal function of Treg cells leads to an over-immune response, which in turn leads to a cytokine storm that drives chemokines and pro-inflammatory factors to other sites, such as the kidneys, respiratory system, cardiovascular system, nervous system, gastrointestinal system, and the liver, resulting in a variety of complications70. However, the strength of TGF-β-mediated immune cell interactions increased from mild to severe, and the expression of TGF-β1 was upregulated with increasing severity. The level of TGF-β1 was significantly correlated with the level of SARS-CoV-2-specific IgA, and the elevated TGF-β levels increased virus-specific IgA, which may have a devastating effect on severe COVID-19 and is the cause of common organ damage observed in COVID-19, such as acute pulmonary embolism and kidney injury71. And TGF-β1 has also been shown to induce fibroblast proliferation in SARS and MERS, leading to pulmonary fibrosis72. Elevated levels of TGF-β1 in critically ill patients with COVID-19 may also indicate that TGF-β1 is a potential target for clinical intervention. Future studies are necessary to evaluate the clinical potential of TGF-β1 in COVID-19 prognosis prediction and treatment73.
By developing the predictive model for COVID-19 severity and drug sensitivity, we identified several clinically relevant non-chemotherapy drugs associated with COVID-19 disease, including Vilobelimab, Baricitinib, Paxlovid, Anakinra, and Tocilizumab. Our analysis also highlighted SB-225,002, a CXCR2 chemokine receptor antagonist and holds theoretical potential for mitigating neutrophil-associated inflammation; however, current evidence supporting its application in the context of COVID-19 remains insufficient. Additionally, our analysis identified SB-225,002 as a potential therapeutic candidate for moderate and severe COVID-19 cases. This CXCR2 chemokine receptor antagonist demonstrates mechanistic potential for attenuating neutrophil-driven inflammatory responses, a key pathological feature of severe COVID-1974. However, robust clinical evidence supporting its therapeutic application in COVID-19 management remains to be further validated in subsequent studies. These findings provide important reference and guidance for future medical research and clinical practice.
Our study not only analyzed differences in immune cells in disease severity at the single-cell level, but also combined TCR and BCR data to analyze their heterogeneity in PBMCs from COVID-19 patients. Specifically, we queried VDJdb and McPAS-TCR databases and found no matching records for the clonotypes in our study. These results suggest that the identified clonotypes may represent novel antigen-specific TCRs not yet cataloged in public databases. It was found that severe samples were characterized by a reduced number and diversity of TCR and BCR clonotypes as well as a bias towards longer CDR3 region lengths. The lower TCR diversity in patients with severe COVID-19 suggests a more focused and restricted T cell response in patients with severe COVID-19, which may be due to the specific targeting of SARS-CoV-2 antigens75. In addition, it was found that there was little sharing of TCR and BCR clonotypes among COVID-19 patients of different severity. This heterogeneity suggests that the TCR repertoire may reflect disease severity and could have potential as a diagnostic test based on high-throughput sequencing of peripheral blood76.
This study also has several limitations. For example, our study analyzed only peripheral blood mononuclear cell data and no broncoalveolar lavage fluid data. In addition, the limited sample size was due to the stringent requirements of our analysis: we needed paired scRNA-seq and scV(D)J-seq data from the same individual, along with coverage across different COVID-19 severity levels, to ensure accurate immune repertoire and transcriptome correlation. Therefore, future studies with longitudinal samples from a larger number of COVID-19 patients may help to determine the causal relationship between the immune profile of different cell types and disease outcomes.
Taken together, this comprehensive multicellular description in our study lays the foundation for future characterization of the complex, dynamic immune response to SARS-CoV-2 infection. The transcriptomic data, together with detailed TCR- and BCR-based profiling information, can serve as a rich resource for a deeper understanding of peripheral lymphocytes in COVID-19 patients and pave the way for a more in-depth understanding of the COVID-19 virus as well as the development of SARS-CoV-2-specific potent drugs in the future.
Conclusions
In our study, we observed that the percentages of CD8 + T cells and Treg cells were lower in severe patients than in mild patients. Of note, patients with severe COVID-19 had an increased proportion of plasma cells, suggesting that this may be related to a stronger humoral immune response. In addition, CD4 + T cell subsets tended to expand in the severe samples, suggesting that they were activated in severely ill samples. Furthermore, we found that ZFP36 expression was increased in activated CD8 + T cells from critically ill patients, which can regulate the mRNA levels of key cytokines and other important genes related to proinflammatory and host innate immune responses, leading to the dysregulation and excessive release of proinflammatory cytokines, which in turn lead to cytokine storm and disease exacerbation. Second, we observed that the “regulation of interleukin-10 production” and “regulation of interleukin-32 production” pathways were highly enriched in critically ill patients. Excess IL-10 and IL-32 arecauses of cytokine storm.
Finally, we can obtain the strength of immune cell interactions by constructing cell interaction networks that weaken as the disease progresses. However, the intensity of TGF-β-mediated immune cell interactions gradually increases from mild to severe, and with the aggravation of the disease, the expression of TGF-β1 was up-regulated, leading to the increase of IgA expression, which has a serious impact on critically ill patients. Elevated levels of TGF-β1 in critically ill patients with COVID-19 may also suggest that TGF-β1 is a potential target for clinical intervention. By constructing a predictive model of the correlation between COVID-19 severity and drug sensitivity, several drugs that are related to the severity of COVID-19 disease were identified. Subsequently, scV(D)J-seq analysis showed a decreasing trend in the number of clonotypes, repertoire diversity, and clonotype overlap of clonal BCR and TCR as the severity of COVID-19 increases. The CDR3 sequence length in COVID-19-specific clonotypes showed a bias towards being longer as the disease progressed.
Data availability
All data and codes in this study are available under proper request, please contact wangli@hrbmu.edu.cn.
References
Umakanthan, S. et al. Origin, transmission, diagnosis and management of coronavirus disease 2019 (COVID-19). Postgrad. Med. J. 96 (1142), 753–758 (2020).
Ruan, Q. et al. Correction to: Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from wuhan, China. Intensive Care Med. 46 (6), 1294–1297 (2020).
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet 395 (10223), 497–506 (2020).
Fathi, N. & Rezaei, N. Lymphopenia in COVID-19: therapeutic opportunities. Cell. Biol. Int. 44 (9), 1792–1797 (2020).
Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in china: summary of a report of 72 314 cases from the Chinese center for disease control and prevention. Jama 323 (13), 1239–1242 (2020).
Zhang, J. L. et al. Azvudine is a thymus-homing anti-SARS-CoV-2 drug effective in treating COVID-19 patients. Signal. Transduct. Target. Ther. 6 (1), 414 (2021).
Lu, J. et al. scRNA-seq data analysis method to improve analysis performance. IET Nanobiotechnol. 17 (3), 246–256 (2023).
Liu, W. et al. Delineating COVID-19 immunological features using single-cell RNA sequencing. Innov. (Camb). 3 (5), 100289 (2022).
Alexovič, M. et al. Human peripheral blood mononuclear cells: A review of recent proteomic applications. Proteomics 22 (15-16), e2200026 (2022).
Rabaan, A. A. et al. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez Med. 28 (2), 174–184 (2020).
Mahallawi, W. H. et al. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 104, 8–13 (2018).
Langgartner, D. et al. COVID-19 vaccination exacerbates ex vivo IL-6 release from isolated PBMCs. Sci. Rep. 13 (1), 9496 (2023).
Potts, M. et al. Proteomic analysis of Circulating immune cells identifies cellular phenotypes associated with COVID-19 severity. Cell. Rep. 42 (6), 112613 (2023).
Ren, Y. et al. Single-cell sequencing reveals effects of chemotherapy on the immune landscape and TCR/BCR clonal expansion in a relapsed ovarian cancer patient. Front. Immunol. 13, 985187 (2022).
Zheng, F. et al. Immune cell and TCR/BCR repertoire profiling in systemic lupus erythematosus patients by single-cell sequencing. Aging (Albany NY). 13 (21), 24432–24448 (2021).
Luo, L. et al. Dynamics of TCR repertoire and T cell function in COVID-19 convalescent individuals. Cell. Discov. 7 (1), 89 (2021).
Minervina, A., Pogorelyy, M. & Mamedov, I. T-cell receptor and B-cell receptor repertoire profiling in adaptive immunity. Transpl. Int. 32 (11), 1111–1123 (2019).
Bai, H. et al. Identification of TCR repertoires in asymptomatic COVID-19 patients by single-cell T-cell receptor sequencing. Blood Cells Mol. Dis. 97, 102678 (2022).
Kim, I. S. et al. Tracking antigen-specific TCR clonotypes in SARS-CoV-2 infection reveals distinct severity trajectories. J. Med. Virol. 95 (11), e29199 (2023).
Zhang, F. et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal. Transduct. Target. Therapy. 5 (1), 156 (2020).
Huo, L. et al. Single-cell multi-omics sequencing: application trends, COVID-19, data analysis issues and prospects. Brief. Bioinform, 22(6). (2021).
Luo, G. et al. Probing infectious disease by single-cell RNA sequencing: progresses and perspectives. Comput. Struct. Biotechnol. J. 18, 2962–2971 (2020).
Parimoo, A. et al. Dynamics of inflammatory markers in predicting mortality in COVID-19. Cureus 13 (10), e19080 (2021).
Unterman, A. et al. Single-cell multi-omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19. Nat. Commun. 13 (1), 440 (2022).
Jin, S. et al. Inference and analysis of cell-cell communication using cellchat. Nat. Commun. 12 (1), 1088 (2021).
Chiffelle, J. et al. T-cell repertoire analysis and metrics of diversity and clonality. Curr. Opin. Biotechnol. 65, 284–295 (2020).
Sun, H. et al. Signatures of B cell receptor repertoire following Pneumocystis infection. Front. Microbiol. 12, 636250 (2021).
Matassoli, F. et al. High frequency of HIV precursor-target-specific B cells in sub-Saharan populations. Cell. Rep. 42 (12), 113450 (2023).
Sureshchandra, S. et al. Single-cell profiling of T and B cell repertoires following SARS-CoV-2 mRNA vaccine. JCI Insight, 6(24). (2021).
Hu, C. et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 51 (D1), D870–d876 (2023).
Jafarzadeh, A. et al. Lymphopenia an important immunological abnormality in patients with COVID-19: possible mechanisms. Scand. J. Immunol. 93 (2), e12967 (2021).
Kumar, N. R. et al. Convalescent plasma therapy in Late-State, severe COVID-19 infection. South. Med. J. 116 (5), 427–433 (2023).
Ramasamy, S. & Subbian, S. Critical determinants of cytokine storm and type I interferon response in COVID-19 pathogenesis. Clin. Microbiol. Rev., 34(3). (2021).
Shin, J. J. et al. MIF is a common genetic determinant of COVID-19 symptomatic infection and severity. Qjm 116 (3), 205–212 (2023).
Westmeier, J. et al. Macrophage migration inhibitory factor receptor CD74 expression is associated with expansion and differentiation of effector T cells in COVID-19 patients. Front. Immunol. 14, 1236374 (2023).
Witkowski, M. et al. Untimely TGFβ responses in COVID-19 limit antiviral functions of NK cells. Nature 600 (7888), 295–301 (2021).
Moreau, J. M. et al. Transforming growth factor-β1 in regulatory T cell biology. Sci. Immunol. 7 (69), eabi4613 (2022).
Ye, Z. et al. Tumor purity-related genes for predicting the prognosis and drug sensitivity of DLBCL patients. Elife, 13. (2024).
Corsello, S. M. et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer. 1 (2), 235–248 (2020).
Maeser, D., Gruener, R. F. & Huang, R. S. OncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform, 22(6). (2021).
Chhipa, M. A., Sanayhie, S. A. & Sabatinos, S. A. Assessing drug sensitivity in fission yeast using Half-Maximal inhibitory concentration (IC50) assays. Methods Mol. Biol. 2862, 241–253 (2025).
Wang, Y. et al. Paxlovid for the treatment of COVID-19: a systematic review and meta-analysis. J. Infect. Dev. Ctries. 18 (8), 1169–1178 (2024).
Yang, C. & Liu, M. Tocilizumab in treatment for patients with COVID-19. JAMA Intern. Med. 181 (7), 1017–1018 (2021).
Vlaar, A. P. J. et al. Anti-C5a antibody (vilobelimab) therapy for critically ill, invasively mechanically ventilated patients with COVID-19 (PANAMO): a multicentre, double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Respir Med. 10 (12), 1137–1146 (2022).
Baricitinib in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial and updated meta-analysis. Lancet, 400(10349): pp. 359–368. (2022).
Amani, B. & Amani, B. Efficacy and safety of nirmatrelvir/ritonavir (Paxlovid) for COVID-19: A rapid review and meta-analysis. J. Med. Virol. 95 (2), e28441 (2023).
Huet, T. et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2 (7), e393–e400 (2020).
Klopfenstein, T. et al. Tocilizumab in COVID-19 pneumonia: practical proposals based on a narrative review of randomised trials. Rev. Med. Virol. 32 (1), e2239 (2022).
Catusse, J. et al. Characterization of the molecular interactions of interleukin-8 (CXCL8), growth related oncogen alpha (CXCL1) and a non-peptide antagonist (SB 225002) with the human CXCR2. Biochem. Pharmacol. 65 (5), 813–821 (2003).
Gutierrez, L., Beckford, J. & Alachkar, H. Deciphering the TCR repertoire to solve the COVID-19 mystery. Trends Pharmacol. Sci. 41 (8), 518–530 (2020).
Zhou, Y. et al. Profiling of the immune repertoire in COVID-19 patients with mild, severe, convalescent, or retesting-positive status. J. Autoimmun. 118, 102596 (2021).
Jo, H. Y. et al. Establishment of the large-scale longitudinal multi-omics dataset in COVID-19 patients: data profile and biospecimen. BMB Rep. 55 (9), 465–471 (2022).
Kim, J. W. et al. T-Cell receptor repertoire characteristics associated with prognostic significance in High-Grade serous ovarian carcinoma. Genes (Basel), 14(4). (2023).
Chang, C. M. et al. Profiling of T cell repertoire in SARS-CoV-2-Infected COVID-19 patients between mild disease and pneumonia. J. Clin. Immunol. 41 (6), 1131–1145 (2021).
Qi, F. et al. Single-cell analysis of the adaptive immune response to SARS-CoV-2 infection and vaccination. Front. Immunol. 13, 964976 (2022).
Wang, G. et al. Comprehensive analysis of TCR repertoire of COVID-19 patients in different infected stage. Genes Genomics. 44 (7), 813–822 (2022).
Xiao, J. et al. The characteristics of BCR-CDR3 repertoire in COVID-19 patients and SARS-CoV-2 vaccinated volunteers. J. Med. Virol. 96 (3), e29488 (2024).
Hoffmann, M. et al. Blood transcriptomics analysis offers insights into variant-specific immune response to SARS-CoV-2. bioRxiv, (2023).
Majumder, J. & Minko, T. Recent developments on therapeutic and diagnostic approaches for COVID-19. Aaps J. 23 (1), 14 (2021).
Yan, S. & Wu, G. Is lymphopenia different between SARS and COVID-19 patients? Faseb J. 35 (2), e21245 (2021).
Zhang, Z. et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 185 (14), 2434–2451e17 (2022).
Havkin-Solomon, T. et al. Translation regulation of specific mRNAs by RPS26 C-terminal RNA-binding tail integrates energy metabolism and AMPK-mTOR signaling. Nucleic Acids Res. 51 (9), 4415–4428 (2023).
Zhao, H. et al. LncTarD 2.0: an updated comprehensive database for experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res. 51 (D1), D199–d207 (2023).
Lu, H. J. et al. The impact of ALDH7A1 variants in oral cancer development and prognosis. Aging (Albany NY). 14 (10), 4556–4571 (2022).
Hu, B., Huang, S. & Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 93 (1), 250–256 (2021).
Batah, S. S. & Fabro, A. T. Pulmonary pathology of ARDS in COVID-19: A pathological review for clinicians. Respir Med. 176, 106239 (2021).
Han, H. et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 9 (1), 1123–1130 (2020).
Zamani, B. et al. Predicting roles of IL-27 and IL-32 in determining the severity and outcome of COVID-19. Int. J. Immunopathol. Pharmacol. 36, 3946320221145827 (2022).
Law, C. C. et al. Clinical implications of IL-32, IL-34 and IL-37 in atherosclerosis: speculative role in cardiovascular manifestations of COVID-19. Front. Cardiovasc. Med. 8, 630767 (2021).
Galván-Peña, S. et al. Profound Treg perturbations correlate with COVID-19 severity. Proc. Natl. Acad. Sci. U S A, 118(37). (2021).
Santa Cruz, A. et al. Post-acute sequelae of COVID-19 is characterized by diminished peripheral CD8(+)β7 integrin(+) T cells and anti-SARS-CoV-2 IgA response. Nat. Commun. 14 (1), 1772 (2023).
Wang, E. Y. et al. Serum levels of the IgA isotype switch factor TGF-β1 are elevated in patients with COVID-19. FEBS Lett. 595 (13), 1819–1824 (2021).
Vaz de Paula, C. B. et al. COVID-19: immunohistochemical analysis of TGF-β signaling pathways in pulmonary fibrosis. Int. J. Mol. Sci., 23(1). (2021).
Cheng, Y. et al. Targeting CXCR2 inhibits the progression of lung cancer and promotes therapeutic effect of cisplatin. Mol. Cancer. 20 (1), 62 (2021).
Buhler, S. et al. HLA variants and TCR diversity against SARS-CoV-2 in the pre-COVID-19 era. Hla 102 (6), 720–730 (2023).
Peng, X. et al. Increase in HIV reservoir and T cell immune response after coronavac vaccination in people living with HIV. Heliyon 10 (9), e30394 (2024).
Acknowledgements
We thank all the funding that provided financial support for this study.
Funding
This work was supported by the National Natural Science Foundation of China (62372144) and the Outstanding Youth Foundation of Heilongjiang Province of China (YQ2023F004).
Author information
Authors and Affiliations
Contributions
LW and HYZ designed the study, implemented the algorithm, and performed the analysis. HYZ, MTF, and LW wrote and revised the manuscript. HYZ, MTF, WTY, ZCG and JB help to collect the data and prepare the figures and tables. All authors read, reviewed, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, H., Fei, M., Yu, W. et al. Severity-specific immune landscape of COVID-19 revealed by single-cell sequencing. Sci Rep 15, 29596 (2025). https://doi.org/10.1038/s41598-025-13888-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13888-2