Abstract
Mitochondrial dysfunction drives intervertebral disc degeneration, and mitochondrial dynamics are regulated by mitofusins (MFNs). In this study, we evaluated the roles of MFN1 and MFN2 in mitochondrial quality control and their responses to inflammation and antioxidant treatment in grade I and III disc nucleus pulposus cells (NPCs). Human NPCs were isolated from intervertebral disc tissues of patients. Tumor necrosis factor-α (TNF-α)-induced inflammation was treated with vitamin E (Vit E) or saponin. Mitochondrial quality control was evaluated via quantitative polymerase chain reaction, western blotting, and immunocytochemistry. Sulfated glycosaminoglycan levels were quantified to assess extracellular matrix (ECM) integrity. Mitochondrial morphology and function were assessed via transmission electron microscopy and a xenograft model using MFN1-knockout NPCs. TNF-α significantly upregulated MFN1 and MFN2, with MFN1 showing heightened sensitivity in grade III disc NPCs, leading to mitochondrial fragmentation and ECM degradation. Antioxidants mitigated these effects, with Vit E proving more effective than saponin in reducing MFN1 expression, preserving mitochondrial structure, and stabilizing ECM composition. Vit E maintained mitochondrial integrity, whereas TNF-α induced mitochondrial swelling. In vivo, MFN1-knockout NPCs exhibited reduced ECM proteoglycan levels, reinforcing its role in disc homeostasis. These findings suggest that although MFN1 and MFN2 respond to TNF-α, MFN1 reacts more robustly, making it a more promising target under inflammatory stress.
Similar content being viewed by others
Introduction
In healthy intervertebral discs (IVDs), maintaining the balance between extracellular matrix (ECM) anabolism and catabolism is crucial for preserving structural integrity1. However, aging and mechanical stress can disrupt this balance, leading to ECM degradation and IVD degeneration. These stressors affect the mitochondria, which are essential for energy production and apoptosis regulation, thereby resulting in mitochondrial dysfunction. Specifically, this dysfunction is characterized by disrupted membrane potential and accumulation, which intensifies apoptosis and contributes to IVD degeneration. Therefore, preserving mitochondrial function is key to delaying the degenerative processes in IVD (Fig. 1)2,3.
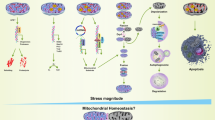
Schematic summary of the proposed mechanism. In human nucleus pulposus cells, TNF-α stimulation induces MFN1/2 expression, leading to mitochondrial fragmentation and extracellular matrix (ECM) degradation. These effects are more pronounced in grade III degenerated discs, particularly via MFN1. Antioxidants such as vitamin E and saponin attenuate MFN expression and preserve mitochondrial and ECM integrity, with vitamin E showing greater efficacy. MFN: mitofusin, TNF-α: tumor necrosis factor-α, Vit E: vitamin E, SPN: saponin. (Created using OriginPro 2025b (OriginLab Corporation, Northampton, MA, USA). Figure was drawn by Dr. Dong-Soo Jang, Yonsei University College of Medicine).
Mitochondria regulate their number, size, and distribution through continuous fission and fusion. Mitochondrial fission, where one mitochondrion is divided into two mitochondria, involves several steps facilitated by proteins, such as dynamin-related protein 1, fission 1 (Fis1), and mitochondrial fission factor (MFF). In contrast, fusion involves merging of the outer membranes of two mitochondria, mediated by mitofusin (MFN) proteins (MFN1 and MFN2), and progresses to the inner mitochondrial membrane through optic atrophy 1 (OPA1)4,5. In addition to mitochondrial function, balanced fission and fusion are associated with cellular metabolism and nutritional status. Mitochondrial dynamics also include the repair of damaged mitochondria; however, if repair is unsuccessful, dysfunctional mitochondria are selectively removed via the autophagic mechanism of mitophagy6.
During IVD degeneration, mechanical loading, pro-inflammatory cytokines, oxidative stress, and glucose stress contribute to mitochondrial dysfunction, creating a vicious cycle of ROS production and damage. Mitochondrial dynamics, particularly the balance between fusion and fission, are disrupted under stress conditions7. Oxidative stress induces mitochondrial fission while inhibiting fusion, thereby altering the expression of key proteins such as MFF, Fis1, MFN1, MFN2, and OPA1. For instance, preventing fission with mitochondrial division inhibitor 1 reduces mitochondrial damage and apoptosis, highlighting the critical roles of MFN1 and MFN2 in maintaining mitochondrial integrity during IVD degeneration8.
Research has focused on the use of antioxidant agents to reduce inflammation, lower pro-inflammatory cytokine levels, and alleviate stress to prevent IVD degeneration. Although studies have shown the potential of antioxidant agents to mitigate inflammation, the exact mechanisms by which they affect mitochondrial function in nucleus pulposus cells (NPCs) are not fully understood9,10,11. This knowledge gap highlights the need for further investigation into the specific mitochondrial pathways involved in the protection of NPCs from degeneration via antioxidant therapy.
Although MFN1 and MFN2 are important for mitochondrial fusion, their precise mechanisms and roles in membrane fusion are not fully understood. A fusion step that is dependent on the GTP-binding domain of MFN has been identified12,13; however, whether this step is related to GTPase activity remains unclear. MFN1 exhibits a homotypic interaction in the GTP-dependent mitochondrial tethering reaction that is 100-fold stronger than that of MFN2, indicating that these proteins have distinct functions during the fusion process12,13. Therefore, the aim of this study was to understand mitochondrial quality control in disc NPCs by investigating the roles of mitofusion, mitofission, and mitophagy during inflammation. We also explored the distinct roles of MFN1 and MFN2 in these processes.
Results
Patient and cell characteristics
This study involved seven male and eight female patients with a mean age of 38.41 ± 4.21 years. The expression of ovo-like zinc finger 2 (OVOL2) and paired box-1 (PAX1) in cultured cells was confirmed through reverse transcription quantitative polymerase chain reaction (RT-qPCR), verifying these cells as IVD cells used in the experiment (Supplementary Fig. 1). MFN1 expression in NPCs was significantly higher in Pfirrmann grade III (2.14 ± 0.67) and IV discs (2.64 ± 0.34) than in Pfirrmann grade I discs (0.21 ± 0.01, p = 0.0105). As IVD degeneration progresses, MFN1 expression increases, indicating a correlation between the progression of disc degeneration and elevated MFN1 level (Supplementary Fig. 2).
Effects of tumor necrosis factor-α on MFN proteins and the effects of antioxidants
RT-qPCR revealed that tumor necrosis factor-α (TNF-α)-induced inflammation resulted in a significant increase in the expression of MFN mRNA, including MFN1, compared with that in the control (control: 0.29 ± 0.00 vs TNF-α: 1.00 ± 0.02, p = 0.0127). Subsequent treatment with vitamin E (Vit E) significantly reduced its expression (TNF-α: 1.00 ± 0.02 vs Vit E: 0.70 ± 0.01, p = 0.0463). However, Western blot analysis revealed no significant differences in MFN1 and MFN2 protein expression among TNF-α, Vit E, and SPN treatment groups in grade I disc NPCs.
Conversely, in grade III disc NPCs, MFN1 expression significantly increased in response to TNF-α treatment compared with the control (control: 0.52 ± 0.01 vs TNF-α: 1.21 ± 0.00, p = 0.0221) and decreased significantly with Vit E treatment (TNF-α: 1.21 ± 0.00 vs Vit E: 0.79 ± 0.03, p = 0.0234). SPN treatment did not result in a significant change compared with TNF-α treatment (TNF-α: 1.21 ± 0.00 vs SPN: 1.18 ± 0.03, p = 0.9381). These significant changes in MFN1 were more pronounced compared with those observed in MFN2 in grade III disc NPCs (Fig. 2).
Expression levels of MFN1 and MFN2 in grade I and Ⅲ disc NPCs. The expression levels of MFN1 and MFN2 in grade I and Ⅲ disc NPCs were analyzed using RT-qPCR (A) and western blotting (B). Control, TNF-α-induced inflammation, anti-inflammation by Vit E, and anti-inflammation by SPN groups were compared. Equal amounts of protein (20 μg per lane) were loaded for western blotting, and the band intensity of MFN1 was semiquantitatively analyzed after normalization to β-actin as the loading control. Data are presented as mean ± standard deviation (n = 3) from paired t-tests. *p < 0.05. MFN: mitofusin, TNF-α: tumor necrosis factor-α, Vit E: vitamin E, SPN: saponin, RT-qPCR: reverse transcription quantitative polymerase chain reaction, NPC: nucleus pulposus cell.
Immunocytochemistry indicated a greater increase in MFN1 expression than in MFN2 expression post-TNF-α treatment, followed by a more pronounced sequential decrease with Vit E treatment (Fig. 3).
MFN1 and MFN2 expression in grade III disc NPCs. (a) Immunofluorescence showing MFN1 (red), actin (phalloidin, green), and nuclei (DAPI, blue) under different conditions: TNF-α (−), TNF-α (+, 10 ng mL−1), TNF-α + Vit E(50 µM), and TNF-α + SPN(25 µg mL−1). Antioxidant treatments reduced MFN1 expression, with Vit E showing greater efficacy. (b) Immunofluorescence staining for MFN2 (red), actin (green), and nuclei (blue) under the same conditions. Similar to MFN1, antioxidants decreased MFN2 expression, with Vit E being more effective. Control, TNF-α-induced inflammation, anti-inflammation by Vit E, and anti-inflammation by SPN groups were compared. MFN, Quantification of MFN1 and MFN2 fluorescence intensity normalised to DAPI-positive cell count. Data represent mean ± SD of three independent experiments performed in triplicate (n = 9), All images were captured in a single confocal session with identical laser and exposure settings. Scale bars = 50 µm. : mitofusin, TNF-α: tumor necrosis factor-α, Vit E: vitamin E, SPN: saponin, NPC: nucleus pulposus cell.
Effects of TNF-α on mitofission and autophagy receptor-related proteins and the role of antioxidants in mitigating these effects
Quantitative messenger RNA (mRNA) expression analysis revealed distinct patterns of calcium-binding and coiled-coil domain 2 (NDP52), optineurin (OPTN), and MFF expression in grade I and III disc NPCs under different experimental conditions, suggesting differential responses to inflammation and antioxidant treatments.
In grade I disc NPCs, NDP52 mRNA expression increased significantly post-TNF-α treatment compared with that in the control condition (control: 0.31 ± 0.01 vs TNF-α: 1.00 ± 0.01, p = 0.0006). The administration of Vit E (p = 0.1203) and SPN (p = 0.1821) did not result in a decrease in NDP52 mRNA expression. Conversely, in grade III disc NPCs, TNF-α treatment led to an increase in NDP52 mRNA expression (control: 0.65 ± 0.03 vs TNF-α: 1.00 ± 0.01, p = 0.0481), whereas Vit E (TNF-α: 1.00 ± 0.01 vs Vit E: 0.61 ± 0.08, p = 0.0493) and SPN (TNF-α: 1.00 ± 0.01 vs SPN: 0.52 ± 0.20, p = 0.0431) reduced its expression. These trends were consistent with the findings of the western blot analysis, which mirrored the mRNA results.
For OPTN, in grade I disc NPCs, TNF-α treatment did not substantially alter its mRNA expression (control: 1.25 ± 0.00 vs TNF-α: 1.02 ± 0.01, p = 0.1209) nor did the administration of Vit E (p = 0.0915) and SPN (p = 0.5792). However, SPN treatment alone led to a decrease in OPTN mRNA expression in grade III disc NPCs (TNF-α: 1.00 ± 0.01 vs SPN: 0.62 ± 0.00, p = 0.0041). The western blot analysis showed a distinct pattern: OPTN protein expression increased in response to TNF-α treatment in grade I disc NPCs (control: 0.31 ± 0.01 vs TNF-α: 1.02 ± 0.31, p = 0.0001), whereas no significant decrease in its expression was observed with Vit E or SPN in grade I (p = 0.9570, p = 0.2680) or grade III disc NPCs (p = 0.6138, p = 0.1777).
MFF exhibited an increase in mRNA expression in both grade I and Ⅲ disc NPCs post-TNF-α treatment (grade I: control: 0.61 ± 0.00 vs TNF-α: 1.01 ± 0.03, p = 0.0015; grade III: control: 0.66 ± 0.00 vs TNF-α: 0.92 ± 0.06, p = 0.0011). When treated with Vit E or SPN, MFF mRNA expression decreased in both disc NPCs of both grades. However, the western blot analysis results revealed some variations: no significant change was observed in MFF protein expression across TNF-α (p = 0.5310), Vit E (p = 0.5513), and SPN (p = 0.1754) treatments in grade I disc NPCs. In grade III disc NPCs, MFF protein expression significantly increased under TNF-α treatment (control: 0.78 ± 0.00 vs TNF-α: 1.10 ± 0.08, p = 0.0293) and decreased significantly with Vit E treatment (TNF-α: 1.10 ± 0.08 vs Vit E: 0.56 ± 0.08, p = 0.0162). Although SPN reduced MFF expression in grade III disc NPCs, the change was not statistically significant (p = 0.0772) (Fig. 4).
NDP52, MFF, and OPTN expression in grade I and Ⅲ disc NPCs. The expression of NDP52, MFF, and OPTN in grade I disc NPCs was analyzed using RT-qPCR (A) and western blotting (B). In grade Ⅲ disc NPCs, the expression of NDP52, MFF, and OPTN was analyzed using RT-qPCR (A) and western blotting (B). Control, TNF-α-induced inflammation, anti-inflammatory by Vit E, and anti-inflammatory by SPN groups were compared. For western blotting, each band intensity was normalized to that of β-actin. Equal loading of 20 μg total protein per lane was ensured. Data are presented as mean ± standard deviation (n = 3) from paired t-tests. *p < 0.05. NDP52: calcium binding and coiled-coil domain 2, OPTN: optineurim, MFF: mitochondrial fission factor, RT-qPCR: reverse transcription quantitative polymerase chain reaction, TNF-α: tumor necrosis factor-α, MFF: mitochondrial fission factor, NPC: nucleus pulposus cell.
Effects of TNF-α on changes in the ECM and the effects of antioxidants
The sulfate glycosaminoglycan (GAG) levels decreased significantly in response to TNF-α treatment in grade III disc NPCs (control: 0.95 ± 0.07 μg/mg vs TNF-α: 0.88 ± 0.22 μg/mg, p = 0.0296). No statistically significant changes were observed post-treatment with Vit E (p = 0.1078) and SPN (p = 0.3729). Additionally, the immunocytochemistry analysis revealed a tendency for proteoglycan expression to decrease, consistent with the GAG level measurements. Vit E and SPN treatments did not significantly affect its expression (Fig. 5).
GAG and proteoglycan expression in grade I and Ⅲ disc NPCs. (A) Sulfate and chondroitin-sulfate GAG expression in grade I and Ⅲ disc NPCs as analyzed using ELISA. (B) Proteoglycan expression levels as analyzed using immunocytochemistry. TNF-α (-) condition served as negative control, with comparison to TNF-α-induced inflammation, anti-inflammation by Vit E, and anti-inflammation by SPN groups. Nonspecific binding and autofluorescence in the negative control were negligible. Data are presented as mean ± standard deviation (n = 3) from paired t-tests. *p < 0.05. GAG: glycosaminoglycan, ELISA: enzyme-linked immunosorbent assay, Vit E: vitamin E, SPN: saponin, TNF-α: tumor necrosis factor-α, NPC: nucleus pulposus cell.
Expression levels of MFN1, MFN2, and proteoglycan in grade III NPCs from in vivo experiments
The expression levels of MFN1, MFN2, and proteoglycan in grade III disc NPCs from in vivo experiments were analyzed using western blotting, dividing the samples into control (mock), MFN1 knockout, and positive control groups. Evaluations were performed on days 1 and 7 post-transduction. On day 1, MFN1 expression was significantly reduced in the MFN1-knockout group compared with that in the mock group (mock: 2.55 ± 0.02 vs knockout: 1.23 ± 0.00, p < 0.0001), whereas the positive control group maintained relatively higher expression levels of MFN1 (1.23 ± 0.00). By day 7, MFN1 expression further decreased in the MFN1-knockout group (mock: 1.57 ± 0.00 vs knockout: 0.53 ± 0.00, p < 0.0001), and even the mock group showed a decline in MFN1 level compared with that on day 1 (day 1: 1.23 ± 0.00 vs day 7: 0.53 ± 0.00, p < 0.0001). The MFN2 level was higher in the MFN1-knockout group than in the mock group on both days 1 and 7 (day 1: 0.92 ± 0.02 vs 1.23 ± 0.00; day 7: 1.19 ± 0.03 vs 0.53 ± 0.00, p = 0.0703). Notably, on day 7, the MFN1-knockout group maintained MFN2 expression, indicating a potential adaptive response to MFN1 absence. The proteoglycan expression showed no statistically significant difference between the MFN1-knockout and mock groups on either day 1 or day 7 (day 1: mock 0.62 ± 0.01 vs knockout 0.68 ± 0.21, p = 0.7604; day 7: mock 0.64 ± 0.01 vs knockout 0.32 ± 0.01, p = 0.6185). Despite the apparent decrease in the knockout group on day 7, this difference was not statistically significant (Fig. 6).
Expression of MFN1, MFN2, and proteoglycan in grade Ⅲ disc NPCs. Expression was evaluated in vivo using western blot analysis. Samples were categorized into negative control (mock), MFN1-knockout, and positive control groups. (a) Grade II disc NPCs; (b) MFN1, (c) MFN2, and (d) proteoglycan in grade Ⅲ disc NPCs. In all experiments, the samples were categorized into negative control (mock), MFN1-knockout, and positive control groups. Western blot analysis was performed using 20 μg of protein per lane. The band intensity was normalized to that of β-actin for semiquantitative comparison across groups and time points. Expression was evaluated on days 1 and 7 post-transplantation to assess temporal changes among the groups. Data are presented as mean ± standard deviation (n = 3) from paired t-tests. *p < 0.05. MFN: mitofusin, KO: knockout mice, GFP: green fluorescent protein, NPC: nucleus pulposus cell.
The immunofluorescence analysis performed on day 7 post-transduction showed that MFN1 expression was absent in MFN1-knockout cells, confirming the successful knockout. In contrast, MFN2 expression was maintained in the knockout group. Proteoglycan expression in MFN1-knockout cells was lower than that in the mock group (Fig. 7).
Immunofluorescence analysis of MFN1, MFN2, proteoglycan, and GFP in the mock and MFN1-knockout cells. (a) Expression and co-localization of MFN1 (green) and MFN2 (red) in the mock and MFN1-knockout cells. MFN1 is absent in the knockout group; however, MFN2 is more prominently expressed in the knockout group than in the mock group. (b) Distribution of proteoglycan (green) and GFP (yellow) in mock and MFN1-knockout cells. Proteoglycan expression in the MFN1-knockout group was lower than that in the mock group.
Discussion
Our investigation revealed that inflammation triggers a multifaceted response in IVD NPCs, activating mitofusion, mitofission, and autophagy, thereby preserving cellular homeostasis14,15,16. This intricate process involves the interplay of key proteins, including MFN1, MFN2, NDP52, OPTN, and MFF, which regulate mitochondrial dynamics and quality control. Notably, MFN1 exhibited heightened sensitivity to inflammation, particularly in grade III disc NPCs, suggesting its crucial role in the pathogenesis of disc degeneration. Furthermore, Vit E demonstrated significant anti-inflammatory effects by effectively modulating the expression of these proteins and mitigating inflammation-induced damage. These findings underscore the critical roles of mitochondrial dysfunction and inflammation in IVD degeneration, highlighting the potential of targeting mitochondrial dynamics, particularly MFN1, as a novel therapeutic strategy for degenerative disc diseases. This coordinated response involving mitofission and mitophagy is essential for the survival of disc NPCs under inflammatory conditions14,17,18.
NPCs exhibit a complex cellular response aimed at managing inflammation, cellular stress, and tissue remodeling in response to inflammation. Mitophagy activation helps maintain mitochondrial health by removing damaged components and optimizing mitochondrial function. Concurrently, the increase in ECM suggests that NPCs actively stabilize and repair tissue structures, which is a crucial step in maintaining IVD integrity. Collectively, these processes reflect the mechanisms by which NPCs are activated to preserve homeostasis and protect against cellular damage under inflammatory conditions. This balance between cellular repair and tissue remodeling indicates the adaptive response of NPCs to maintain functionality and prevent degeneration14.
Based on TEM images, the morphology of mitochondria can vary depending on the cells’ condition. In grade I disc NPCs, mitochondria tend to be smaller and more numerous, which is indicative of a metabolically active and healthy state. However, they tend to be larger and fewer in grade III disc NPCs, which may be associated with degenerative changes or a damaged cellular state. These changes may reflect mitochondrial dysfunction or impaired cellular energy metabolism. In grade I and Ⅲ disc NPCs, the number of mitochondria decreased during inflammation, whereas it increased upon the introduction of anti-inflammatory agents, which also appeared more pronounced or dense. During inflammation, cellular energy metabolism and function are impaired, leading to a decreased number of mitochondria. Inflammatory conditions increase oxidative stress and decrease mitochondrial function, thereby affecting the number and structural integrity of mitochondria (Fig. 8).
Mitochondrial morphology of grade I and Ⅲ disc NPCs using TEM. (a) In grade I NPCs, the mitochondria are small, numerous, and structurally intact, reflecting a healthy state with active metabolism. In all experiments, control, TNF-α-induced inflammation, anti-inflammation by Vit E, and SPN-induced inflammation groups were compared. (b) Grade Ⅲ disc NPCs exhibit larger and fewer mitochondria with structural damage and reduced density, indicative of degenerative changes and impaired mitochondrial function. In all experiments, control, TNF-α-induced inflammation, anti-inflammation by Vit E, and SPN-induced inflammation groups were compared. Vit E: vitamin E, SPN: saponin, TNF-α: tumor necrosis factor-α, TEM: transmission electron microscopy, NPC: nucleus pulposus cell.
The RT-qPCR results demonstrated that MFN1 and MFN2 expression increased in response to inflammation in disc NPCs, highlighting their roles in managing mitochondrial dynamics under inflammatory stress. Although MFN1 showed a slightly more pronounced initial response, MFN2 exhibited comparable regulatory effects on mitochondrial fusion and protection of mitochondrial function. Following inflammation, MFN1 and MFN2 expression decreased, even after Vit E treatment, with MFN1 showing a slightly greater reduction in grade III disc NPCs. This decrease in expression correlated with reduced ECM levels, suggesting that MFN1 and MFN2 jointly contribute to mitochondrial dynamics and ECM regulation under inflammatory conditions, albeit with minor differences in sensitivity to inflammatory signals14,19,20.
The in vivo study results highlight the critical role of MFN1 in maintaining ECM integrity and regulating proteoglycan levels under physiological conditions. Additionally, the significant reduction in proteoglycan levels in the MFN1-knockout group on days 1 and 7 underscores MFN1’s essential function in preserving ECM stability. MFN2 alone was insufficient to fully preserve proteoglycan levels, indicating that MFN1 has a unique and indispensable role in ECM maintenance. These findings emphasize the interplay between MFN1 and MFN2 in regulating mitochondrial dynamics and ECM stability, and they provide a foundation for exploring therapeutic strategies targeting mitochondrial function in degenerative disc diseases.
Although we observed a compensatory increase in MFN2 expression in MFN1-knockout NPCs, we did not assess whether this upregulation functionally restored mitochondrial fusion capacity or ECM synthesis, which is a limitation of our study. Furthermore, the in vivo subcutaneous xenograft model that we used did not fully replicate the avascular and hypoxic conditions of the native IVD niche. Thus, although our findings provide preliminary insights, orthotopic models (e.g., intradiscal injection into rat tail discs) may yield more translationally relevant results. Moreover, our assessment of ECM components was limited to sulfated GAG quantification and proteoglycan immunocytochemistry. We did not analyze collagen II or aggrecan mRNA/protein levels, which is another limitation of this study. Future studies should incorporate these additional ECM markers to provide a more comprehensive understanding of matrix homeostasis and degeneration processes.
Vit E, SPN, curcumin, omega-3, glucosamine, and chondroitin are well-known anti-inflammatory agents recognized for their ability to reduce pro-inflammatory cytokines. In this study, we conducted experiments using Vit E and SPN to modulate the inflammatory pathway in disc NPCs. We observed that Vit E exhibited a more pronounced effect on expression changes in disc NPCs than SPN, with significant differences in the expression of MFN1, MFF, and NDP52 proteins in grade III disc NPCs. Specifically, Vit E demonstrated a greater ability to modulate the expression of genes associated with inflammation than SPN, leading to significant reductions in inflammatory marker levels and enhanced anti-inflammatory responses. Moreover, this finding suggests that Vit E plays an important role in regulating the inflammatory environment in disc NPCs, thereby providing better protection against inflammation-induced damage21,22.
The inflammatory and anti-inflammatory effects on MFN1, MFN2, NDP52, OPTN, and MFF expression observed via western blotting were more pronounced in grade III disc NPCs than in grade I disc NPCs. This finding could be because more advanced disc degeneration (as observed in grade III) leads to greater cellular stress and mitochondrial dysfunction. As disc degeneration progresses, cells may become more sensitive to inflammatory stimuli, causing a heightened response in mitochondrial dynamics. Consequently, the anti-inflammatory effects of agents such as Vit E are also more pronounced in mitigating inflammation-induced stress, particularly in more severely degenerated discs.
Comparison with previous studies
Although prior research has established that mitochondrial dysfunction plays a central role in IVD degeneration, particularly through oxidative stress and disrupted mitochondrial dynamics, our study adds significant new dimensions. Previous studies have primarily focused on mitochondrial fission and autophagy as key mechanisms underlying degenerative processes2,4,7. The current study highlights the distinct and critical roles of MFN1 and MFN2 in regulating mitochondrial fusion under inflammatory conditions. In grade III disc NPCs, MFN1 demonstrated effects comparable to those of MFN2, suggesting that both proteins play important roles in responding to inflammatory stimuli and maintaining mitochondrial dynamics. This is an important distinction, as the heightened sensitivity of MFN1 may make it a more relevant therapeutic target in conditions characterized by chronic inflammation, such as degenerative disc disease. Contrary to the earlier focus on general mitochondrial dysfunction, our results indicate that selective modulation of MFN1 enhances mitochondrial fusion and protects cellular function. Although the use of antioxidants to combat inflammation is well established, our data provide detailed mechanistic insights into the modulation of proteins related to mitochondrial dynamics, including MFN1 and its associated pathways, a connection that has been previously underexplored22.
Clinical implications
As degenerative disc disease continues to be a major contributor to chronic pain, developing therapies that specifically target mitochondrial dynamics may improve patient outcomes. The ability of Vit E to downregulate MFN1 expression in response to inflammation suggests that antioxidant therapies serve as a viable, non-invasive treatment option for slowing or reversing disc degeneration. This finding holds particular relevance in the context of aging populations, where the prevalence of degenerative conditions is increasing.
Targeting MFN1 to modulate mitochondrial fusion and fission may represent a novel therapeutic strategy beyond traditional anti-inflammatory and surgical interventions. This could delay the need for surgical treatments, such as spinal fusion and disc replacement, thereby reducing the risk of surgical complications and improving the long-term quality of life of patients.
From a policy perspective, our findings suggest that early intervention with antioxidant therapies could be incorporated into clinical guidelines for managing early-stage disc degeneration. Such an approach could help reduce healthcare costs by delaying disease progression and reducing the need for invasive procedures. In clinical practice, these findings could support the development of personalized treatment protocols where patients with high inflammatory profiles may benefit from antioxidant therapies specifically targeting mitochondrial dysfunction. However, further clinical trials should explore the optimal dosing and timing of Vit E and other antioxidants to maximize their protective effects on disc NPCs.
Future research directions
In the future, therapeutic strategies that combine antioxidants with agents that specifically modulate mitochondrial fusion and fission should be explored. Clinical trials that evaluate the efficacy of antioxidant therapies in preventing or delaying disc degeneration would also provide direct evidence for integrating these treatments into clinical practice. Mitochondrial quality control is involved in various pathways beyond those discussed, and its roles in inflammation and anti-inflammation should be considered in future research23.
Conclusions
In disc NPCs, inflammation activates mitofusion, mitofission, and autophagy, which help maintain cellular homeostasis. Vit E has anti-inflammatory effects on NPCs. Notably, MFN1 tended to be activated as the pathogenesis of NPCs progressed, with significant differences observed between inflammatory and anti-inflammatory conditions. These findings highlight the critical roles of mitochondrial dysfunction and inflammation in IVD degeneration. Furthermore, the modulation of mitochondrial dynamics by antioxidants, such as Vit E, may provide a potential therapeutic strategy for treating and preventing degenerative disc disease.
Methods
Ethics statements
This study was approved by the Institutional Review Board of the College of Medicine at Yonsei University approved all experimental procedures (approval number: 4-2022-0167). All experimental procedures in this study were approved by the Institutional Animal Care and Use Committee of Yonsei University Health System (approval number: 2024-0281). All methods were performed in accordance with the relevant guidelines and regulations, including the ARRIVE guidelines (https://arriveguidelines.org). All methods were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained from all individuals participating in the study. This study complied with the standard reporting guidelines outlined in the Strengthening the Reporting of Observational Studies in Epidemiology statement to ensure the transparency, accuracy, and reproducibility of its findings.
Patient samples
NPC samples were obtained from 15 patients diagnosed with disc-related conditions, including scoliosis. The degree of IVD degeneration was assessed using magnetic resonance imaging and classified according to a modified version of the Pfirrmann grading system24. Samples from patients with grades I (n = 5), II (n = 3), III (n = 5), and IV (n = 2) IVD were obtained. The experiments were primarily conducted using grade I and Ⅲ disc NPCs (Supplementary Table 1). It was challenging to obtaining samples of grade 2 and grade 4 was challenging. In our initial experiments using western blot analysis with the tissue samples, we observed that grade 2 exhibited similar MFN1 and MFN2 expression levels to grade 1, while whereas grade 4 showed expression patterns similar to those of grade 3. Due Owing to the difficulty in obtaining additional tissue samples and the clear distinctions observed between grades 1 and 3, we focused our experiments on these two grades. The exclusion criteria were a history of spinal surgery, active infection, inflammatory disorders, malignancies, or systemic diseases that could influence the inflammatory process or mitochondrial function. Patients who received antioxidant treatments before surgery were also excluded.
Isolation and expansion of human NPCs
The IVD tissues obtained during the surgical procedure were washed with DPBS, and then the nucleus pulposus NP and annulus fibrosus. AF regions were separated using forceps. The tissues were chopped cut using scalpel. Samples that were difficult to separate due to the mixture of annulus fibrosus AF and nucleus pulposus NP regions were excluded. The minced tissues were digested for 2 h at 37 °C with gentle agitation in a medium comprising equal parts Dulbecco’s modified Eagle medium and Ham’s nutrient mixture F-12 (DMEM/F12, 1:1; Gibco, Life Technologies Corporation, Grand Island, NY, USA) supplemented with collagenase type II (2.5 mg/mL; Sigma, St. Louis, USA). After digestion, the cells were passed through a sterile nylon mesh filter with a pore size of 100 μm (Falcon, Durham, NC, USA) and seeded in T25 flasks (EasYFlask; Nunc, Sjælland, Denmark) at a density of approximately 1 × 105 cells/mL. Primary cultures were maintained for 2–3 weeks in maintenance medium, which is comprised DMEM/F12 medium containing 10% fetal bovine serum (FBS), 1% v/v penicillin, streptomycin, and nystatin (Gibco, Life Technologies Corporation), and ascorbic acid (250 mg/mL) at 37 °C in a humidified incubator (Thermo Fisher Scientific, Marietta, OH, USA) with 5% carbon dioxide. The culture medium was refreshed three times per week (Supplementary Fig. 3). Ascorbic acid was added to promote extracellular matrixECM synthesis and support collagen production, maintaining a physiologically relevant phenotype of NPCs during in vitro culture.
Cell culture
Only NPCs, with the annulus tissue excluded, were used in this study. The cells exhibited typical nucleus pulposus NP morphology, and the expression of paired box-1 (PAX1) and ovo-like zinc finger 2 (OVOL2) was analyzed using reverse transcription quantitative polymerase chain reaction (RT-qPCR). The expression of PAX1 and OVOL2, both known molecular markers for confirming the identity of nucleus pulposus cellNPCs, was analyzed using RT-qPCR25. Next, the cells were used at passage two-fivetwo through five. The cells were seeded in 100-mm culture dishes (TPP, Trasadingen, Switzerland) at a density of 2 × 105 cells/mL during culture incubation. They were treated with 4 ng/mL tumor necrosis factor-α (TNF-α) for 24 h after 16 h of serum starvation. Additionally, the cells were also treated with 200 mM saponin (SPN) and vitamin E (Vit E) for 24 h after TNF-α treatment. The following groups were studied: control, TNF-α-induced inflammation, anti-inflammation by Vit E, and SPN-induced inflammation.
Sulfated glycosaminoglycan assay
Sulfated glycosaminoglycan (GAG) was isolated from cells lysed using a papain-based buffer composed of 0.1 M sodium acetate, 0.01 M sodium ethylenediaminetetraacetic acid (EDTA) (pH 6.4), and 5 mM cysteine hydrochloride. The extracted GAG was concentrated using the sulfated GAG assay protocol and quantified at 650 nm using a microplate reader, according to the Blyscan GAG assay kit instructions (Biocolor Ltd., Carrickfergus, UK). Finally, the concentration of sulfated GAG was calculated based on a standard curve generated with bovine tracheal chondroitin 4-sulfate provided in the assay kit (Biocolor Ltd.).
RNA extraction and real-time polymerase chain reaction analysis
Total RNA was extracted from chondrocyte-like cells using the QIAGEN RNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized using the Maxime RT Premix Kit (QIAGEN). Genes of interest, including MFF, MFN1, MFN2, calcium-binding and coiled-coil domain 2 (NDP52), optineurin (OPTN), vimentin (VIM), tissue inhibitor of metalloproteinases-1 (TIMP-1), phosphatase and tensin homolog-induced kinase 1 (PINK-1), and type II collagen (Col II), were amplified using the CFX96 Touch RT-qPCR Detection System (Bio-Rad, Hercules, CA, USA). The relative gene expression levels were normalized to β-actin level as the reference gene (Supplementary Table 2). The full-length, uncropped RT-PCR gel images corresponding to the data presented in Supplementary Fig. 2 are provided (Supplementary Fig. 6).
Protein extraction and western blot analysis
Cultured NPCs were lysed in lysis buffer containing 150 mM NaCl, 5 mM EDTA (pH 8.0), 10 mM Tris–Cl (pH 7.4), 1% Triton X-100, and 0.2% sodium dodecyl sulfate (SDS), supplemented with protease and phosphatase inhibitors (Pierce Mini Tablets; Thermo Fisher Scientific). The culture medium was collected to detect soluble proteins. Cells treated with TNF-α and vit E or TNF-α and SPN for 24 h were lysed in radioimmunoprecipitationRIPA lysis buffer (ATTO Corporation, Tokyo, Japan) containing protease and phosphatase inhibitors. Next, the lysates were loaded at 20 μg each, and the medium were was loaded at 35 μLl for each and separated using SDS–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Pierce; Thermo Fisher Scientific) with the Mini Trans-Blot Cell transfer system (Bio-Rad).
The membranes were probed with primary antibodies against TIMP-1 (anti-TIMP1 antibody-carboxyterminal end, ab38978, 1:10000, Abcam, Cambridge, UK), Col II (Anti-Collagen II antibody, ab34712, 1:10000, Abcam, Cambridge, UK), P38 (p38 MAPK Antibody, #9212, 1:10000, Cell Signaling Technology Inc., Danvers, MA, USA), phospho-P38 (Phospho-p38 MAPK (Thr180/Tyr182) (D3F9), #4511, 1:10000, Cell Signaling Technology Inc.), MFF, MFN1 (Mitofusin-1 (D6E2S) Rabbit mAb, #14739, 1:10000, Cell Signaling Technology Inc.), MFN2 (Mitofusin-2 (D1E9) Rabbit mAb, #11925, 1:10000, Cell Signaling Technology Inc.), NDP52 (NDP52 (D1E4A) Rabbit mAb, #60732, 1:10000, Cell Signaling Technology Inc.), OPTN (Optineurin (D2L8S) Rabbit mAb, #58981, 1:10000, Cell Signaling Technology Inc.), VIM (Vimentin (D21H3) XP® Rabbit mAb, #5741, 1:10000, Cell Signaling Technology Inc.), PINK-1 (PINK1 (D8G3) Rabbit mAb, #6946, 1:10000, Cell Signaling Technology Inc.)., and tThe primary antibodies were incubated for overnight at 4℃. Immunoreactive bands were visualized using the EzWestLumiPlus western blot detection system (ATTO Corp., Tokyo, Japan) after incubation with the secondary antibodies for 2 h at room temperature. The membranes were stripped and re-probed with b-actin (anti-actin antibody, ab3280, 1:10000, Abcam).). For semiquantitative analysis, the band intensities intensity of the proteins of interest were was normalized to that of the loading control (b-actin). The full-length, uncropped original western blot images corresponding to the data presented in Figs. 2, 4, and 6 are provided in Supplementary Figs. 7, 8, and 9.
Immunofluorescence
The cells were seeded in glass-bottomed culture dishes at a density of 3 × 104 cells per well (MarTek Corporation, Ashland, MA, USA) and incubated for 24 h. After treatment, the cells were fixed in 4% formaldehyde for 10 min, permeabilized with 0.5% Triton X-100 (v/v) in phosphate-buffered saline (PBS) for another 10 min, and finally blocked with 10% FBS in PBS for 1 h. Subsequently, the cells were incubated overnight at 4 °C with primary antibodies against MFN1 and MFN2 (Cell Signaling Technology Inc.), as well as with chondroitin sulfate proteoglycan (anti-NG2 Chondroitin Sulfate Proteoglycan, AB5320 Millipore, Temecula, MA, USA). Next, the cells were washed and treated with anti-rabbit Alexa Fluor 488-conjugated secondary antibodies (green) at a 1:200 dilution (Invitrogen; Thermo Fisher Scientific) and 10 μM 4′,6-diamidino-2-phenylindole (DAPI; Abcam, ab228549, Milton, UK) for nuclear staining at room temperature (20–22 °C) for 1 h. The samples were examined using a confocal microscope (Zeiss LSM700; Carl Zeiss, Berlin, Germany).
The cells for in vivo assay were seeded in hydrogel (TrueGel3D Hydrogel Kot; Merck, St. Louis, MO, USA) and implanted in BALB/c nude mice for 1 and 7 days. After sacrificing the experimental animals, the hydrogels were fixed in 4% formaldehyde for 1 day, permeabilized with 0.5% Triton X-100 (v/v) in PBS for another 10 min, and blocked with 10% FBS in PBS for 1 h. Subsequently, the cells were incubated overnight at 4 °C with primary antibodies against MFN1 and MFN2 (Cell Signaling Technology Inc.), as well as with chondroitin sulfate proteoglycan (Merck, Temecula, MA, USA). Next, the cells were washed and treated with anti-rabbit Alexa Fluor 488-conjugated secondary antibodies (green) at a 1:200 dilution (Invitrogen; Thermo Fisher Scientific), anti-mouse Texas red conjugated secondary antibodies (red) at a 1:200 dilution (Vector Laboratories, Inc. Burlingame, CA, USA), and 10 μM DAPI (Abcam) for nuclear staining at room temperature (20–22 °C) for 1 h. Finally, the samples were examined using a confocal microscope (Zeiss LSM700; Carl Zeiss, Berlin, Germany).
Transmission electron microscopy
The cells treated with SPN or Vit E for 24 h after TNF-α treatment were collected using 0.5% trypsin EDTA (Gibco; Life Technologies Corporation) in a 1.5-mL Eppendorf tube, centrifugated, and washed with Dulbecco’s PBS (Gibco; Life Technologies Corporation). Next, the cells were washed with PBS and fixed for one 1 h with 2% glutaraldehyde (#G5882, Sigma-Aldrich, Taufkirchen, Germany) in 0.2 mol/L cacodylate buffer (#AGR1503, Aagar Sscientific, EssexStansted, UK) (pH 7.2) at room temperature. This was followed by incubation with 2% osmium tetroxide (#0972B-6, Polyscience Inc., Hirschberg, Germany) for 90 min. The cells were then collected using a cell scraper and fixed in agar blocks. Samples were then dehydrated in an ascending ethanol series, starting with 50% ethanol, followed by incubation in 70% ethanol, 1% uranyl acetate (#21447, Polyscience Inc.), Hirschberg, Germanyand 1% phosphotungstic acid (#455970, Sigma-Aldrich, Taufkirchen, Germany) overnight at 4 °C. Dehydration continued the next day with an ascending ethanol series (80–100%). The samples were carefully transferred into epoxy resin (consisting of glycidether (#21045.02, Serva, Heidelberg, Germany), methylnadic anhydride (#29452.02, Serva, Heidelberg, Germany), 2-dodecenyl succinic anhydride (#20755.01, Serva, Heidelberg Germany), and 2,4,6-tris (dimethylaminomethyl)phenol (#36975.01, Serva Heidelberg, Germany) and incubated in propylene oxide (#807027, Sigma-Aldrich, Taufkirchen, Germany). After polymerization, ultrathin sections were cut using an LKB Ultratome (LKB, Broma, Sweden) and collected on formvar-coated grids. Finally, the sections were stained with uranyl acetate and lead citrate and evaluated using transmission electron microscopy (TEM) (HT7800; Hitachi, Tokyo, Japan) operated at 80 kV (Supplementary Fig. 4).
Establishment of a xenograft model subcutaneously implanted with MFN 1-knockout cells (in vivo study)
Candidate single guide RNAs (sgRNAs) targeting human MFN1 were designed using the Cas-designer tool. Seven sgRNAs were evaluated for their knockdown efficiency in HEK293T cells. These sgRNAs were individually cloned into the LentiArray Lentiviral sgRNA system (#35533; Thermo Fisher Scientific) to generate the lenti-Cas9-sgRNA vectors. Subsequently, HEK293T cells were co-transfected with these lenti-Cas9-sgRNA vectors and lentivirus packaging plasmids (MDLg, VSVG, and Rev). After 72 h, the lentiviral particles were harvested and used to infect the HEK293T cells. Genomic DNA was subsequently extracted from infected cells and analyzed using Surveyor assays. Among the seven sgRNAs tested, one sgRNA (5'-GTTATATGGCCAATCCCACT-3') demonstrated the highest targeting efficiency and was selected for further experiments. NPCs were transduced with lenti-Cas9-sgRNA or lenti-Cas9 alone, followed by puromycin selection over 7 days, resulting in the establishment of stable MFN1-knockdown human NPCs and corresponding negative control cells.
BALB/c nude mice (27–30 g, 10 weeks of age) were housed under specific pathogen-free conditions in the barrier facility of the Department of Laboratory Animal Resources at Yonsei Biomedical Research Institute, Yonsei University College of Medicine. After 1 week of acclimatization, 1.3 × 10⁶ MFN1-knockout cells were subcutaneously implanted into the central region of the right flank of each mouse anesthetized with isoflurane. The mice were observed at 1 day and 1 week post-implantation, after which they were euthanized using a carbon dioxide euthanasia apparatus (LC500; Yuyan Scientific Instruments Co.). Next, the implanted cells were collected for analysis (Fig. 8). All animal experiments were approved by the Yonsei University Health System Institutional Animal Care and Use Committee and adhered to the Guide for the Care and Use of Laboratory Animals (National Research Council, Washington, D.C., USA) (Approval No. 2023-0292) (Supplementary Fig. 5).
Statistical analysis
Data were obtained from three independent experiments, each conducted in triplicate, using chondrocyte-like cell cultures derived from 15 different donors. Results are expressed as mean ± standard deviation. The Shapiro–-Wilk test was used to assess normality. Statistical analyses were performed using data from three independent experiments, each conducted in triplicate. Quantitative results from RT-qPCR, wWestern blot densitometry, and GAG assays were are expressed as mean ± standard deviation (SD). Parametric data were analyzed using paired t-tests, and nonparametric data were evaluated using the Mann–Whitney U test. All statistical analyses were conducted using IBM SPSS Statistics for Windows, version 23.0 (IBM Corp., Armonk, NY, USA), with statistical significance set at p < 0.05.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- IVDs:
-
Intervertebral discs
- ECM:
-
Extracellular matrix
- TNF-α:
-
Tumor necrosis factor-α
- MFN:
-
Mitofusin
- OPA1:
-
Optic atrophy 1
- Fis1:
-
Fission 1
- NPCs:
-
Nucleus pulposus cells
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- SPN:
-
Saponin
- Vit E:
-
Vitamin E
- PAX1:
-
Paired box-1
- OVOL2:
-
Ovo-like zinc finger 2
- GAG:
-
Glycosaminoglycan
- NDP52:
-
Oiled-coil domain 2
- OPTN:
-
Optineurin
- VIM:
-
Vimentin
- TIMP-1:
-
Tissue inhibitor of metalloproteinases-1
- PINK-1:
-
Phosphatase and tensin homolog-induced kinase 1
- Col II:
-
Type II collagen
- PBS:
-
Phosphate-buffered saline
- TEM:
-
Transmission electron microscopy
- sgRNAs:
-
Single guide RNAs
- mRNA:
-
Messenger RNA
References
Roberts, S., Evans, H., Trivedi, J. & Menage, J. Histology and pathology of the human intervertebral disc. J. Bone Joint Surg. Am. 88(Suppl 2), 10–14. https://doi.org/10.2106/JBJS.F.00019 (2006).
Sakai, D. & Andersson, G. B. J. Stem cell therapy for intervertebral disc regeneration: Obstacles and solutions. Nat. Rev. Rheumatol. 11, 243–256. https://doi.org/10.1038/nrrheum.2015.13 (2015).
Urban, J. P. G. & Roberts, S. Degeneration of the intervertebral disc. Arthritis Res. Ther. 5, 120–130. https://doi.org/10.1186/ar629 (2003).
Chan, D. C. Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. 15, 235–259. https://doi.org/10.1146/annurev-pathmechdis-012419-032711 (2020).
Gottlieb, R. A. & Carreira, R. S. Mitochondrial dynamics in apoptosis: Fusion, fission, and the role of DRP1. Antioxid. Redox Signal. 13, 1249–1258 (2010).
Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884. https://doi.org/10.1038/nrm3013 (2010).
Zhang, C. et al. Update on the correlation between mitochondrial dysfunction and intervertebral disk degeneration. DNA Cell Biol. 41, 257–261. https://doi.org/10.1089/dna.2021.1012 (2022).
Lin, Z. et al. The role of mitochondrial fission in intervertebral disc degeneration. Osteoarthr. Cartil. 31, 158–166. https://doi.org/10.1016/j.joca.2022.10.020 (2023).
Hua, W. et al. Icariin protects human nucleus pulposus cells from hydrogen peroxide-induced mitochondria-mediated apoptosis by activating nuclear factor erythroid 2-related factor 2. Biochim. Biophys. Acta Mol. Basis Dis. 1866, 165575. https://doi.org/10.1016/j.bbadis.2019.165575 (2020).
Ezeriņa, D., Takano, Y., Hanaoka, K., Urano, Y. & Dick, T. P. N-Acetyl cysteine functions as a fast-acting antioxidant by triggering intracellular H2S and sulfane sulfur production. Cell Chem. Biol. 25, 447-459.e4. https://doi.org/10.1016/j.chembiol.2018.01.011 (2018).
Tardiolo, G., Bramanti, P. & Mazzon, E. Overview on the effects of N-acetylcysteine in neurodegenerative diseases. Molecules 23, 3305. https://doi.org/10.3390/molecules23123305 (2018).
Chen, H. & Chan, D. C. Mitochondrial dynamics in mammals. Curr. Top. Dev. Biol. 59, 119–144. https://doi.org/10.1016/S0070-2153(04)59005-1 (2004).
Li, Y. J. et al. Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset. Nat. Commun. 10, 4914. https://doi.org/10.1038/s41467-019-12912-0 (2019).
Zhang, Y., Chen, X., Gueydan, C. & Han, J. Mitophagy and mitochondrial dynamics in cell death and stress responses. Cell. Mol. Life Sci. 75, 2077–2098 (2018).
Picca, A., Calvani, R., Coelho-Junior, H. J. & Marzetti, E. Cell death and inflammation: The role of mitochondria in health and disease. Cells 10, 537. https://doi.org/10.3390/cells10030537 (2021).
Tan, W., Zou, J., Yoshida, S., Jiang, B. & Zhou, Y. The role of inflammation in age-related macular degeneration. Int. J. Biol. Sci. 16, 2989–3001. https://doi.org/10.7150/ijbs.49890 (2020).
Pickles, S., Vigié, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185. https://doi.org/10.1016/j.cub.2018.01.004 (2018).
Ashrafi, G. & Schwarz, T. L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42. https://doi.org/10.1038/cdd.2012.81 (2013).
Detmer, S. A. & Chan, D. C. Complementation between mouse Mfn1 and Mfn2 protects mitochondrial fusion defects caused by CMT2A disease mutations. J. Cell Biol. 176, 405–414. https://doi.org/10.1083/jcb.200611080 (2007).
Chen, H. et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200. https://doi.org/10.1083/jcb.200211046 (2003).
Tan, B., Wu, X., Yu, J. & Chen, Z. The role of saponins in the treatment of neuropathic pain. Molecules 27, 3956. https://doi.org/10.3390/molecules27123956 (2022).
Smith, L. J., Nerurkar, N. L., Choi, K. S., Harfe, B. D. & Elliott, D. M. Vitamin E as a modulator of oxidative stress in disc NP cells: Implications for therapeutic strategies. J. Orthop. Res. 36, 2434–2440 (2018).
Rajasekaran, S. et al. Novel Biomarkers of health and degeneration in human intervertebral discs: In-depth proteomic analysis of collagen framework of fetal, healthy, scoliotic, degenerate, and herniated discs. Asian Spine J. 17, 729–738. https://doi.org/10.31616/asj.2021.0535 (2023).
Pfirrmann, C. W. A., Metzdorf, A., Zanetti, M., Hodler, J. & Boos, N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine 26, 1873–1878. https://doi.org/10.1097/00007632-200109010-00011 (2001).
Minogue, B. M., Richardson, S. M., Zeef, L. A. H., Freemont, A. J. & Hoyland, J. A. Transcriptional profiling of bovine intervertebral disc cells: Implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther. 12, R22. https://doi.org/10.1186/ar2929 (2010).
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
J.W.S. conducted the research, performed data analysis, and drafted the majority of the manuscript. Y.M.K. contributed substantially to conducting experiments and assisted in data interpretation. H.S.K., as the corresponding author, provided overall guidance, supervision, and direction for the study. S.H.M. provided critical advice on the manuscript and contributed to revisions. K.S.S., S.Y.P., B.H.L. and J.W.K. primarily provided advice and guidance on the study design and manuscript development. All authors approved the final manuscript and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The Institutional Review Board of the College of Medicine at Yonsei University approved all experimental procedures (Approval Number: 4-2022-0167), and written informed consent was obtained from all individuals participating in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shin, J.W., Kang, Y.M., Kim, H.S. et al. Mitofusin 1 in mitochondrial quality control and anti-inflammatory responses in nucleus pulposus cells during disc degeneration. Sci Rep 15, 38530 (2025). https://doi.org/10.1038/s41598-025-19540-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-19540-3