Abstract
Enhancing diagnostic capability of microprobe endoscopic ultrasonography (MEUS) for GISTs is clinical significance. Despite the promise of artificial intelligence (AI) in aiding diagnosis, challenges remain in model interpretability, generalization and diagnostic specificity. To address this, lesion characteristics were integrated into MEUS images to align AI inference with clinical diagnostic workflow, leading to the development of seven deep learning models. The top model, named ECMAI-ME, was assessed in two external validation sets for generalizability and compared with endoscopists in multicenter test for effectiveness. Using 9,229 MEUS images from 873 SELs across five Chinese hospitals, training involved 522 SELs, with 95 and 93 SELs from different sources for external validation, and 163 SELs for a multicenter test. ECMAI-ME achieved an AUC of 0.972 for case classification internally. In two external validation sets, ECMAI-ME maintained consistency diagnostic performance. In multicenter test, ECMAI-ME significantly outperformed endoscopists in accuracy (85.28% vs. 56.44–77.91%, p<0.05) and specificity (89.19% vs. 52.25–72.92%, p<0.001) with comparable sensitivity, and demonstrating high accuracy in distinguishing lesion echogenicity (96.93%), originating layer (88.34%), and echo heterogeneity (77.30%). This interpretable model offers high specificity, adaptability across hospitals and equipment, and strong potential for integration into clinical workflows.
Similar content being viewed by others
Introduction
In recent years, detection rates for subepithelial lesions (SELs) in the digestive tract have notably improved. Accurate diagnosis of these lesions is the basis for clinical decision-making, especially for gastrointestinal stromal tumors (GISTs), the most common and potentially malignant type of SELs. Endoscopic ultrasonography (EUS) is the primary modality for classifying SELs, with particular focus on evaluating the originating layer, echogenicity, and echo heterogeneity1,2,3. However, standalone EUS imaging is insufficient for diagnosing GISTs1,2, with diagnostic accuracy for distinguishing benign from malignant SELs hovering around 43–50%, sensitivity from 64 to 80%, and specificity from 77–80%1,4,5,6. Interobserver variability further indicates that agreement among observers for SELs, excluding cysts and lipomas, ranges from poor to moderate7. Enhancing the non-invasive diagnostic capability and consistency of EUS for GISTs without relying on tissue pathology biopsy is a key research focus.
Artificial intelligence-enhanced endoscopic ultrasonography, termed EUS-AI models, has shown promise in the non-invasive differentiating GISTs8,9. However, current EUS-AI models face several limitations. Firstly, EUS-AI models often lack interpretability, creating a “black box effect“10,11,12,13,14,15,16,17,18which may hamper clinical understanding and raise ethical concerns. Secondly, improved generalizability of EUS-AI models across various clinical scenarios is needed. Many existing studies rely on small, single-center datasets11,14,18lack external test10,11,12,13,14,18or exhibit significant differences in diagnostic accuracy depending on the type of EUS devices employed, ranging from approximately 28.6–91.3%15. Thirdly, the diagnostic performance of EUS-AI models, particularly specificity, needs improvement, with the lowest reported specificity being 64.3%14. Improving diagnostic specificity can help avoid unnecessary pathological biopsies, lesion resections, and excessive follow-ups, thereby reducing the overall healthcare burden.
Microprobe endoscopic ultrasonography (MEUS) is widely used in primary healthcare settings in China due to its affordability and ease of use, particularly in regions where access to linear-array or radial EUS is limited. Additionally, the varying levels of expertise among endoscopists in in primary healthcare settings further highlight the need to enhance the diagnostic accuracy of EUS-AI models designed for MEUS. Against this backdrop, our research group is developing AI studies for SELs, involving endoscopic ultrasonography-centric multimodal artificial intelligence (ECMAI) models. Our initial goal is to simulate the diagnostic workflow of endoscopists and develop an interpretable AI model based on MEUS images, named ECMAI-ME.
Results
Basic information
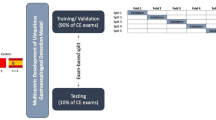
A total of 522 SELs (152 GISTs and 370 non-GISTs) with 5278 MEUS images were chosen for training and internal validation set from 4 hospitals. External validation set I from a different hospital included 95 SELs (30 GISTs and 65 non-GISTs) with 1754 MEUS images, while external validation set II from a different MEUS probe included 93 SELs (32 GISTs and 61 non-GISTs) with 904 MEUS images. The multicenter test set included 163 SELs (52 GISTs and 111 non-GISTs) with 1293 MEUS images. Study flowchart and patient disposition was shown in Fig. 1. Detailed patient information and clinicopathological features were presented in Table 1 and Supplementary Table 1s.
Study flowchart and patient disposition. Patients with SELs from five hospitals were included. After quality control, MEUS images were divided into five subsets. Data from Hospitals 1–4 supported model development and validation. External validation sets from Hospital 5 and a different MEUS probe assessed generalizability. A multicenter test set was used to compare AI and endoscopist performance. Abbreviations: AI, artificial intelligence; SELs, subepithelial lesions. † Excluded low-quality images.
Performance and selection of AI models in the validation sets
Seven AI models were developed in this study (Fig. 2). In the internal validation set, AI model-7 exhibited the highest area under the curve (AUC) of 0.966 (95% CI: 0.957–0.975) by image and 0.972 (95% CI: 0.948–0.997) by lesion, surpassing the other six AI models with AUC values ranging from 0.810 to 0.881 by image and 0.853 to 0.895 by lesion. Similarly, in the external validation set I, AI model-7 maintained its leading position, achieving an AUC of 0.810 (95% CI: 0.789–0.830) by image and 0.826 (95% CI: 0.745–0.907) by lesion. While it performed equally to model-5 in this set, it outperformed the remaining five AI models. In the external validation set II, the AUC values of the seven AI models ranged from 0.597 to 0.820 by image and 0.561 to 0.781 by lesion. Once again, AI model-7 demonstrated superior performance, attaining the highest AUC of 0.820 (95% CI: 0.792–0.849) by image and 0.781 (95% CI: 0.627–0.889) by lesion (Fig. 2). Due to its consistent high performance, AI model-7 was selected as the ECMAI-ME model and developed into an AI system with a graphical user interface (GUI) using the Python programming language (GUI structure and clinical workflow are presented in Supplementary Fig. 1 s).
Receiver operating characteristic curves of seven artificial intelligence models for diagnosing gastrointestinal stromal tumors. Each panel compares the diagnostic performance of Model 1–7 across different datasets. Curves are plotted both at the image level (top row) and lesion level (bottom row). Panel a: Results from the internal validation set). Panel b: Results from external validation set (I) Panel c: Results from external validation set (II) Abbreviations: AUC, area under the curve.
Comparison of ECMAI-ME model in different external validation sets
In comparison between external validation set I and II, the ECMAI-ME model achieved an AUC of 0.826 (95% CI: 0.745, 0.908) compared to 0.781 (95% CI: 0.671, 0.891) (p = 0.516). The accuracy was 74.74% (95% CI: 66.00%, 83.47%) versus 75.27% (95% CI: 66.50%, 84.04%) (p = 0.759), sensitivity was 83.33% (95% CI: 70.00%, 96.67%) versus 78.13% (95% CI: 63.80%, 92.45%) (p = 0.604), and the specificity was 70.77% (95% CI: 59.71%, 81.83%) versus 73.77% (95% CI: 62.73%, 84.81%, p = 0.707) in distinguishing GISTs, respectively. These results suggest consistent performance across both external validation sets without statistically significant variation (Table 2).
In the subgroup of tumors smaller than 20 mm, ECMAI-ME exhibited an accuracy of 73.42% (95% CI: 63.68%, 83.16%) in external test set I and 75% (95% CI: 65.95%, 84.05%) in external test set II without statistically significant difference (p = 0.781). Similarly, for lesions with a size of 20 mm or larger, the accuracy of ECMAI-ME remained comparable between external test sets I and II (81.25% vs. 80.00%, p = 0.292). Additionally, no significant differences in diagnostic accuracy, sensitivity, or specificity were observed between lesion size subgroups within the same test set (all p >0.05, Supplementary Table 2 s).
Performance of ECMAI-ME model in multicenter test set
In the multicenter test set, the ECMAI-ME model achieved an AUC of 0.889 (95% CI: 0.868, 0.911) by image and 0.885(95% CI: 0.827, 0.944) by lesion(Fig. 3). Other measures included accuracy of 85.28% (95%CI: 79.84%, 90.72%), specificity of 89.19% (95%CI: 83.41%, 94.97%), sensitivity of 76.92% (95%CI: 65.47%, 88.37%) (Table 3). False positives were most common in leiomyomas (9/63, 14.29%), which aligns with the similarity of their EUS features to those of GISTs. NETs (96.7% specificity) and other subtypes (100% specificity) performed well (Supplementary Table 3 s).
ROC and confusion matrix–based comparison of ECMAI-ME and endoscopists in diagnosing gastrointestinal stromal tumors. (a) ROC curve of the ECMAI-ME model in multicenter test set. The colored symbols indicate the diagnostic performance of six endoscopists. (b) Confusion matrices comparing diagnostic outcomes of ECMAI-ME and the six endoscopists against the pathological gold standard. Abbreviations: ROC, receiver operating characteristic; AUC, area under the curve; GISTs, gastrointestinal stromal tumors.
ECMAI-ME showed comparable diagnostic performance across tumor size subgroups. For lesions < 20 mm, the model achieved an accuracy of 85.14%, sensitivity of 74.42%, and specificity of 89.52%. For lesions ≥ 20 mm, the corresponding values were 86.67%, 88.89%, and 83.33%, respectively. No significant differences were observed in accuracy, sensitivity, or specificity between the two subgroups (all p >0.05, Supplementary Table 2 s).
Meanwhile, the ECMAI-ME model demonstrated accuracy of 88.34% (95%CI: 83.42%, 93.27%) in distinguishing originating layer, 96.93% (95%CI: 94.29%, 99.58%) for lesion echogenicity and 77.30% (95%CI: 70.87%, 83.73%) for echo heterogeneity. Confusion matrixes were presented in Supplementary Fig. 2s.
Comparison of ECMAI-ME model and endoscopists in multicenter test set
The ECMAI-ME model achieved significantly higher accuracy (85.28% vs. 56.44–77.91%, p<0.05) and specificity (89.19% vs. 52.25–72.92%, p<0.001) compared to endoscopists, with comparable sensitivity (76.92% vs. 65.38–90.38%, p>0.05). When compared to initial diagnoses in the original reports, the ECMAI-ME model demonstrated significantly higher accuracy (84.87% vs. 78.95%, p<0.001) and specificity (88.24% vs. 72.55%, p = 0.026), while demonstrating comparable sensitivity (78.00% vs. 92.00%, p=0.118). The details were shown in Table 3; Figs. 3 and 4.
Statistical comparison of diagnostic performance between ECMAI-ME and endoscopists. Pairwise comparisons were performed between ECMAI-ME and six endoscopists. (a) Accuracy. (b) Specificity for non-GISTs. (c) Sensitivity for GISTs. Values above the bars indicate performance differences; ↑ indicates higher and ↓ lower performance relative to ECMAI-ME. Corresponding p-values are shown above each bar. Abbreviations: GISTs, gastrointestinal stromal tumors.
Discussion
Differentiating GISTs from non-GISTs remains a significant clinical challenge. In this study, we present ECMAI-ME, an interpretable deep learning model based on MEUS images. The model incorporates structured lesion characteristics to guide training, aligning its diagnostic logic with clinical reasoning. It achieved high diagnostic specificity of nearly 90%, consistently outperformed endoscopists across all experience levels. Moreover, its robust performance across centers and devices underscores its potential for clinical deployment as an adjunctive diagnostic tool.
Interpretability is essential in clinical AI applications, as it fosters trust and facilitates adoption among endoscopists to use AI models17. This study introduces several innovations to enhance both global and instance-levels interpretability. First, unlike previous models focusing solely on lesion area10,17,18we incorporated surrounding context to provide a more comprehensive diagnostic perspective. A pretreatment technique using edge and circle detection was applied to improve lesion localization and guide attention to clinically relevant regions. Second, ECMAI-ME was developed using a clinically guided multi-task learning strategy, with three diagnostic features (originating layer, echogenicity, and echo heterogeneity) as parallel learning targets. This design simulates endoscopic diagnostic reasoning and grounds predictions in quantifiable, meaningful features. We constructed seven convolutional neural network (CNN) variants incorporating different combinations of diagnostic features (Fig. 5). Unlike models relying on abstract representations, our method emphasizes explicit diagnostic elements to improve robustness. Third, the model outputs instance-level semantic predictions of GIST probability and structured lesion features, offering transparent, case-specific reasoning aligned with clinical decision-making. Compared to black-box models, this facilitates user validation through familiar diagnostic dimensions. To support structured prediction despite incomplete annotations, we employed a self-labeling semi-supervised learning strategy (Supplementary Appendix 1 s, Supplementary Fig. 3 s), enabling the model to infer missing features. While our prior work used attention heatmaps19we found semantic outputs more clinically actionable, as they correspond directly to decision-relevant attributes.
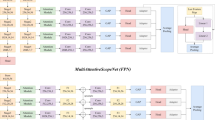
Development process of the ECMAI-ME model simulating endoscopists’ diagnostic reasoning. Multi-center MEUS images and structured lesion data were used to train CNN-based models with branches learning key features: originating layer, echogenicity, and echo heterogeneity. Seven models simulating different diagnostic strategies were developed. The best-performing model, ECMAI-ME, was selected and compared with endoscopists. Abbreviations: AI, artificial intelligence; MEUS, microprobe endoscopic ultrasonography; CNNs, convolutional neural networks.
While achieving interpretability, ECMAI-ME demonstrates excellent performance and generalizability, highlighting its potential for real-world clinical use. In the multicenter test, it achieved a specificity of 89.19% and outperformed endoscopists with 7.4–28.8% gains in accuracy and 17.1–36.9% in specificity, reducing 19 to 41 false-positive GIST cases (Fig. 3). This exceeds the reported specificity of previous models (64.3–85.7%))10,11,12,13,14,15,16,17 and approaches the performance of contrast-enhanced harmonic EUS systems (90.9%)18. Furthermore, when compared with the initial clinical diagnoses documented in the original reports, which reflect real-world diagnostic practices incorporating lesion location, white-light endoscopy (WLE) findings, and dynamic MEUS details. ECMAI-ME demonstrated significantly superior diagnostic accuracy (84.87% vs. 78.95%) and specificity (88.24% vs. 72.55%). Importantly, ECMAI-ME maintained stable performance across two independent external validation sets from different hospitals and MEUS probes, with no statistically significant differences observed. This contrasts with previous studies, such as Yang X et al.15where diagnostic accuracy dropped to 28.6% when using different probes or 50.0% with radial scanning EUS. Such robustness underscores the model’s suitability for diverse clinical environments, including primary care settings.
ECMAI-ME, as a noninvasive tool, is especially valuable in settings where biopsy is technically difficult, contraindicated, declined by patients, or resources-limited. This applies notably to small SELs (< 20 mm), where biopsy yields are often low. Biopsy techniques such as EUS-guided fine-needle aspiration or biopsy (EUS-FNA/B) and mucosal incision-assisted biopsy (MIAB) show a diagnostic accuracy range of 74–88.2%, and they carry procedural risks of 0.9–7.9%20,21. By improving specificity, ECMAI-ME may reduce unnecessary interventions, such as EUS-FNA/B, surgical resection, and close surveillance, thereby reducing patient burden and healthcare costs. International guidelines recommend follow-up over biopsy for low-risk and small SELs1,2,22,23and ECMAI-ME aligns with this principle. Importantly, ECMAI-ME is designed to assist clinical decision-making, not replace physician judgment or guidelines. Prospective studies are needed to further assess its effectiveness and optimize diagnostic strategies.
Another key advantage of the ECMAI-ME model is its comprehensive depiction of primary lesion characteristics, distinguishing it from previous studies10,11,12,13,14,15,16,17,18. The model demonstrated high diagnostic accuracy for lesion echogenicity (96.93%) and originating layer (88.34%), although lower for internal echo heterogeneity (77.30%). This variability likely stems from the diverse pathological subtypes of SELs and the heterogeneous internal echoes observed within the same lesion type. For example, internal echo heterogeneity in GISTs can present as hyperechoic spots, anechoic cystic spots, hypoechoic marginal halos, or increased echogenicity compared to the surrounding muscle layer24. While the current model classifies internal echoes as either homogeneous or inhomogeneous, it remains limited in addressing their complexity. Future improvements could incorporate more detailed heterogeneity features or advanced methods like radiomics and attention mechanisms to enhance the characterization and diagnostic accuracy of internal echoes.
This study has several limitations. First, the current ECMAI-ME model supports binary classification and lacks WLE input, which may limit its applicability. We are expanding the model to support multi-class classification (e.g., GIST, leiomyoma, and NET) and integration of WLE data. Still, its accuracy (85.28%) matches the reported WLE–EUS multimodal models (83.5%)25. Second, the current version cannot process video in real time. To address this, we are developing a multi-step key-frame selection algorithm (Supplementary Appendix 2 s) to automatically extract high-quality frames from MEUS videos, enabling future video-based model integration. Third, deployment currently relies on manual image input by GUI. We have developed a real-time deployment pipeline (Supplementary Fig. 4 s) in which an attachable AI terminal enables real-time data input, analysis, diagnosis, and structured reporting without requiring hardware modifications. These developments lay the foundation for prospective clinical validation.
In conclusion, ECMAI-ME offers an interpretable and generalizable solution for GIST diagnosis and lesion characterization using MEUS images. It holds strong potential for clinical integration and future studies will focus on developing multi-class, multimodal frameworks to support comprehensive SEL management.
Methods
Patients
Patients with SELs were retrospectively derived from five Chinese hospitals from Jan.2013 to Dec.2023 (listed in Table 1, Supplementary Table 1 s). Inclusion criteria: (a) patients with SELs diagnosed by MEUS; and (b) cases with definitive pathological diagnosis based on surgical or endoscopic resection specimens of the entire lesion. For key diagnostic categories such as GISTs, leiomyomas, schwannomas, and NETs, immunohistochemical (IHC) staining was routinely performed. For benign lesions with typical histologic features (e.g., cysts or lipomas), IHC was applied only when H&E staining was insufficient for a definitive diagnosis. Exclusion criteria included: (a) cases without a pathological diagnosis or with uncertain pathological diagnosis; (b) SELs lacking MEUS images or having incomplete information; and (c) low-quality images, including those with poor focus or resolution, motion or acoustic shadow artifacts, incomplete lesion visualization, inappropriate gain settings, unrelated content, measurement line occlusion, or duplicate images. Data collected included MEUS images, age, sex, tumor location, maximum diameter, pathological diagnosis, originating layer, echogenicity, echo heterogeneity, and initial diagnosis stated in the original reports. The deliberated diagnoses of originating layer, echogenicity, and echo heterogeneity from three experienced endoscopists (J.L, J.S, Z-K Z) served as the diagnostic gold standard.
Study design
The study comprised three sections: development of AI models, validation and selection of ECMAI-ME, and testing. A total 522 SELs in 517 patients were sourced from four Chinese hospitals utilizing microprobe EUS devices of Olympus (Japan) (Supplementary Table 4 s). The lesions were randomly stratified by hospital and divided into a training set and an internal validation set with a ratio of 7:3. The training set developed the AI models, while the internal validation set optimized and selected the suitable model. Supplementary Appendix 3 s presents the estimation of the required sample size, and at least 93 SELs with histologically-confirmed diagnoses should be enrolled. External validation set I comprised 95 SELs in 93 patients from the Fifth Hospital using Olympus devices. External validation set II included 93 SELs in 92 patients using a different microprobe EUS device from InnerMedical (China). Multicenter test set included 163 SELs in 155 patients from all the five hospitals across a different time period. The study flowchart is in Fig. 1, and MEUS images from various devices are in Supplementary Fig. 5s.
This study followed Declaration of Helsinki, and it was approved by the ethics committee of the third people’s hospital of Chengdu on April 10, 2023 (IRB No. 2023-S-48). Due to the retrospective nature of the study informed consent was waived by the ethics committee of the third people’s hospital of Chengdu. It was registered in the China Clinical Trial Registry (https://www.chictr.org.cn) as number ChiCTR2400080928 on February 18, 2024.
Development of AI models
Two experienced endoscopists (J.L. and Z.K Z.) independently assessed image quality. Images were retained only if both reviewers deemed the quality acceptable; in cases of disagreement, a third experienced endoscopist (J.S) served as the adjudicator. In the training set, images were cropped to square with patient identifiers removed, and tumor boundaries were manually annotated using VGG Image Annotator (show in Supplementary Fig. 6 s).
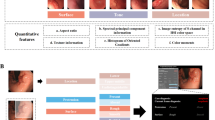
Endoscopists classify SELs based on visual characteristics such as the originating layer, echogenicity, and echo heterogeneity, which collectively form their diagnostic logic1,2,3. This study aims to simulate this diagnostic reasoning by integrating global and local visual information from MEUS images to enhance diagnostic interpretability, generalizability, and accuracy.
Based on MEUS images and different characteristics combinations, we developed seven AI models for GIST diagnosis: Model-1 includes the originating layer, Model-2 incorporates echogenicity, Model-3 incorporates echo heterogeneity, Model-4 combines the originating layer and echo heterogeneity, Model-5 combines the originating layer and echogenicity, Model-6 combines echogenicity and echo heterogeneity, and finally, Model-7 leverages all three lesion characteristics for classification. Figure 5 summarizes the AI development process, and supplementary Appendix 1 s and supplementary Fig. 3 s outlines the AI architecture. Further algorithm details are in our publication19.
Validation and selection of ECMAI-ME model
During validation, AI models used unlabeled MEUS images without requiring manual input of lesion characteristics. For each image, the trained models provided a predictive score for GISTs, ranging from 0 to 1, with a score of 1 indicating a high likelihood of GIST. For each lesion, the models outputted the mean predictive score of all images belonging to that specific lesion. Additionally, the models also provide diagnostic information of lesion characteristics including originating layer, echogenicity, and echo heterogeneity. The model with the highest area under the receiver operating characteristic curve (AUC) in the internal validation set was chosen as ECMAI-ME.
Testing of ECMAI-ME model
The ECMAI-ME model was evaluated in two external validation sets to assess its generalizability, alongside compared with endoscopists of varying experience levels in multicenter test to measure effectiveness. Six endoscopists, including two experts (Liu.L, Li. L) with over 1000 MEUS procedures’ experience, two novices (X-H C, Z-X S) with less than 500 MEUS procedures’ experience and two Trainees (M.Z, Y-R L) with less than 100 MEUS procedures’ experience, independently classified SELs as either GISTs or non-GISTs during the multicenter test. Furthermore, the model’s performance was compared to the endoscopists’ initial diagnoses in the original reports. The lesions without exact initial diagnosis of SEL types in the original reports will be excluded from the comparison test.
Statistical analysis
Categorical variables were expressed as number (percentage), while continuous variables were presented as mean ± standard deviation (SD). The primary evaluation metrics used to assess the performance of the AI models was AUC. Sensitivity, specificity, accuracy, positive predictive value (PPV), and negative predictive value (NPV) were calculated with 95% confidence intervals (CI).
Diagnostic accuracy, sensitivity, and specificity were compared using McNemar’s test (for paired comparisons) and the chi-squared test or Fisher’s exact test (for unpaired comparisons). Statistical analysis was conducted using SPSS 29.0 (IBM, CA, USA), with p-values < 0.05 indicating significance.
Data availability
Due to patient privacy concerns, the image datasets are not publicly available but can be obtained from the corresponding author upon reasonable request. The complete source code is available at https://github.com/cylijiao/ECMAI-ME.
References
Jacobson, B. C. et al. ACG clinical guideline: diagnosis and management of Gastrointestinal subepithelial lesions. Am. J. Gastroenterol. 118, 46–58. https://doi.org/10.14309/ajg.0000000000002100 (2023).
Deprez, P. H. et al. Endoscopic management of subepithelial lesions including neuroendocrine neoplasms: European society of Gastrointestinal endoscopy (ESGE) guideline. Endoscopy 54, 412–429. https://doi.org/10.1055/a-1751-5742 (2022).
Qi, Z. P., Li, Q. L., Zhong, Y. S. & Zhou, P. H. [Interpretation of the Chinese consensus on endoscopic diagnosis and management of Gastrointestinal submucosal tumors (version 2018)]. Zhonghua Wei Chang. Wai Ke Za Zhi. 22, 609–612. https://doi.org/10.3760/cma.j.issn.1671-0274.2019.07.002 (2019).
Hwang, J. H. et al. A prospective study comparing endoscopy and EUS in the evaluation of GI subepithelial masses. Gastrointest. Endosc. 62, 202–208. https://doi.org/10.1016/s0016-5107(05)01567-1 (2005).
Karaca, C., Turner, B. G., Cizginer, S., Forcione, D. & Brugge, W. Accuracy of EUS in the evaluation of small gastric subepithelial lesions. Gastrointest. Endosc. 71, 722–727. https://doi.org/10.1016/j.gie.2009.10.019 (2010).
Reddymasu, S. C. et al. Are endoscopic ultrasonography imaging characteristics reliable for the diagnosis of small upper Gastrointestinal subepithelial lesions? J. Clin. Gastroenterol. 46, 42–45. https://doi.org/10.1097/MCG.0b013e318226af8e (2012).
Gress, F. et al. Interobserver agreement for EUS in the evaluation and diagnosis of submucosal masses. Gastrointest. Endosc. 53, 71–76. https://doi.org/10.1067/mge.2001.111384 (2001).
Ye, X. H., Zhao, L. L. & Wang, L. Diagnostic accuracy of endoscopic ultrasound with artificial intelligence for Gastrointestinal stromal tumors: A meta-analysis. J. Dig. Dis. 23, 253–261. https://doi.org/10.1111/1751-2980.13110 (2022).
Zhang, B., Zhu, F., Li, P. & Zhu, J. Artificial intelligence-assisted endoscopic ultrasound in the diagnosis of Gastrointestinal stromal tumors: a meta-analysis. Surg. Endosc. 37, 1649–1657. https://doi.org/10.1007/s00464-022-09597-w (2023).
Kim, Y. H. et al. Application of A convolutional neural network in the diagnosis of gastric mesenchymal tumors on endoscopic ultrasonography images. J. Clin. Med. 9 https://doi.org/10.3390/jcm9103162 (2020).
Minoda, Y. et al. Efficacy of endoscopic ultrasound with artificial intelligence for the diagnosis of Gastrointestinal stromal tumors. J. Gastroenterol. 55, 1119–1126. https://doi.org/10.1007/s00535-020-01725-4 (2020).
Oh, C. K. et al. Convolutional neural network-based object detection model to identify Gastrointestinal stromal tumors in endoscopic ultrasound images. J. Gastroenterol. Hepatol. 36, 3387–3394. https://doi.org/10.1111/jgh.15653 (2021).
Hirai, K. et al. Artificial intelligence-based diagnosis of upper Gastrointestinal subepithelial lesions on endoscopic ultrasonography images. Gastric Cancer. 25, 382–391. https://doi.org/10.1007/s10120-021-01261-x (2022).
Seven, G., Silahtaroglu, G., Seven, O. O. & Senturk, H. Differentiating Gastrointestinal stromal tumors from leiomyomas using a neural network trained on endoscopic ultrasonography images. Dig. Dis. 40, 427–435. https://doi.org/10.1159/000520032 (2022).
Yang, X. et al. An artificial intelligence system for distinguishing between Gastrointestinal stromal tumors and leiomyomas using endoscopic ultrasonography. Endoscopy 54, 251–261. https://doi.org/10.1055/a-1476-8931 (2022).
Zhu, C. et al. A multimodal multipath artificial intelligence system for diagnosing gastric protruded lesions on endoscopy and endoscopic ultrasonography images. Clin. Transl Gastroenterol. https://doi.org/10.14309/ctg.0000000000000551 (2022).
Lu, Y. et al. Artificial intelligence in the prediction of Gastrointestinal stromal tumors on endoscopic ultrasonography images: development, validation and comparison with endosonographers. Gut Liver. https://doi.org/10.5009/gnl220347 (2023).
Tanaka, H. et al. Value of artificial intelligence with novel tumor tracking technology in the diagnosis of gastric submucosal tumors by contrast-enhanced harmonic endoscopic ultrasonography. J. Gastroenterol. Hepatol. 37, 841–846. https://doi.org/10.1111/jgh.15780 (2022).
Fan, L., Gong, X., Zheng, C. & Li, J. Data pyramid structure for optimizing EUS-based gists diagnosis in multi-center analysis with missing label. Comput. Biol. Med. 169, 107897. https://doi.org/10.1016/j.compbiomed.2023.107897 (2023).
Facciorusso, A. et al. Comparative diagnostic yield of different endoscopic techniques for tissue sampling of upper Gastrointestinal subepithelial lesions: a network meta-analysis. Endoscopy 56, 31–40. https://doi.org/10.1055/a-2156-0063 (2024).
Verloop, C. A. et al. Diagnostic yield of endoscopic and EUS-guided biopsy techniques in subepithelial lesions of the upper GI tract: a systematic review. Gastrointest. Endosc. 99, 895–911e813. https://doi.org/10.1016/j.gie.2024.02.003 (2024).
Sharzehi, K., Sethi, A. & Savides, T. A. G. A. Clinical practice update on management of subepithelial lesions encountered during routine endoscopy: expert review. Clin. Gastroenterol. Hepatol. 20, 2435–2443e2434. https://doi.org/10.1016/j.cgh.2022.05.054 (2022).
Hirota, S. et al. English version of Japanese clinical practice guidelines 2022 for Gastrointestinal stromal tumor (GIST) issued by the Japan society of clinical oncology. Int. J. Clin. Oncol. 29, 647–680. https://doi.org/10.1007/s10147-024-02488-1 (2024).
Kim, G. H. et al. Is it possible to differentiate gastric gists from gastric leiomyomas by EUS? World J. Gastroenterol. 15, 3376–3381. https://doi.org/10.3748/wjg.15.3376 (2009).
Zhu, C. et al. A multimodal multipath artificial intelligence system for diagnosing gastric protruded lesions on endoscopy and endoscopic ultrasonography images. Clin. Transl Gastroenterol. 14, e00551. https://doi.org/10.14309/ctg.0000000000000551 (2023).
Acknowledgements
We extend the appreciation to Li Liu, Zhong-Xin Sun, Ya-Ru Li from Third People’s Hospital of Chengdu, and Xiao-Hong Chen from Chengdu Office hospital of the people’s Government of Tibet Autonomous Region for their assistance in the test of the AI model. We also greatly appreciate the help of Tao Shu of the Third People’s Hospital of Chengdu for his selfless and hard-working of data acquisition.
Author information
Authors and Affiliations
Contributions
Conceptualization: JL, XG, X-B S; Data acquisition: JL, X-J J, QZ, X-X W, LW, JS, Z-K Z, D-D J, Y-F Y, LL, MZ; Development of AI models: XG, LF, C-Y Z; Data interpretation, statistical analysis, and manuscript editing: JL, XG, X-B S, JS.All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Financial support
The study was supported by the National Natural Science Foundation of China (62376231), the National Institute of Hospital Administration (YLXX24AIA011), and the Health Family Planning Commission of Chengdu (2024077).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, J., Jing, X., Zhang, Q. et al. Interpretable deep learning model diagnoses gastrointestinal stromal tumors and lesion characteristics with microprobe endoscopic ultrasonography. Sci Rep 15, 34366 (2025). https://doi.org/10.1038/s41598-025-17018-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-17018-w