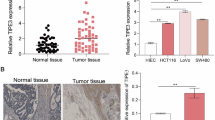
Abstract
Tumor metastasis is an important risk factor for death in patients with colorectal cancer (CRC). This study aims to explore the effect of CXCL9 and SPP1 (CS) polarity alteration of tumor-associated macrophages (TAMs) on CRC progression and its associated molecular mechanisms. The heterogeneity of cellular subsets between the CRC and Normal groups was analyzed using single-cell RNA sequencing (scRNA-seq) technology. Developmental trajectories of the cellular subsets were plotted using pseudotime analysis, and the differences in enrichment scores among the cellular subsets were analyzed by combining gene set variation analysis (GSVA). Mouse CRC models were constructed, and SPP1− TAMs or SPP1+ TAMs were isolated from mouse tumor tissues by flow cytometry sorting. MC38 cells were treated with the JAK/STAT3 inhibitor WP1066 and co-cultured with SPP1+ TAMs. MC38 cell viability was detected by the cell counting kit-8 (CCK-8) assay. The apoptosis rate of MC38 cells was detected by TdT-mediated dUTP nick end labeling (TUNEL) staining. The expression of JAK/STAT3 pathway proteins was detected using western blot (WB), and the expression of epithelial-mesenchymal transition (EMT)-related proteins was detected using immunofluorescence. There were significant differences in SPP1+ TAMs between the CRC and Normal groups by scRNA-seq, and JAK/STAT3 signaling pathway had significant scores in each cell subset. In vitro assays showed that compared with the Negative group, MC38 cells in the Positive group exhibited higher viability and lower apoptosis rate, protein levels of p-JAK2 and p-STAT3 were significantly up-regulated, and the EMT levels were increased. In contrast, after the cells were co-treated with WP1066 on the basis of co-culture with SPP1+ TAMs, MC38 cell viability was decreased, and apoptosis was increased; the levels of JAK2, STAT3, and their phosphorylation were decreased, and the EMT process was inhibited. Therefore, SPP1+ TAMs promote CRC cell proliferation and EMT by activating JAK2/STAT3 signaling pathway.
Similar content being viewed by others
Introduction
Approximately 9.2% of deaths worldwide are associated with colorectal cancer (CRC), which ranks fourth among cancer-related causes of death1,2. Although the overall incidence of CRC has declined since the twenty first century, there is evidence suggesting that the population with CRC is becoming younger3. Cancer statistics for the United States in 2024 exhibit that although the overall mortality rate for CRC has declined significantly, CRC still ranks first and second, respectively, as a cause of death for men and women under 50 years of age4. Furthermore, the International Agency for Research on Cancer (IARC) estimates that CRC incidence and mortality are expected to double by 20245, making CRC a major public health issue for all of society.
CRC is a type of cancer with metastatic properties. The 5-year survival rate for patients with primary CRC is about 65%, but for patients with metastatic CRC, it is only about 14%6,7. Immune escape is an important way to facilitate tumor proliferation and metastasis, involving cell-tumor cell interactions in the tumor microenvironment (TME). TME is a complex network of cells containing tumor cells, stromal cells, and immune cells, as well as extracellular matrix and signaling molecules, which can dynamically adjust to the metabolic and energetic demands of neighboring tumor cells8,9. Studies have indicated that TME can increase systemic inflammation and oxidative stress and promote CRC progression by participating in the activation and recruitment of immune cells10,11. In addition, cytokines (including IL-1β, IL-6, and TNF-α) in TME have been recognized as key mediators of cancer malignancy12,13. However, there is still a gap in research on the mechanisms of TME in CRC.
Tumor-associated macrophages (TAMs) are the most abundant component of the TME and can be recruited by a variety of chemokines and cytokine colony-stimulating factor 1 (CSF1)14. In CRC, TAMs have been found to promote tumor cell immune escape, trigger epithelial-mesenchymal transition (EMT), and enhance tumor cell growth and invasion15,16. Previous studies have shown that TAMs can be polarized into M1 or M2 types with different phenotypic characteristics, exhibiting pro-inflammatory and anti-inflammatory properties, respectively17,18. A recently published study has redefined macrophage polarity through the expression of CXCL9 and SPP1 (CS)19. This finding supports the idea that the M1/M2 phenotype can define the extreme state of cells in vitro but does not necessarily reflect the complexity of TAMs in vivo19. Moreover, CS polarity has a more pronounced prognostic relevance compared to the M1/M2 polarization taxonomy, with SPP1+ TAMs being the key pro-carcinogenic and pro-metastatic subtypes associated with poor prognosis20,21.
After bibliographic retrieval, it is found that the current studies of macrophage (Mac) CS polarity in CRC are mainly conducted by single-cell and spatial transcriptomics analysis22,23. In this study, the heterogeneity of SPP1+ TAMs in CRC will be explored by single-cell RNA sequencing (scRNA-seq) analysis. Meanwhile, in vitro assays will be combined to verify the effects of SPP1+ TAMs on CRC malignant phenotype and explore molecular mechanisms of alterations associated with SPP1 over-expression in TAMs.
Materials and methods
Data sources
Single-cell data for CRC were obtained from the dataset GPL21697_GSE200997 of the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/), including 16 CRC samples and 7 normal samples.
scRNA-seq analysis
The GSE200997 dataset was analyzed using the R package Seurat (v4.1.0). The number of cell genes was 200–2000, the UMI number was 200–6000, and the mitochondrial gene expression rate of each cell was < 10% as the threshold value. Low-quality cells and low-expression genes were filtered out. The scRNA-seq dataset was normalized using the NormalizeData function, and 2000 highly variable genes were identified using the vst method in the FindVariableFeatures function. The data were normalized using the function ScaleData. The principal component analysis (PCA) was carried out by the RunPCA function, and the number of principal components (PCs) was selected by the visual plot of the ElbowPlot function. The drawing was visualized using the RunUMAP function, setting n.eighbors to 30 and dims to 1:30. Batch correction was performed through the R package Harmony. The shared nearest neighbor (SNN) graph was constructed with the first 30 PCs using the FindClusters function, and the cells were clustered by setting resolution = 0.3. Depending on the presence of known marker genes the clusters were grouped as: Endothelial cells (VWF and CD31+), fibroblasts (COL1A1, DCN, COL1A2 and ACTA2), NK cells (NKG7, GNLY and KLRD1), T cells (CD3D, CD3E and IL7R), epithelial cells (EPCAM, KRT8 and KRT18), myeloid cells (LYZ) and B cells. Differentially expressed genes (DEGs) between each cell type were identified using the FindAllMarkers function in Seurat, with min.pct = 0.1 and logfc.threshold = 0.25, retaining genes with a p-value < 0.05. Developmental trajectories of cell subsets were depicted using the R package Monocle2, and differences in enrichment scores between cell subsets were analyzed by gene set variation analysis (GSVA).
Induction of CRC mouse models by azoxymethane (AOM)/Dextran sodium sulfate (DSS)
Twenty-four C57BL/6J male mice aged 6–8 weeks, 18–22 g, were purchased from GemPharmatech Co. Ltd, with free access to diet and water. The mice were divided into the Control group and the Model group, with 12 mice in each group. Mice in the Control group were fed normally. Mice in the Model group were intraperitoneally injected with AOM(10 mg/kg) on day 0, and fed with drinking water for 7 days. The drinking water was replaced with a 2.5% DSS aqueous solution, after which it was switched back to regular drinking water and continued for an additional 14 days. The feed was fed for 3 consecutive cycles according to this method. During this period, the survival and weight changes of mice were observed and recorded every 7 days. After the modeling was completed, pentobarbital sodium was used for anesthesia. Then, the mice were sacrificed by cervical dislocation, the intestines were removed and the tumor tissues were isolated.
The animal experiments were approved by the Ethical Committee of Ningbo University Experimental Animal Center (Ethics approval number: 13938). All methods were carried out in accordance with relevant guidelines and regulations, and all methods are reported in accordance with ARRIVE guidelines.
Hematoxylin-eosin (H&E) staining
Tumor tissues and normal tissues were fixed in a 4% paraformaldehyde solution for paraffin embedding. Sections were dried, dewaxed, and hydrated. Subsequently, the sections were stained with a hematoxylin staining solution for 3–5 min. After washing with water, the sections were stained with an eosin staining solution for 5 min. Finally, the sections were dehydrated and sealed. Images were observed under a microscope and photographed for recording.
Isolation and identification of Macs
The extracted mouse tumor tissues or paracancerous tissues were washed twice with 1640 medium and filtered through sterile nylon mesh. After centrifugation, cells were resuspended in 90% RPMI-l640 + 10% FBS medium containing GM-CSF (10ng/ml) and inoculated in culture dishes at a cell density of 1 × 106 cells/ml to culture for 3 days. Macs were digested and centrifuged to obtain the Mac precipitate, and the cells were resuspended by adding Buffer. Next, 5 µl each of CD11b (101211, Biolegend, USA) and F4/80 antibody (123115, Biolegend, USA) were added to the cell suspension, mixed well, and reacted for 5–15 min at room temperature away from light. Subsequently, SPP1 antibody was added. After 5 min of reaction, the cells were washed twice with PBS and centrifuged. Finally, the cells were resuspended with PBS, and the cell suspension was transferred into a flow tube for identification on the machine.
Cell culture and grouping
MC38 cells (C7399, Beyotime) were cultured in RPMI-1640 medium containing 10% FBS and 1% penicillin-streptomycin mixture and placed in an incubator at 37 °C with 5% CO2. The cells were divided into the following six groups: Control group, where MC38 cells were cultured in RPMI-1640 medium alone; Negative or Positive group, where MC38 cells were cultured in SPP1− TAMs or SPP1+ TAMs conditioned medium, respectively; DMSO group and WP1066 group, DMSO or JAK/STAT3 pathway inhibitor WP1066 (10 ng/ml, HY-15312, MCE) were added to MC38 cell medium, respectively. The medium was replaced with the original medium after 48 h of incubation; Pos + WP1066 group, WP1066-treated MC38 cells were cultured in SPP1+ TAMs conditioned medium.
Co-culture of TAMs and MC38 cells
SPP1− TAMs or SPP1+ TAMs were placed in a Mac complete medium and incubated for 48 h at 37 °C in an incubator with 5% CO2. The collected supernatant was centrifuged again and then taken as Mac conditioned medium. Next, 5 × 106 freshly adhered MC38 cells were taken, and the conditioned medium was diluted using RPMI-1640 medium at a ratio of 1:1. The TAMs and MC38 cells were co-cultured in an incubator at 37 °C, 5% CO2. After 72 h, the original medium was aspirated and discarded, and the cells were washed with PBS and digested with 0.25% trypsin. Finally, the cells were assayed according to the experimental requirements.
Cell counting Kit-8 (CCK-8) assay
Cell viability was detected using the CCK-8 kit (C0037, Beyotime). MC38 cells were inoculated into 96-well plates at 2000 cells per well, and the medium was added to a total volume of 100 µL in the wells. After the cells were attached to the wall, 10 µL of CCK-8 solution was added to each well, and the optical density (OD) values were detected using an enzyme marker at 450 nm at 0, 24, 48, and 72 h after culture, respectively.
Western blot (WB)
Cells were collected and washed with PBS. After cell lysis, the lysates were quantified using a BCA protein assay kit. The lysates were loaded and separated on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to membranes. Subsequently, the membranes were closed with 5% bovine serum albumin (BSA) for 1 h and incubated with primary antibodies overnight. After washing, the membranes were incubated with horseradish peroxidase (HRP)-labeled secondary antibody (A0208, Beyotime) for 1 h to detect protein bands. The following primary antibodies were used: JAK2 (AF1489, Beyotime), p-JAK2 (AF1486, Beyotime), STAT3 (AF1492, Beyotime), p-STAT3 (AF1276, Beyotime), and β-actin (AF5003, Beyotime).
Immunofluorescence
Tissues were fixed with 4% paraformaldehyde after rinsing with pre-cooled PBS. After dehydration with 30% sucrose, the tissues were transferred to OCT for embedding and then sectioned to 10 μm. The sections were then dried and immersed in a citric acid repair solution. Subsequently, the sections were blocked with PBS blocking buffer containing 0.3% TritonX-100 for 1 h at room temperature and dropwise added with SPP1 (ab218237, Abcam) primary antibody for overnight incubation at 4 °C. On the next day, the sections were rinsed with PBS and incubated with fluorescent secondary antibody (A0288, Beyotime) for 1 h at room temperature away from light. Next, the sections were washed with PBS and re-stained by dropwise adding DAPI staining solution to seal the sheet. Finally, the images were observed under a confocal microscope.
Cells were fixed at 4 °C and permeabilized with 0.1% Triton X-100. Next, the cells were incubated with a blocking buffer for 1 h and then incubated with the primary antibody at 4 °C overnight. After washing with PBS, the cells were incubated for 1 h with the appropriate biotinylated secondary antibodies (A0286, Beyotime). Subsequently, the cells were incubated for 1 h with triple antibodies Alexa Fluor 488 (Molecular Probes, 1:500) and Alexa Fluor 594 goat anti-mouse (Molecular Probes, 1:500). Finally, the cells were restained with DAPI for coverslip preparation, and the images were analyzed via a confocal microscope. The following primary antibodies were used: anti-E-cadherin (ab231303, Abcam), N-cadherin (ab98952, Abcam), and Vimentin (ab8978, Abcam).
TdT-mediated dUTP nick-end labeling (TUNEL) assay
MC38 cell apoptosis was detected using the TUNEL apoptosis detection kit. Cells were inoculated into 6-well plates at a density of 2 × 105 cells/well and incubated overnight. TUNEL staining was performed according to the protocols, and the nuclei were stained with DAPI for 5 minutes at room temperature. The cells were observed and recorded under a fluorescence microscope.
Statistical analysis
All analyses were performed using GraphPad Prism 6.0. Data were presented as mean ± SD. Comparisons between two groups were conducted using the unpaired t-test, and comparisons among multiple groups were conducted using the one-way ANOVA test. P < 0.05 was considered statistically significant.
Results
CRC cell annotation
According to the corresponding cell type marker genes, the clusters obtained from clustering were annotated into Epithelial cells (Ephs), T cells, B cells, Fibroblasts (Fibs), NK cells, Endothelial cells (ECs), Mast cells, and Myeloid cells (Fig. 1A,B). The UMAP plots presented the distribution of different cell types in the Normal and CRC groups (Fig. 1C). Although overall Ephs, NK cells, Fibs, and Myeloid cells were somewhat different between the CRC and Normal groups, there was a significant increase in the proportion of Myeloid cells and Fibs when combined with the individual CRC samples, suggesting that there was strong heterogeneity between the groups for these two cell types (Fig. 1D,E). Finally, the present study compared the differences in the proportions of Myeloid cells and Fibs in the microsatellite stability (MSS) group with those in the high-frequency microsatellite instability (MSI-H) group, and found that both types of cells were present at relatively high proportions in the MSS group (Fig. 1F).
scRNA-seq analysis of the TME in CRC. (A) Cells in the CRC group were annotated using marker genes; (B) UMAP plot of each cell type; (C) UMAP plots of cell-type distribution in the CRC and Normal groups; (D) Bar stacking plot of the proportion of each cell type in each sample; (E) Proportion of each cell type between the CRC and Normal groups; (F) The relationship between the proportion of Fibs and Myeloid cells and MSS.
Myeloid cell subset identification
Myeloid cells are the major cellular component of the body’s innate immune activity, including Macs (and monocyte precursors) and dendritic cells (DCs). According to the corresponding cell type marker genes, the clusters obtained from clustering were annotated into four cell subsets: SPP1+ Mac, MRC1+ Mac, S100A8+ Mono, and LAMP3+ DC (Fig. 2A,B). The UMAP plots and bar graph demonstrated the distribution of different cell subsets in the Normal and CRC groups, with a significantly higher proportion of SPP1+ Mac in the CRC group (Fig. 2C,D). Subsequently, this study compared the IFNγ scores, immune evasion-related gene scores, and immune checkpoint scores strongly correlated with ICI response among various cell subsets (Fig. 2E–G), and constructed the pathway for differentiation from monocytes to macrophages. SPP1+ Mac gradually expanded along the trajectory, MRC1+ Mac differentiated uniformly with the trajectory from the starting point, LAMP3+ DCs were distributed on both branches, and S100A8+ Mono diverged along the trajectory from the starting point (Fig. 2H,I). In addition, we also analyzed the correlation between SPP1 + Mac cells and clinical characteristics such as age, TMB (tumor mutational burden), and tumor stages (stages) (Fig. S1), and constructed a survival analysis based on the SPP1 + MAC-specific gene feature signature (Fig. S2). Finally, SPP1+ TAM-related signaling pathways were identified using GSVA. The GSVA plots showed that pathways such as JAK/STAT3 signaling, IL2/STAT5 signaling, and WNT/β-catenin signaling were enriched to varying degrees in each cell subset (Fig. 2J). Among them, the JAK/STAT3 signal was significantly activated in SPP1+ TAMs but silenced in SPP1− TAMS (Fig. 2K). Based on this result, this study selects the JAK/STAT3 signal as the key discussion object.
Subset clustering and enrichment analysis of Myeloid cells. (A) Myeloid cell subsets were annotated using marker genes; (B) UMAP plot of each cell subset; (C) UMAP plots of each cell subset between the CRC and Normal groups; (D) Proportion of each cell subset between the CRC and Normal groups; (E) IFNγ scores among various cell subsets; (F) Immune escape scores among various cell subsets; (G) Immune checkpoint scores among various cell subsets; (H,I) Trajectory plots of each cell subset; (J) GSVA results of each cell subset; and (K) GSVA results of SPP1+ TAMs and SPP− TAMs.
There were a large number of SPP1+ TAMs in mouse CRC tissues
In this study, mouse CRC models were constructed using the AOM/DSS method. With the advance of modeling time, the body weight of mice in the Control group maintained a steady increase, and the DAI score was relatively stable. However, with the change of DSS feeding cycle, the body weight of mice in the Model group decreased first and then increased, and the DAI score increased first and then decreased (Fig. 3A,B). In this study, the tumor induction rate of AOM/DSS was 100%. H&E staining revealed that the intestinal structure of mice in the Control group was normal, and no inflammatory cell infiltration was observed. The overall structure of the intestinal tissue of the Model group mice was abnormal, with intramucosal neoplasia. Local recess structure necrosis and disappearance were observed in the mucous lamina propria. The recess spacing was increased, and inflammatory cell infiltration was observed in the interstitium (Fig. 3C–E). q-PCR results showed that SPP1 expression level was higher in the Model group than in the Control group (Fig. 3F). Ki-67 staining showed a significant increase in cell proliferation in the Model group compared to the Control group (Fig. 3G). Moreover, the results of multiple immunofluorescence staining showed that SPP1 was significantly highly expressed in the Model group, and was mainly localized in macrophages (Fig. 3H). The number of total Macs in the Normal and Model groups was detected by flow analysis. SPP1− TAMs and SPP1+ TAMs were isolated using the SPP1 antibody. The results showed that most of the macrophages isolated from the Control group were SPP1− TAMS, while most of the macrophages isolated from the Model group were SPP1+ TAMs (Fig. 3I).
Flow sorting of SPP1+ TAMs in CRC model mice. Mice in the Control group were fed normally, and CRC was induced in the Model group by the AOM/DSS method. After the modeling, the mice were euthanized. The intestines were removed, and the tumor tissues were separated for follow-up experiments. (A) Body weight changes of mice in the Control and Model groups during modeling; (B) DAI changes of mice in Control and Model groups during modeling; (C) H&E staining to detect histopathological changes in colorectal tissues of mice in the Normal and Model groups, red arrows indicate inflammatory cell infiltration; (D) Pathological scores of H&E staining, score: 0- No abnormality; 1- Minor changes, such as abnormal crypt lesions; 2- Highly atypical hyperplasia 3- Intramucosal carcinoma; (E) Immune cell infiltration was stained with H&E; (F) q-PCR to detect the mRNA levels of SPP1 in the Control group and Model group; (G) Ki-67 staining to detect cell proliferation in the Control group and Model group; (H) The expression of SPP1 in Control and Model groups were detected by immunofluorescence and were targeted by the macrophage marker F4/80 and the epithelial marker E-cadherin; and (I) Flow cytometry to identify the content of SPP1− TAMs or SPP1+ TAMs in the Normal and Model groups.
SPP1+ TAMs promoted MC38 cell proliferation, invasion, and migration
The ELISA results showed that the expression of SPP1 in the Positive group increased compared with the Ctrl group and the Negative group (Fig. 4A). Compared with SSP1− TAMS, the cell viability and migration ability of SSP1+ TAMs were significantly stronger (Fig. 4B,C). In vitro assays showed that the Control group and the Negative group maintained similar cell proliferation rate, while the proliferative activity of MC38 cells in the Positive group was significantly increased (Fig. 4D). TUNEL staining showed that co-culture with SPP1+ TAMs did significantly inhibit MC38 cell apoptosis, while co-culture with SPP1− TAMs had little effect on the number of apoptotic MC38 cells (Fig. 4E). Immunofluorescence detection showed that the expression levels of E-cadherin were higher in Control group and Negative group, while the expression levels of N-cadherin and Vimentin were lower. The expression level of E-cadherin was lower in the Positive group, while the expression levels of N-cadherin and Vimentin were higher (Fig. 4F).
In vitro assays to validate the promoting effect of SPP1+ TAMs on the malignant phenotype of MC38 cells. MC38 cells in the Ctrl group were cultured with normal medium, and MC38 cells were cultured with SPP1+ TAMs or SPP1− TAMS conditional medium, which were named the Positive group and the Negative group, respectively, and then tested after moderate culture. (A) ELISA to detect the protein level of SPP1; (B) CCK-8 assay to detect the cell viability of SSP1+ TAMs and SSP1− TAMS; (C) CCK-8 assay to detect MC38 cell viability; (D) Transwell experiment tested the migration ability of SSP1+ TAMs and SSP1− Tams; (E) TUNEL staining to detect the apoptosis rate of MC38 cells; (F) Immunofluorescence to detect the expression levels of EMT-related molecules (E-cadherin, N-cadherin, Fibronectin) in MC38 cells.
SPP1+ TAMs promoted CRC by activating JAK/STAT3 signaling pathway
To verify the relationship between SPP1+ TAMs and JAK/STAT3 signaling pathway, JAK2, STAT3, and their phosphorylation levels were detected by WB in this study. The results pointed out that there was no difference in the total protein levels of JAK2 and STAT3 in the Positive group compared with the Control and Negative groups, but the phosphorylation levels of JAK2 and STAT3 were significantly increased (Fig. 5A,B). As shown in Fig. 5C, the DMSO group had the fastest cell proliferation rate, the WP1066 group had the slowest cell proliferation rate, and the Pos + WP1066 group had the middle cell proliferation rate. TUNEL staining showed that compared with the DMSO group, the apoptosis rate of the WP1066 group was significantly increased. Compared with the WP1066 group, the apoptosis rate of the Pos + WP1066 group was decreased (Fig. 5D). Immunofluorescence results showed that the expression levels of E-cadherin in the DMSO group were lower, while the expression levels of N-cadherin and Vimentin were higher. The expression levels of E-cadherin were higher in the WP1066 group and the Pos + WP1066 group, while the expression levels of N-cadherin and Vimentin were lower (Fig. 5E–G). After the inhibitor treatment, the levels of JAK2, STAT3, and their phosphorylation were significantly reduced in both the WP1066 and Pos + WP1066 groups compared with the DMSO group (Fig. 5H,I). In addition, it was found that SPP1+ TAMs failed to promote CRC after inhibiting JAK/STAT3 signaling pathway.
In vitro assays to verify the relationship between SPP1+ TAMs and JAK/STAT3 signaling pathway. The group descriptions of Ctrl, Positive, and Negative are the same as in Fig. 4. The DMSO group added DMSO to the medium to culture MC38 cells as a control. WP1066 group added WP1066 in medium; In the Pos + WP14066 group, WP1066-treated MC38 cells were cultured in SPP1+ TAMs conditioned medium. (A,B) WB detection of JAK/STAT3 pathway protein expression levels in the Control, Negative, and Positive groups. Compared with the Control group, ****P < 0.0001, and compared with the negative group, ####P < 0.0001; (C) CCK-8 assay to detect MC38 cell viability in each group; (D) TUNEL staining to detect the apoptosis rate of MC38 cells in each group; (E–G) Immunofluorescence to detect the expression of EMT-related molecules (E-cadherin, N-cadherin, fibronectin) in each group; H–I) WB detection of JAK/STAT3 pathway proteins in the DMSO, WP1066, and Pos + WP1066 groups, Compared with the DMSO group, ****P < 0.0001.
Discussion
SPP1 is a multifunctional protein involved in the regulation of cancer-associated signaling that promotes malignant phenotypes and treatment resistance in cancer cells24. Previous studies have shown that SPP1 is highly expressed in multiple malignant tumors and correlates with prognosis and immune infiltration25,26,27. With the concept of macrophage CS polarity, a number of studies have linked SPP1+ TAMs to tumor progression and outcome. scRNA-seq analysis has shown that SPP1+ TAMs can promote tumor progression28. There is evidence that SPP1+ TAMs interact with cancer-associated fibroblasts (CAF) in CRC, resulting in poorer patient survival and immunotherapy outcomes22,29. In addition, CRC cells located at the invasive front enhance the anti-tumor immunity and invasion of CRC by inducing leukocyte antigen G (HLA-G) to produce SPP1+ TAMs23. In this study, the heterogeneity of SPP1+ TAMs in the Normal and CRC groups was verified by scRNA-seq analysis, and on this basis, cellular assays were performed. The present study co-cultured SPP1+ TAMs or SPP1− TAMs with MC38 cells and found that SPP1+ TAMs can significantly facilitate MC38 cell proliferation, invasion, and migration and inhibit apoptosis to promote EMT.
The GSVA results demonstrated a reliable signaling pathway: JAK/STAT3 signaling pathway. It has been reported that JAK/STAT3 signaling is involved in the regulation of cell growth, immune function, and other physiological processes. Furthermore, its aberrant expression has been shown to be associated with cancer development and progression. For example, in malignant cancers such as glioma and ovarian cancer, STAT3 is overexpressed and associated with poor prognosis30,31. There is evidence that aberrant activation of JAK/STAT3 signaling pathway can promote EMT, proliferation, invasion, and metastasis in CRC cells32,33. SPP1 has been demonstrated to enhance radiotherapy tolerance in esophageal cancer by rapidly activating JAK2/STAT3 signaling pathway, thereby maintaining esophageal cancer progression34. This finding has revealed the role of SPP1 in regulating the downstream JAK/STAT3 signaling pathway during tumor progression.
Based on the above conclusions and evidence, this study conducted in vitro experiments to verify the relationship between SPP1 and JAK2/STAT3 signaling pathway in CRC. Firstly, WB was used to detect the expression levels of JAK2 and STAT2 in MC38 cells under different culture conditions. The results showed that the expression levels of p-JAK2 and p-STAT3 in the Positive group were significantly increased compared with the Negative group. This indicates that SPP1+ TAMs can activate the JAK2/STAT3 signaling pathway. Subsequently, MC38 cells with or without WP1066 treatment were co-cultured with SPP1+ TAMs. It was found that compared with the DMSO group, the total protein and phosphorylation levels of JAK2 and STAT3 were significantly reduced in the WP1066 group and the Pos + WP1066 group. Our study also found that compared with the DMSO group, the cell proliferation rate was decreased, and the apoptosis rate was increased in the WP1066 group. But there was no significant difference in these results between the WP1066 group and the Pos + WP1066 group. The results suggested that SPP1+ TAMs can promote CRC progression by activating JAK/STAT3 signaling pathway.
Furthermore, patients with advanced CRC still maintain a high chance of metastasis, which is associated with a poor prognosis35. It has been determined that EMT can increase cancer cell motility and invasion, a process essential for promoting tumor metastasis36. EMT is regulated by many mechanisms, including the activation of genes and signaling pathways37. Previously, it has been shown that activation of JAK/STAT3 signaling pathway in CRC cells promotes the expression of EMT markers and tumor migration and invasion38,39,40. There is also evidence that SPP1+ TAMs can promote metastasis of CRC cells41. Therefore, the present study examined the expression levels of EMT markers in MC38 cells using immunofluorescence. It was found that SPP1+ TAMs significantly decreased E-cadherin expression and up-regulated N-cadherin and Fibronectin expression in MC38 cells. This result highlighted that SPP1+ TAMs can promote EMT in MC38 cells. The study later discovered that the expression levels of E-cadherin, N-cadherin, and Vimentin in MC38 cells remained unchanged following treatment with the inhibitor WP1066, regardless of whether the cells were co-cultured with SPP1+ TAMs or not. Therefore, it can be inferred that SPP1+ TAMs promote the EMT in CRC cells by activating JAK2/STAT3 signaling pathway. However, to better translate from basic research to clinical applications, it is essential to explore the similarities and differences between human and mouse SPP1+ TAMs. In both human and mouse CRC, SPP1+ TAMs show significant infiltration in the TME. They are recruited to the tumor site and interact with tumor cells, stromal cells, and other immune cells. Furthermore, they play a pro-tumor role and are associated with poor prognosis. However, they differ in their cell phenotypes and gene expression profiles, which may lead to different responses in cell function. Moreover, limitations of existing techniques have led to relatively little understanding of the origin and differentiation of SPP1+ TAMs in humans. Additionally, there may be some regulatory pathways that are different from those in mice.
In summary, this study has revealed the heterogeneity of SPP1+ TAMs in the Normal and CRC groups and verified the relationship between SPP1+ TAMs and CRC progression and their activation of JAK2/STAT3 signaling pathway by in vitro assays. Indeed, this study demonstrates that SPP1+ TAMs can promote CRC cell proliferation and EMT by activating JAK2/STAT3 signaling pathway through in vitro assays. However, there are still several limitations. First, the trend of apoptosis in Fig. 4B was not consistent with the theory. After the interference of some external factors was excluded through repeated experiments, this study speculated that the lack of SPP1 in the conditioned medium of SPP1− TAMS might lead to the activation of other apoptosis-related signaling pathways, resulting in the increase of apoptosis rate in the Negative group. But its specific mechanism still needs to be further explored. In future studies, the role of JAK2/STAT3 signaling pathway as a bridge between SPP1+ TAMs and CRC cells should be further clarified. Moreover, it is essential to investigate whether other cell types and molecular mechanisms affect SPP1 expression in normal and tumor tissues. In addition, the regulatory effect of SPP1+ TAMs on JAK2/STAT3 signaling pathway and its effect on CRC metastasis should be further investigated through in vivo experiments. Previous studies on CXCL9+ TAMs, which contrast with SPP1+ TAMs, have provided valuable insights. This cell phenotype is often associated with favorable clinical outcomes. In future studies, we will further explore the dominant differences between SPP1+ TAMs and CXCL9+ TAMs in CRC, as well as the mechanism of this phenotypic difference on CRC outcomes.
Data availability
Single-cell data for CRC were obtained from the dataset GPL21697\_GSE200997 of the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/).
References
Shivappa, N. et al. Dietary inflammatory index and colorectal cancer risk—A meta-analysis. Nutrients 9 (9), 1043 (2017).
Dekker, E., Tanis, P. J., Vleugels, J. L., Kasi, P. M. & Wallace, M. B. Colorectal cancer. Lancet 394 (10207), 1467–1480 (2019).
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2018. Cancer J. Clin. 68 (1), 7–30 (2018).
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. Cancer J. Clin. 74 (1), 12–49 (2024).
Morgan, E. et al. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 72 (2), 338–344 (2023).
Siegel, R. L. et al. Colorectal cancer statistics, 2017. CA: Cancer J. Clin. 67(3), 177–193 (2017).
Rumpold, H. et al. Prediction of mortality in metastatic colorectal cancer in a real-life population: A multicenter explorative analysis. BMC Cancer. 20, 1–9 (2020).
Duan, Q., Zhang, H., Zheng, J. & Zhang, L. Turning cold into hot: Firing up the tumor microenvironment. Trends Cancer. 6 (7), 605–618 (2020).
Nenkov, M., Ma, Y., Gaßler, N. & Chen, Y. Metabolic reprogramming of colorectal cancer cells and the microenvironment: Implication for therapy. Int. J. Mol. Sci. 22 (12), 6262 (2021).
Huot, J. R., Novinger, L. J., Pin, F. & Bonetto, A. HCT116 colorectal liver metastases exacerbate muscle wasting in a mouse model for the study of colorectal cancer cachexia. Dis. Models Mech. 13 (1), dmm043166 (2020).
Lima, J. D. et al. Tumour‐derived transforming growth factor‐β signalling contributes to fibrosis in patients with cancer cachexia. J. Cachexia Sarcopenia Muscle. 10 (5), 1045–1059 (2019).
Voronov, E. & Apte, R. N. Targeting the tumor microenvironment by intervention in interleukin-1 biology. Curr. Pharm. Design. 23 (32), 4893–4905 (2017).
Webster, J. M., Kempen, L. J., Hardy, R. S. & Langen, R. C. Inflammation and skeletal muscle wasting during cachexia. Front. Physiol. 11, 597675 (2020).
Chen, C. et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat. Commun. 9 (1), 3826 (2018).
Wu, Y. et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discov. 12 (1), 134–153 (2022).
Shin, A. E., Giancotti, F. G. & Rustgi, A. K. Metastatic colorectal cancer: Mechanisms and emerging therapeutics. Trends Pharmacol. Sci. 44 (4), 222–236 (2023).
Cassetta, L. & Pollard, J. W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discovery. 17 (12), 887–904 (2018).
Kasprzak, A. The role of tumor microenvironment cells in colorectal cancer (CRC) cachexia. Int. J. Mol. Sci. 22 (4), 1565 (2021).
Bill, R. et al. CXCL9: SPP1 macrophage Polarity identifies a network of cellular programs that control human cancers. Science 381 (6657), 515–524 (2023).
Tang, W. et al. Revealing the role of SPP1 + macrophages in glioma prognosis and therapeutic targeting by investigating tumor-associated macrophage landscape in grade 2 and 3 gliomas. Cell. Bioscience. 14 (1), 37 (2024).
Yang, C-L. et al. Integrating single-cell and bulk RNA sequencing reveals CK19 + cancer stem cells and their specific SPP1 + tumor-associated macrophage niche in HBV-related hepatocellular carcinoma. Hep. Intl. 18 (1), 73–90 (2024).
Qi, J. et al. Single-cell and Spatial analysis reveal interaction of FAP + fibroblasts and SPP1 + macrophages in colorectal cancer. Nat. Commun. 13 (1), 1742 (2022).
Ozato, Y. et al. Spatial and single-cell transcriptomics Decipher the cellular environment containing HLA-G + cancer cells and SPP1 + macrophages in colorectal cancer. Cell. Rep. 42 (1), 111929 (2023).
Nallasamy, P. et al. Pancreatic tumor microenvironment factor promotes cancer stemness via SPP1–CD44 axis. Gastroenterology 161 (6), 1998–2013 (2021). e7.
Wang, X. et al. Secreted phosphoprotein 1 (SPP1) contributes to second-generation EGFR tyrosine kinase inhibitor resistance in non-small cell lung cancer. Oncol. Res. 27 (8), 871 (2019).
Gao, W. et al. SPP1 is a prognostic related biomarker and correlated with tumor-infiltrating immune cells in ovarian cancer. BMC Cancer. 22 (1), 1367 (2022).
Deng, G. et al. BET inhibitor suppresses melanoma progression via the noncanonical NF-κB/SPP1 pathway. Theranostics 10 (25), 11428 (2020).
Liu, Y., Zhang, L., Ju, X., Wang, S. & Qie, J. Single-cell transcriptomic analysis reveals macrophage–tumor crosstalk in hepatocellular carcinoma. Front. Immunol. 13, 955390 (2022).
Sathe, A. et al. Colorectal cancer metastases in the liver establish immunosuppressive spatial networking between tumor-associated SPP1 + macrophages and fibroblasts. Clin. Cancer Res. 29 (1), 244–260 (2023).
Liang, B. et al. Clinicopathological and prognostic roles of STAT3 and its phosphorylation in glioma. Disease Markers 2020, (2020).
Yoshikawa, T. et al. JAK2/STAT3 pathway as a therapeutic target in ovarian cancers. Oncol. Lett. 15 (4), 5772–5780 (2018).
Pennel, K. A. et al. JAK/STAT3 represents a therapeutic target for colorectal cancer patients with stromal-rich tumors. J. Exp. Clin. Cancer Res. 43 (1), 64 (2024).
Wang, F. et al. Overexpression of GSTP1 promotes colorectal cancer cell proliferation, invasion and metastasis by up-regulating STAT3. (2021).
Wang, M., Sun, X., Xin, H., Wen, Z. & Cheng, Y. SPP1 promotes radiation resistance through JAK2/STAT3 pathway in esophageal carcinoma. Cancer Med. 11 (23), 4526–4543 (2022).
Bertocchi, A. et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer cell. 39 (5), 708–724 (2021). e11.
Brabletz, T., Kalluri, R., Nieto, M. A. & Weinberg, R. A. EMT in cancer. Nat. Rev. Cancer. 18 (2), 128–134 (2018).
Tang, Q. et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J. Exp.Clin. Cancer Res. 39, 1–17 (2020).
Lee, Y. I. et al. WNT5A drives interleukin-6-dependent epithelial–mesenchymal transition via the JAK/STAT pathway in keloid pathogenesis. Burns Trauma. 10, tkac023 (2022).
Shen, M. et al. Inhibition of ATM reverses EMT and decreases metastatic potential of cisplatin-resistant lung cancer cells through JAK/STAT3/PD-L1 pathway. J. Exp. Clin. Cancer Res. 38, 1–14 (2019).
Xue, J. et al. LncRNA AB073614 induces epithelial-mesenchymal transition of colorectal cancer cells via regulating the JAK/STAT3 pathway. Cancer Biomarkers. 21 (4), 849–858 (2018).
Liu, X. et al. ANGPTL2 + cancer-associated fibroblasts and SPP1 + macrophages are metastasis accelerators of colorectal cancer. Front. Immunol. 14, 1185208 (2023).
Acknowledgements
Not applicable.
Funding
This research received no specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Shaohui Yang contributed to the study concepts, study design, and definition of intellectual content; Chenyang Ma contributed to the literature research the experimental studies and data acquisition; Shaohui Yang, Chenyang Ma and Yibin Zhao contributed to the manuscript editing and review. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent to participate
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, S., Ma, C. & Zhao, Y. SPP1+ macrophages promote colorectal cancer progression by activating JAK2/STAT3 signaling pathway. Sci Rep 15, 37502 (2025). https://doi.org/10.1038/s41598-025-21420-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-21420-9