Abstract
The medical community demands accurate predictive models for early heart disease diagnosis because heart disease remains a significant worldwide health concern. Deep learning research presents a predictive system for heart disease that uses K-mode clustering to optimize data preparation and implements Harris Hawks Optimization (HHO) for essential feature selection. The Cardiovascular Disease dataset of 70,000 patient records with many clinical parameters was used to develop model training and validation. The accuracy of cardiovascular disease prediction depends on neural networks and other deep learning architectures which analyze patient risk factors to determine disease development. The model operates efficiently using precision, recall, accuracy and the AUC score to evaluate its performance while utilizing the ROC curve. The developed feature selection method applying HHO increases model efficiency while maintaining prediction capabilities by eliminating unneeded features. The Gated Recurrent Unit (GRU) model achieved the highest accuracy of 88.03% among all the tested frameworks. Deep Learning methodologies integrated with advanced feature selection demonstrate high effectiveness in early detection of cardiovascular disease. It leads to efficient diagnostic solutions for health applications that scale across various systems.
Similar content being viewed by others
Introduction
Coronary heart disease (CHD) itself is one of the major global health threats by being responsible for 17.9 million yearly deaths, which constitute 32% of worldwide fatalities. Quarter of all Indian fatalities stem from cardiovascular diseases at present while cardiac problems are increasing among youth age groups1. More frequent occurrences of congenital heart conditions demonstrate that urgent identification and proper medical treatment are essential2. The main cause of CHD arises from atherosclerosis, which leads to blocked blood flow through arteries because of plaque accumulation, thereby creating severe medical complications3. Smoking, together with obesity, hypertension, diabetes, high cholesterol levels and leading a sedentary lifestyle, multiplicatively increases the chances of developing CHD. The critical aspect of sustained surveillance together with preventive measures, results from research showing both hereditary background and persistent inflammation foster CHD evolution4.
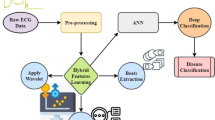
A deep learning methodology serves as the foundation for heart disease prediction through effective accuracy and operational efficiency enhancement5. The framework combines clustering and optimisation functions to optimise the feature selection procedure. K Modes clustering enables preprocessing functions where the method detects viral patterns within categorical values to enhance input feature representation. Applying the Harris Hawks Optimization (HHO) algorithm based on Harris Hawks hunting behaviour optimises feature selection within the proposed framework. The approach creates a harmonious connection between exploration and exploitation processes so that it removes unimportant attributes while preserving key attributes. The improved medical dataset supports model training to extract hidden complex patterns from medical data, thus enhancing heart disease prediction accuracy6.
A model evaluation uses the Cardiovascular Disease Dataset to train and validate its performance using 70,0000 clinical patient records. The model achieves population-wide applicability because of its access to extensive health information, including patient age, blood pressure readings, and cholesterol numbers. BMI values, glucose test results and personal lifestyle choices7. Deep learning methods with a special focus on neural networks perform precise risk assessments when they investigate the multiple variables present in the data. Feature selection strategies in advanced stages enhance the dataset by identifying the most clinically essential characteristics for improving model prediction accuracy. Applying deep learning together with an extensive dataset simultaneously improves efficiency levels and scalability, which helps to facilitate disease recognition and appropriate medical assistance8.
Key performance measures of evaluation, such as precision, accuracy, recall, area under the curve (AUC) measure, and the receiver operating characteristic (ROC) curve, make for a comprehensive performance of predictive model evaluation. These indicators provide essential information regarding how well the model differentiates CHD-positive subjects from CHD-negative subjects with limited false predictions9. The Harris Hawks Optimization (HHO) implementation in experimental tests proves successful because it identifies and removes surplus features while maintaining prediction accuracy. The model uses vital clinical variables to improve its ability to generalise and its computational effectiveness for medical implementations. According to the research findings, deep learning combined with advanced feature selection techniques yields more successful and prompt cardiovascular disease detection while enabling prompt medical response10.
The dataset employed in this work is significantly larger (70,000 records) than traditional datasets like the Cleveland and Hungarian Heart Disease datasets, which typically comprise only 200–1000 samples. This increases the model’s generalizability by guaranteeing that it is trained on a varied population. Figure 1 depicts the graphical abstract of this work. The contribution of this work is in the integration of feature engineering with K-mode clustering, a type of clustering that is optimised for categorical data. This combination improves feature representation and enables the extraction of significant patterns within the dataset.
-
The researchers evaluate basic predictive models by combining them with Harris Hawks Optimization (HHO) techniques for advanced feature selection to develop an improved selected feature set.
-
The research incorporates K-Modes clustering for categorical data while performing feature engineering, distinguishing this study from others. Such a combination delivers improved feature representation that identifies essential patterns in the dataset.
-
The study performs an extensive statistical and qualitative review of the dataset, which identifies critical coronary heart disease risk factors. The research evaluates the network effects between different clinical characteristics and their role in producing accurate predictions.
-
The study applies deep learning approaches to detect advanced nonlinear patterns between risk variables, thus producing more dependable and accurate predictive results. Such accuracy surpasses traditional statistical approaches because they fail to identify complex patterns.
Literature survey
The healthcare sector increasingly requires automated diagnostic systems as data science and medical technology advance. Deep learning methods and machine learning are now critical components for advancing medical diagnosis since they assist in predicting coronary heart disease (CHD). The technical sector faces significant challenges when attempting to achieve high accuracy levels for all detections.
Several research studies have compared heart disease classification with several machine learning algorithms. Shorewala11 has researched ensemble learning for the identification of CHD. The study could not achieve 90% accuracy with ensemble methods with the identification that model dependability should be achieved. Sinha et al.12 created iCardo as an intelligent healthcare system that utilizes machine learning to identify cardiovascular disease. The system unveiled new approaches, but its real accuracy rate was below 90% since real-time healthcare identification remains an issue. Arooj et al.13 employed deep convolutional neural networks to identify successful instances of early heart disease. The positive outcome of their solution was unable to achieve a 90% accuracy benchmark and prove the necessity of better feature selection techniques or hybrid models to be developed. Heart disease prediction has been performed using various machine-learning techniques.
Authors of14 proposed an AI-powered heart disease diagnosis system that utilised machine learning techniques. The model’s accuracy could not be even 98%, which establishes that conventional machine learning techniques must be further developed. Boukhatem et al.15 explored this approach but encountered accuracy limitations that highlighted the need for improved optimization methods to enhance performance. As proposed by Dutta et al.16, a convolutional neural network (CNN) is proposed to predict CHD. Although this model was encouraging, deep learning accuracy in this application was only 77%, which secured the boundaries of deep learning.
In the study of Alotaibi et al.17, the performance of different algorithmic techniques and data mining algorithms for predicting heart disease is compared to the task. According to their research findings, traditional classification techniques could not identify complex medical data patterns with low accuracy rates. Cenitta et al.18 developed Hybrid Residual Attention-Enhanced LSTM (HRAE-LSTM) model to predict ischemic heart disease, whereby on UCI they have reported 97.71% and 97.41% accuracy and sensitivity respectively. The method performed better than the conventional ML and baseline deep learning models, with potential in early diagnosis and clinical support. Nevertheless, this study has been found to have a limitation of the data variety, readability, and validation in larger populations. The authors19 performed the prediction of heart attacks using nature-inspired feature selection with explainable AI (XAI) with the use of 1,763 patient records and 12 clinical features. Mutual Information (90% accuracy, 94% precision) was the top performer (out of 14 algorithms) with such classifiers as Random Forest, XGBoost, and LightGBM. Five XAI techniques (SHAP, LIME, Eli5, QLattice, Anchors) have pointed to the important risk items of ST slope, Oldpeak, type of chest pain, and exercise induced angina. The results show that XAI-based feature selection based on optimization is better at predictive performance and interpretability than standard feature selection methods, making it useful in clinical decision-making.
Santhosh et al.20 presented a model of heart disease prediction named CARDIACX, which was evaluated on an Indian hospital data set. The best performance was demonstrated by the Random Forest that was optimized with the help of the Grid Search, the Random Search, and the Bayesian Optimization (98.5% accuracy, AUC 0.99). CARDIACX made interpretable predictions based on XAI (SHAP, LIME, QLattice, Anchor), but needs to be validated on larger and more diverse datasets.
The study, conducted by Abdulwahab Ali Almazroi et al.21, aims to improve the diagnosis of heart disease by leveraging a Keras-based deep learning model with a dense neural network. It demonstrated that the proposed deep learning model achieves superior accuracy, sensitivity, and specificity compared to individual models and alternative ensemble approaches.
For feature selection and extraction of HRV features, Ritu Aggarwal et al.22 targeted clinical participants with congestive heart failure (CHF) and normal sinus rhythm (NSR). By utilising several different feature selection techniques, it is found that the HRV features were the most effective. The feature selection methods are used to reduce complexity of the system. From the experimental results it is evident that the filtering method is the most effective approach for feature selection. The filtering approach achieves a 95.40% accuracy rate and the average accuracy rate without Feature selection is 86.35%.
According to Lili Jiang et al.23, the random forest and XGBoost models performed better in terms of recall and accuracy, whereas stepwise logistic regression and SVM produced more great AUC values. A prediction-based approach using K-fold cross-validation along with scaled conjugate gradient backpropagation artificial neural networks was proposed by Banibrata Paul et al.24. The proposed method results in a maximum accuracy rate in comparison to various alternatives.
Table 1 presents a comparison of the work conducted by various authors. However, the research studies demonstrate that machine learning and deep learning methods can significantly augment heart disease prediction, but there are still accuracy problems. These issues need to be overcome by future research studies that address mixture modelling and sophisticated feature engineering methods with improved methods. Employing cutting-edge data processing reinforcement learning techniques and deep learning techniques would result in more precise heart disease diagnosis systems.
Proposed methodology
Dataset description
The Cardiovascular Disease dataset30 on Kaggle comprises 70,000 patient records and is widely used for heart disease prediction and analysis. It has 11 features and one target variable. The dataset was unstructured using a comma-separated values (CSV) file format with separate column records. The dataset is clean, with no missing values and each variable is classified as either discrete or numerical.
Data preprocessing
Two crucial preprocessing issues arose in the preparation phase. Since they had high standard deviations, certain continuous variables had extreme measurement values and possible global anomalies. The analysis performed value trimming on all the features, showing notable deviations that excluded both top and bottom two percent values. There were various errors in the dataset since measurements showed systolic blood pressure levels lower than diastolic blood pressure when such a physiological condition is improbable. Data reliability improved due to the exclusion of incorrect data entries that assisted in generating improved model performance31. The dataset relationships are evident through the visualisation of variable relationships presented in Fig. 2. The heatmap analysis facilitates patterns and feature relationship identification that aid in identifying features for possible deletion to improve dataset quality and predictability. The normalisation process normalised all non-categorical numerical variables into a range of values from 0 to 1. The target variable with data related to coronary heart disease was given a balanced distribution so that analysts could conduct their analysis without making any weighting adjustments. Preprocessing resulted in the evolution of continuous variables, as depicted in Fig. 3.
Feature analysis
Pearson’s correlation coefficient helped analyse feature relationships with the target variable while producing a heatmap. The examination involved grouping data points for determining cluster performance. An investigation of age and systolic blood pressure in relation to the target variable allowed a better comprehension of its distribution pattern32.
The data visualisation in Fig. 4 shows two principal continuous variables, which demonstrated significant Pearson correlation values. The analysis presents these variables in relation to cholesterol, using a random sample of 400 data points to avoid confirmation bias. Patients with coronary heart disease had elevated total cholesterol levels combined with increased blood pressure qualities. Age did not demonstrate a distinct pattern with the target variable when the clustering approach was implemented.
Due to its classification nature, we selected clustering algorithms that would help us investigate patterns in data—our K-Modes analysis groups data points with comparable relations to our target dimension. The positions of centroids were determined by experts for the continuous data set in which one graph depicts cluster results with respect to centroids, and the other represents the main target. The chart in Fig. 4 illustrates the performance of the data analysis method in relation to blood pressure values. We signify the performance of the data analysis method with blood pressure values. We calculated these values to find centroid locations using the formula described below.
X(dataset)={x1, x2, x3,…, xc} V (cluster set) ={v1, v2, v3,., vn} Σ c i = 1 Σ ci j = 1 ||xi – vj||^2.
Where:
-
‘ci’ symbol represents the count of elements in cluster i.
-
‘c’ represents the count of cluster centre found in data.
‘||xi - vj||’ represents the distance between point xi and vj as per the Euclidean system.
Figure 5 shows that clustering processing rendered input blood pressure values more valuable due to their high degree of statistical association uncovered by HHO. The findings suggest that patients suffering from coronary heart disease are primarily concentrated in clusters 5, 6, and 7. Clustered tests using age, height, and weight data showed infinitesimal yet valuable ways of structuring the patients based on coronary heart disease diagnosis. We built the most ideal mathematical models for our five continuous variables. The trend between age was a straightforward straight line but other variables followed a polynomial or curved pattern. The data shows that logistic regression would be ideal in classifying data.
We tested the impact of all variables on the target response through Z-score calculation and their respective p-values. The process of evaluation was effective for logistic regression models but did not show how to assess clustering-based models. Data analysis showed gender had minimal impact in the logistic model because its Z-value was 0.655 and p-values came out at 0.512. The available variables had a strong impact on our model, with Z-scores ranging from 4.21 to 60.68.
Feature selection using Harris Hawks optimization (HHO)
For the identification of better features and enhancing the quality of the model, the HHO algorithm was used. It follows a nature inspired approach which emulates group behaviour of Harris Hawks seeking the solutions. It searches for better features using exploration-exploitation movements that do not fall into bad solutions and chooses better subsets of features. HHO employs a sequence of initial solution sets to achieve its mission and subsequently refines these solutions dynamically. During global search hawks select target locations randomly but once they move to the next phase, they employ alternative attack modes such as soft or hard siege to narrow their options. HHO excels in identifying prominent features since its dynamic approach allows it to make fine decisions and reduces results for improved classification performance33,34. Using the HHO technique allowed us to identify the most significant feature to employ in the identification of cardiovascular disease. By removing redundant features, the filter improved both the interpretability and runtime efficiency of the model. We trained our system and further optimised it based on the selected final features. Algorithm 1 elaborates the steps in the HHO algorithm.
Oversampling using SMOTE
SMOTE generated synthetic samples by employing intermediate points of the original data to balance imbalanced data following feature selection. The initial balance of our target variable was broken after we conducted additional filtering operations during feature selection. SMOTE compensates for data imbalance by generating new instances of the minority class from points that make underrepresented classes more prominent.
The SMOTE method chooses minority class instances and determines their nearest neighbours to create new samples on the line connecting them. The method creates natural new samples within the existing feature space instead of copying existing observations. SMOTE balanced the smaller data set that optimised the prediction errors and improved the classification35. The HHO method helped identify the ideal features. SMOTE was then enabled to restore imbalanced data so that deep learning models for predicting cardiovascular disease could be trained.
The suggested system is depicted in Fig. 6, which combines data preprocessing, clustering (K-Modes), feature selection (Harris Hawks Optimization), SMOTE-based data balancing, and a recurrent neural network for classification. Accuracy, precision, recall, F1-score, AUC, and ROC curve are then used to evaluate the model.
Experimental setup and results
The feature selection process is proceeded with Harris Hawks Optimization (HHO). Feature selection is an essential step to eliminate unimportant features that exert pressure on the classification accuracy. The HHO algorithm performed an iterative search for maximally accurate features that would optimise the selected feature set. Features to be kept were cholesterol, smoke, age group, BMI, MAP, and clusters due to their rated importance sources as illustrated in Fig. 7.
Different deep learning models employed to the pre-processed dataset for training to determine their optimal performance capabilities36. The classification task revealed negligible importance of alcohol, smoking, and glucose features they were eliminated from the data analysis based on HHO results. We partitioned the data into training (80%) on test (20%) subsets to determine metrics performance. The dataset was treated with randomisation treatment before its application during training to optimise generalisation and reduce overfitting effects. Different evaluation metrics such as accuracy, precision, recall, F1-score and AUC score were used to determine model performance. The development was performed with python programming language consisting of tensor flow framework for the implementation of DL models.
DL models
Different DL models were developed for coronary disease diagnosis, depending on the evaluation process. The models employed are given below along with deep learning architecture. Figure 8 depicts the generalized deep learning architecture.
MLP
Artificial neural network called Multilayer Perceptron is used to solve complex nonlinear problem with great efficiency. Every neuron of the hidden layers calculated weighted inputs via an addition method and the applies the output via activation functions like SELU and ReLU but in Hyperbolic Tangent form. With this, the AdamW optimiser and binary cross entropy loss is used to train an MLP model. A model’s performance is highly dependent on some basic configurations for the hidden layers, learning rate and batch size37. For the current study we created a two-layer MLP neural network consisting of 128 and 64 units and applied dropout 0.2 and trained the model by iterating through 32 batches of learning rate 0.001.
A Multilayer Perceptron model performed sophisticated nonlinear data pattern recognition from the provider data set and activation function model called SELU improved learning ability in addition to the application of dropout layers to avoid overfitting and improve generalisation. The learning rate parameter of 0.001 was used in AdamW optimiser for optimisation from the training. The MLP performed well in binary classification because it had several hidden layers that enabled it to identify complex patterns. It was trained on 200 epochs with a batch size of 32 to achieve an 87.64% accuracy demonstration that it delivered excellent classification outcomes.
-
Input Layer: 128 neurons, activation = ‘selu’, (input dimension = 6)
-
Dense Layer: 64 neurons, activation = ‘selu’.
-
Dropout: 0.2.
-
Dense Output Layer: 1 neuron with a ‘sigmoid’ activation function.
-
Optimiser: AdamW optimiser with a learning rate of 0.001.
-
Loss Function: Utilises the Binary cross-entropy function.
-
Batch Size: Training performed with a batch size of 32.
-
epochs: 200.
-
Validation Split: 20% (train-test split: 80 − 20).
CNN
The image processing community heavily uses Convolutional Neural Networks (CNNs) while these networks demonstrate equivalent ability to classify structured data. The neural network architecture contains convolutional feature extractors followed by max-pooling reducers and complete connection classifiers. The study incorporated dropout for regularisation with a single convolutional layer and a dropout set to 0.3 while using AdamW optimiser alongside binary cross-entropy loss38,39.
This study, 1D CNN was used to discover interesting patterns within well-structured medical data for disease classification. AdamW as a main optimiser technique was adopted for stable and effective training. To reduce features over a one-dimensional layer, we have a Conv1D layer of 32 filters of kernel size 3 and a MaxPooling1D layer to achieve that. Dropout layers with a dropout rate of 0.3 were incorporated to mitigate overfitting. The model achieved an accuracy of 86.14%, showing it can learn complex feature interactions and improve the classification accuracy.
-
Input Layer: Conv1D (32 filters, kernel_size = 3, activation=’selu’).
-
MaxPooling Layer: Applied pooling with a size of 2 to reduce dimensionality.
-
Flatten Layer.
-
Dense Layer: A fully connected layer with 64 neurons and ‘selu’ as the activation function.
-
Dropout: 0.3.
-
Dense Output Layer: 1 neuron with a ‘sigmoid’ activation function.
-
Optimiser: AdamW optimiser.
-
Loss Function: Utilises the Binary cross-entropy function.
-
Batch Size: Training performed with a batch size of 64.
-
epochs: 200.
-
Validation Split: 20% (train-test split: 80 − 20).
LSTM
To process sequential data, Recurrent Neural Networks (RNNs)40 were designed with internal memory states which stores past information. It turns out, regular RNNs struggle with long term dependency retention which is due to the vanishing gradient problem. LSTM network connects this issue using gated system controls that regulate memory storage and data oblivion. The LSTM model, configured with stacked layers of 50 and 25 units respectively, improved pattern recognition in time-series data. The model used AdamW as its optimiser to achieve stable training since it facilitated effective convergence. The model was trained for 200 epochs with 64 samples per batch to achieve an accuracy of 87.91% for sequence-based pattern understanding.
-
Input Layer: LSTM (50 units, return_sequences = True, activation=’selu’).
-
LSTM Layer: 25 units, activation=’selu’.
-
Dense Output Layer: 1 neuron, activation=’sigmoid’.
-
Optimiser: AdamW optimiser.
-
Loss Function: Utilises the Binary cross-entropy function.
-
Batch Size: Training performed with a batch size of 64.
-
epochs: 200.
-
Validation Split: 20% (train-test split: 80 − 20).
BiLSTM
BiLSTM builds upon regular LSTMs by processing information through the network from both past and future directions to boost context recognition abilities. The ability of BiLSTM to process information in both directions helps it excel at the job, where text requires understanding changes in the sequence41. The research used two directional LSTM layers (50 and 25 units) within a BiLSTM framework with the SELU activation function.
The model acquires enhanced abilities to identify intricate patterns in data through its bidirectional processing mechanism. The model employed SELU activation parameters to enhance its learning abilities with AdamW optimisation that provided an efficient and robust training process. Dropout layers were applied along with the model as a preventive measure against overfitting. BiLSTM model reached an accuracy of 87.78% through the combination of these methods dealing with sequential data. It demonstrated its ability to identify complex relations within the dataset.
-
Input Layer: Bidirectional LSTM (50 neurons, return_sequences = True activation=’SELU’).
-
Bidirectional LTM: 25 neurons, activation=’SELU’.
-
Dense Layer: 1 neuron with a ‘sigmoid’ activation function (Binary Classification Output).
-
Optimiser: AdamW optimiser with a Learning Rate of 0.001.
-
Loss Function: Utilises the Binary Cross-Entropy function.
-
Batch Size: 64.
-
epochs: 200.
GRU
The combination of the original input and forget gates forms an update gate, reducing the LSTM architecture to that of the GRU. The compact GRU architecture minimizes the need for computations without sacrificing its efficiency in order processing. The system suggested combines data preparation with K-Modes clustering and Harris Hawks Optimization for feature selection based on Fig. 6. SMOTE handles imbalanced data, and recurrent neural networks test the result. Accuracy testing, application of precision, recall, F1-score is performed, AUC score is calculated, and the ROC curve is represented to evaluate the model42.
GRUs are an enhanced version of RNNs with less complexity by virtue of their two gate operations that are the update gate and reset gate. The design cuts down on complexity while computing and still has the capability to recognise extended dependencies in sequential data. The study employed a GRU model with two stacked GRU layers that both incorporated batch normalisation and dropout layers for the enhancement of training stability with prevention from overfitting. A dense layer links all the inputs in serial order prior to the output layer to aid in feature extraction. The Adam optimiser worked at a 0.001 learning rate to facilitate effective weight updating that enabled efficiency in convergence. This model demonstrated good classification capabilities by achieving 88.03% accuracy and hence can be effectively used for medical setup diagnosis from sequential data.
-
GInput Layer: GRU (128 neurons. Return_sequences = True, activation=’RELU’).
-
Batch Normalisation, Dropout (0.3).
-
GRU Layer: 64 neurons, activation=’RELU’.
-
Batch Normalisation, Dropout (0.3).
-
Dense Layer: 128 neurons, activation=’RELU’.
-
Dropout (0.3).
-
Dense Output Layer: 1 neuron with a ‘sigmoid’ activation function.
-
Optimiser: Adam optimiser with a Learning Rate of 0.001.
Performance of DL models
This article evaluates the proposed heart disease diagnosis model using key performance metrics such as accuracy, precision, recall, F1-score, AUC Score represented in equation number 1 to 6.
Accuracy: Reflects the proportion of correct predictions out of all predictions
Precision: Represents the reliability of positive predictions.
Recall: Captures the ability to find all true positive cases.
F1-score: Combines precision and recall into a single effectiveness measure.
AUC score: Summarizes the model’s overall class-separation capability.
Plot ROC curve: TPR vs. FPR.
AUC = 1 (perfect), AUC = 0.5 (random guessing).
Where,
TP: The model accurately identifies a positive case.
FP: The model mistakenly labels a negative case as positive.
TN: The model correctly recognizes a negative case.
FN: The model incorrectly classifies a positive case as negative.
The evaluation of various deep learning models’ metrics is given in Tables 3 and 4. The evaluation indicates that all tested models achieved comparable success rates, averaging approximately 87.5%. The Gated Recurrent Unit (GRU) performed outstandingly by achieving a level of accuracy at 88.03% that outplaced other models.
The Bidirectional LSTM (BiLSTM) and the Recurrent Neural Network (RNN-LSTM) performed noteworthy results that closely resembled each other as their accuracy levels reached 87.78% and 87.91% respectively. The tests revealed that the Convolutional Neural Network (CNN) recorded the lowest accuracy at 86.14%, followed by the Multi-Layer Perceptron (MLP) with 87.64%, while the GRU model achieved the highest accuracy at 88.03%. The success of GRU architectures is clear from the results achieved. The high rate of accuracy of the GRU model reveals how it identifies patterns through a sequence better than other models used in the research. Deep Learning models show their capability for class differentiation via the Receiver Operating Characteristic (ROC) curve plot give in Fig. 9. Several deep learning classifiers have their accuracy percentage illustrated via a bar plot presented in Fig. 10. Every bar in the graphical figure represents a classification system that shows unique height values corresponding to their respective accuracy. The various classifiers employed in the research are plotted on the x-axis while the y-axis shows accuracy percentage in the assessment. Figures 11 and 12 show a comparison of all metrics incorporated in this research. A confusion matrix is a performance evaluation tool used to summarize how well a classification model performs on a set of test data. It provides a tabular representation of the predicted vs. actual outcomes. Figures 13, 14, 15, 16 and 17 show the confusion matrix for the Deep Learning Models. Table 5 shows accuracy comparison with other existing works. The accuracy comparison between the proposed and existing works is displayed in Fig. 18.
Ablation study
The ablation analysis in the article is undertaken implicitly by successive analysis of the effect of preprocessing, feature selection, and architecture assessment of model performance in classification. As a pre-processing, K-Modes clustering would be utilized to improve the representation of categorical data, and then it will be followed by Harris Hawks Optimization (HHO) to preselect features within the data, accounting that among those the most important features to be selected by the HHO are cholesterol, smoke, age group, BMI, and MAP, and that others are redundant, and so these are removed to make the data interpretable and computationally efficient. As a solution to the introduction of the class imbalance in the feature filtering process, the SMOTE technique is employed as a method to produce synthetic samples of the minority class. These steps are wiped out by comparing performances in different deep learning models, which include MLP, CNN, LSTM, BiLSTM, and GRU, with the latter overcoming the other models with the highest accuracy of 88.03%, testifying to the additive advantage of each module. This progression helps to underline that feature optimization, data balancing, and model selection are all helpful options to improve the prediction accuracy of heart diseases to a significant extent.
Discussion
The findings of this study illustrate how excellent medical application classification performance is achieved by deep learning models through the integration of the feature selection properties of the Harris Hawks Optimization (HHO) algorithm. Cross-platform applications are made more efficient through HHO since it discards unnecessary and less significant features to guide network learning towards crucial attributes. The algorithmic progress results in higher accuracy levels that at the same time reduce training time and enhance generalisation efficiency. The use of deep learning techniques coupled with proper feature selection methods allows researchers to develop enhanced analytics models that enhance standard workflows while possessing robust resistance to intricate medical information. Time-series medical data processing works best with sequential architectural techniques that involve Gated Recurrent Units (GRU) and RNN-LSTM.
These methods are successful because the understand how situation with time-based pattern recognition necessitate diagnostics such as cardiac irregularities and nervous disorders. This information is retained in system histories, allowing for the assessment of future scenarios, thereby improving the system’s effectiveness in distinguishing differences in medical documentation. There is a diagnostic benefit to health care through the ability of these techniques to detect involved patterns found within sequential health data. Convolutional Neural Networks (CNNs) perform well in both speed and accuracy. The primary advantage of CNNs in medical image analysis and structured data stems from their ability to leverage spatial structures and hierarchical features. The CNN’s automated feature learning process renders the networks superior to conventional methods in identifying information necessary to eliminate time-consuming manual data processing tasks and allow simplified data handling processes. Medical applications demand effective tools which render CNNs the best option for fast and accurate medical decision systems. The integration of deep models with optimal feature selection methods yields high improvements in classification performance due to the focused function of neural networks in modern medical analytics.
Advantages
-
The integration of K-Modes clustering with Harris Hawks Optimization (HHO) enables efficient handling of categorical medical features, reducing redundancy and enhancing interpretability compared to standalone HHO or conventional feature selection methods.
-
The proposed K-Modes–HHO–DL framework consistently outperforms baseline models in terms of accuracy, precision, recall, F1-score, and AUC.
-
By grouping categorical risk factors (e.g., chest pain type, fasting blood sugar, gender) through clustering before optimization, the selected features are more aligned with medical reasoning, which enhances trust and adoption by clinicians.
-
The framework is adaptable to other medical diagnostic tasks where categorical attributes dominate, showing potential for broader applications beyond heart disease.
Limitations
-
Experiments are limited to publicly available heart disease datasets, which may not fully represent heterogeneous clinical populations.
-
HHO introduces stochasticity that may lead to slight variations in results across runs.
-
The hybrid approach requires additional preprocessing (clustering + optimization), which increases training complexity compared to simpler models.
Threat to validation
The envisioned K-Modes–HHO–DL framework was tested on publicly available benchmark datasets for heart disease. Dataset-specific factors such as feature distributions, class imbalance, and missing values may reduce the performance. Residual bias cannot be eliminated even after applying conventional preprocessing and stratified sampling. We only use publicly available datasets for our experiments. The variety of clinical populations across geographies and generations may be harder to depict with them. That is why it is essential to validate the model on more diverse and multi-center datasets so that it can be applied to actual clinical situations.Usual measures of performance (F1-score, accuracy, precision, recall, area under the curve, etc.) are used in the assessment. these measures offer a strong technical evaluation. Interpretability and physician trust are also important factors in clinical acceptance. Further research with domain experts is required to evaluate medical usability.
Harris Hawks Optimisation (HHO) is a stochastic metaheuristic that may result in minor performance fluctuations across iterations. We alleviated this by performing several independent trials and presenting average outcomes. Repeatability across diverse computing systems may remain problematic in the absence of standardised parameter configurations.
Conclusion
The research shows that coronary heart disease prediction through deep learning becomes stringer when run in conjunction with Harris Hawks Optimization (HHO) algorithm for identifying the important features. The model’s performance is enhanced through HHO since it eliminates unnecessary medical indicators while preserving the most vital data, resulting in greater efficiency. Gated Recurrent Units scored 88.03% in accuracy, whereas RNN and Bidirectional Long Short-Term Memory recorded 87.91% and 87.78%, respectively, among the models under test. Multi-Layer Perceptron (MLP) was at 87.64%, and Convolution Neural Network (CNN) at 86.14%. The results from research studies confirm that deep learning is better than traditional classification methods in all medical diagnostic and early disease detection applications. The good model performance will also be examined through research that uses real clinical data to identify their capability to generalise to various health contexts.
Research groups can enhance their predictive strategies by carrying out model tests in multiple healthcare settings thereby improving their capability to deal with advanced medical data. The use of various deep learning structures put together to form hybrid models results in more accuracy and reliability of disease prediction. The findings have possible utility in the construction of expert automated diagnosis machinery, which advantages medical professionals through timely disease detection and personalised treatment plan formulation, resulting in better patient outcomes. The study can be further developed into future work by incorporating the Explainable AI (XAI) frameworks, including SHAP, LIME or counterfactual explanations to give more insight into how the model makes its predictions. The combination of such techniques will not only increase the trust in the clinic but will also demonstrate the role of individual characteristics in the assessment of the risks of the disease. This interpretability will help overcome the obstacle between black-box models and medical decision-making, allowing a more informed and reliable application in the real-world healthcare environment.
Data availability
The benchmark research materials used in this research can be downloaded from [https://www.kaggle.com/datasets/sulianova/cardiovascular-disease-dataset].
References
García-Ordás, M. T., Bayón-Gutiérrez, M., Benavides, C., Aveleira-Mata, J. & Benítez-Andrades, J. A. Heart disease risk prediction using deep learning techniques with feature augmentation. Multimedia Tools Appl. 82, 31759–31773 (2023).
Islam, M. M., Tania, T. N., Akter, S. & Shakib, K. H. An improved heart disease prediction using stacked ensemble method. In International Conference on Machine Intelligence and Emerging Technologies, 84–97Springer, Cham, (2022).
Sharma, S. & Parmar, M. Heart diseases prediction using deep learning neural network model. Int. J. Innovative Technol. Exploring Eng. 9, 2244–2248 (2020).
Ahmad, A. A. & Polat, H. Prediction of heart disease based on machine learning using jellyfish optimization algorithm. Diagnostics 13, 2392 (2023).
Tiwari, A., Chugh, A. & Sharma, A. Ensemble framework for cardiovascular disease prediction. Comput. Biol. Med. 146, 105624 (2022).
Al-Safi, H., Munilla, J. & Rahebi, J. Harris hawks optimization (HHO) algorithm based on artificial neural network for heart disease diagnosis. In IEEE International Conference on Mobile Networks and Wireless Communications (ICMNWC), 1–5 (IEEE, 2021)., 1–5 (IEEE, 2021). (2021).
Prasanna, K. S. & Challa, N. P. Deep bi-LSTM with binary Harris Hawkes algorithm-based heart risk level prediction. SN Comput. Sci. 5, 134 (2023).
Mehmood, A. et al. Prediction of heart disease using deep convolutional neural networks. Arab. J. Sci. Eng. 46, 3409–3422 (2021).
Muibideen, M. & Prasad, R. A fast algorithm for heart disease prediction using bayesian network model. ArXiv Preprint arXiv:2012.09429 (2020).
El-Sofany, H., Bouallegue, B. & El-Latif, Y. M. A. A proposed technique for predicting heart disease using machine learning algorithms and an explainable AI method. Sci. Rep. 14, 23277 (2024).
Shorewala, V. Early detection of coronary heart disease using ensemble techniques. Inf. Med. Unlocked. 26, 100655 (2021).
Sinha, N., Jangid, T., Joshi, A. M., Mohanty, S. P. & iCARDO A machine learning based smart healthcare framework for cardiovascular disease prediction. ArXiv Preprint arXiv:2212.08022 (2022).
Arooj, S., Rehman, S. U., Imran, A., Almuhaimeed, A. & Alzahrani, A. K. Alzahrani, A. A deep convolutional neural network for the early detection of heart disease. Biomedicines 10, 2796 (2022).
Chang, V., Bhavani, V. R., Xu, A. Q. & Hossain, M. A. An artificial intelligence model for heart disease detection using machine learning algorithms. Healthc. Analytics. 2, 100016 (2022).
Boukhatem, C., Youssef, H. Y. & Nassif, A. B. IEEE,. Heart disease prediction using machine learning. In 2022 Advances in Science and Engineering Technology International Conferences (ASET), 1–6 (2022).
Dutta, A., Batabyal, T., Basu, M. & Acton, S. T. An efficient convolutional neural network for coronary heart disease prediction. Expert Syst. Appl. 159, 113408 (2020).
Alotaibi, N. & Alzahrani, M. Comparative analysis of machine learning algorithms and data mining techniques for predicting the existence of heart disease. Int. J. Adv. Comput. Sci. Appl. 13, 810 (2022).
Cenitta, D. et al. Ischemic heart disease prognosis: a hybrid residual attention-enhanced LSTM model. IEEE Access. 13, 4281–4289 (2025).
Bairy, M., Chadaga, K., Sampathila, N., Arjunan, R. V. & Bairy, G. M. An explainable analytical approach to heart attack detection using biomarkers and nature-inspired algorithms. Healthcare Analytics 8, Article 100407 (2025).
Santhosh, S., Chadaga, K., Arjunan, R. V. & D’Souza, S. Cardiac clarity: Harnessing machine learning for accurate heart-disease prediction. IEEE Access. 13, 97529–97544 (2025).
Almazroi, A. A., Aldhahri, E. A., Bashir, S. & Ashfaq, S. A clinical decision support system for heart disease prediction using deep learning. IEEE Access. 11, 61646–61659 (2023).
Aggarwal, R. & Kumar, S. HRV based feature selection for congestive heart failure and normal sinus rhythm for meticulous presaging of heart disease using machine learning. Measurement: Sens. 24, 100573 (2022).
Jiang, L., Chen, S., Wu, Y., Zhou, D. & Duan, L. Prediction of coronary heart disease in gout patients using machine learning models. Math. Biosci. Eng. 20, 4574–4591 (2022).
Paul, B. & Karn, B. Heart disease prediction using scaled conjugate gradient backpropagation of artificial neural network. Soft. Comput. 27, 6687–6702 (2023).
Alanazi, S. M. & Khamis, G. S. M. Optimizing machine learning classifiers for enhanced cardiovascular disease prediction. Eng. Technol. Appl. Sci. Res. 14, 12911–12917 (2024).
Dhaka, P., Sehrawat, R. & Bhutani, P. An innovative approach to cardiovascular disease prediction: A hybrid deep learning model. Eng. Technol. Appl. Sci. Res. 13, 12396–12403 (2023).
Bhatt, C. M., Patel, P., Ghetia, T. & Mazzeo, P. L. Effective heart disease prediction using machine learning techniques. Algorithms 16, 88 (2023).
Al-Alshaikh, H. A. et al. Comprehensive evaluation and performance analysis of machine learning in heart disease prediction. Sci. Rep. 14, 7819 (2024).
Muhammad, G. et al. O. Enhancing prognosis accuracy for ischemic cardiovascular disease using K nearest neighbor algorithm: A robust approach. IEEE Access. 11, 97879–97895 (2023).
Sulianova, D. Cardiovascular disease dataset. Kaggle (2020). Available at: https://www.kaggle.com/datasets/sulianova/cardiovascular-disease-dataset
Arunachalam, S. K. & Rekha, R. A novel approach for cardiovascular disease prediction using machine learning algorithms. Concurrency Computation: Pract. Experience. 34, e7027 (2022).
Rabbi, M. S. H. et al. Performance evaluation of optimal ensemble learning approaches with PCA and LDA-based feature extraction for heart disease prediction. Biomed. Signal Process. Control. 101, 107138 (2025).
Peng, L., Cai, Z., Heidari, A. A., Zhang, L. & Chen, H. Hierarchical Harris Hawks optimizer for feature selection. J. Adv. Res. 53, 261–278 (2023).
Elgamal, Z. M., Yasin, N. B. M., Tubishat, M., Alswaitti, M. & Mirjalili, S. An improved Harris Hawks optimization algorithm with simulated annealing for feature selection in the medical field. IEEE Access. 8, 186638–186652 (2020).
Yang, J. & Guan, J. A heart disease prediction model based on feature optimization and SMOTE-XGBoost algorithm. Information 13, 475 (2022).
Xia, B., Innab, N., Kandasamy, V., Ahmadian, A. & Ferrara, M. Intelligent cardiovascular disease diagnosis using deep learning techniques. Sci. Rep. 14, 71932 (2024).
Deepika, D. & Balaji, N. Effective heart disease prediction using novel MLP-EBMDA approach. Biomed. Signal Process. Control. 72, 103318 (2022).
Hussain, S. et al. Novel deep learning architecture for predicting heart disease using CNN. In 2021 19th OITS International Conference on Information Technology (OCIT), 353–357 (IEEE, 2021).
Phoemsuk, A. & Abolghasemi, V. Coronary artery disease classification using one-dimensional convolutional neural network. In IEEE Conference on Artificial Intelligence (CAI), 389–394 (IEEE, 2024)., 389–394 (IEEE, 2024). (2024).
Krishnan, S., Magalingam, P. & Ibrahim, R. Hybrid deep learning model using recurrent neural network and gated recurrent unit for heart disease prediction. Int. J. Electr. Comput. Eng. 11, 5467–5476 (2021).
Hossain, M. M. et al. Cardiovascular disease identification using a hybrid CNN-LSTM model with explainable AI. Inf. Med. Unlocked. 42, 101370 (2023).
Pal, P. & Mahadevappa, M. Prediction of therapy for ischemic heart disease from PPG signals using fuzzy GRU network. Biomed. Signal Process. Control. 87, 105409 (2024).
Tu, M. C., Shin, D. & Shin, D. Effective diagnosis of heart disease through bagging approach. In 2nd International Conference on Biomedical Engineering and Informatics, Tianjin, China, 1–4 (2009). (2009).
Agbemade, E. Predicting Heart Disease Using tree-based Model (Data Science and Data Mining, University of Central Florida, 2023).
Chen, A. H., Huang, S. Y., Hong, P. S., Cheng, C. H. & Lin, E. J. HDPS: Heart disease prediction system. In Computing in Cardiology, 557–560IEEE, (2011).
Funding
No funding was received to assist with the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
The contributions for this research article are as follows: Formal analysis, Conceptualization - D.S., Algorithm implementation - N.M., Methodology and Draft preparation - L.B., Validation and visualization - I.S., Supervision, and project administration - M.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This material is the authors’ own original work, which has not been previously published elsewhere. The paper reflects the authors’ own research and analysis in a truthful and complete manner.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
S, I., B, L., M, N.N. et al. Harris Hawks optimization based deep learning models for heart disease diagnosis. Sci Rep 15, 38395 (2025). https://doi.org/10.1038/s41598-025-22326-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-22326-2