Abstract
Entamoeba (E.) histolytica is a major global health concern as the causative agent of amoebic colitis, yet research is hindered by the lack of an ideal animal model. While E. muris has been traditionally considered non-pathogenic, our study demonstrates that it induces a colitis-like pathology in BALB/c mice, making it a promising model for studying E. histolytica infection. Using a fecal-oral infection route, we confirmed successful E. muris colonization, leading to colonic inflammation characterized by early CD4 + T-cell infiltration and FOXP3 + regulatory T-cell recruitment. We further demonstrate that T-cell immunity is essential for E. muris colonization, as T-cell-depleted BALB/c mice and T-cell-deficient BALB/c nude mice failed to support persistent infection. Additionally, infection disrupts epithelial homeostasis, impairing goblet cell generation, and induces significant shifts in gut microbiota composition, notably reducing beneficial Firmicutes species. Collectively, these findings demonstrate that E. muris infection induces inflammation and microbiota alterations, mirroring critical aspects of amoebic colitis, reinforcing its value as a model for studying E. histolytica pathogenesis and host–parasite interactions.
Similar content being viewed by others
Introduction
Entamoeba is a protozoan parasite that infects humans and animals, including reptiles, birds, amphibians, and others1. This parasite maintains its life cycle within the host and is transmitted via the fecal–oral route1, contaminating the environment2. Among the Entamoeba species, Entamoeba histolytica (E. histolytica) is a zoonotic agent that causes amoebic colitis and dysentery. 3. While most cases are asymptomatic or mild, approximately 10 to 20% of individuals infected with E. histolytica are symptomatic, with variations in severity4. Severe cases present with abdominal pain, bloody stools, and fever4. As such, E. histolytica infection constitutes one of the major etiologies of severe diarrhea worldwide and can be life-threatening, particularly in children under two years old in developing countries5. Despite its clinical significance, the mechanisms by which E. histolytica disrupts intestinal mucosal homeostasis remain incompletely understood3.
Several animal models have been developed, but major limitations persist. In mice, E. histolytica infection rarely establishes persistent colonization in commonly used strains such as C57BL/6 or BALB/c, which rapidly clear trophozoites without lesion formation6. Although strains like CBA and C3H can develop chronic and invasive inflammation resembling severe cases of human amoebic colitis6, these models typically require surgical intracecal inoculation2. This procedure fails to reproduce the natural fecal–oral route of transmission2. In addition, maintaining virulent trophozoites under laboratory conditions is technically unstable and requires further refinement6. As an alternative, E. muris can establish infection via the natural fecal–oral route, but its associated colitis pathology has not yet been fully characterized2,7. Thus, there remains a need for a robust and reproducible model that recapitulates Entamoeba-induced colitis, particularly reflecting the consistent and clinically relevant pathological features observed in the majority of human cases.
E. histolytica disrupts host-cell integrity by adhering to the cell membrane and triggering apoptosis through elevated intracellular calcium levels. The increased calcium levels in host cells promote global protein tyrosine dephosphorylation and caspase-3 activation, ultimately leading to apoptotic cell death3,5. Furthermore, E. histolytica secretes a macrophage migration inhibitory factor that induces inflammatory responses and exacerbates tissue damage in the colonic mucosa. These inflammatory reactions promote tissue destruction and ulcer formation3. To better understand the pathogenesis of amebiasis caused by E. histolytica and the interactions between the amoeba and its host, pigs8, gerbils9, cats10, and mice have been employed as animal models for use to study amebiasis in laboratory settings6. However, the infection efficiency remains low, and this efficiency depends greatly on the condition of the amoeba maintained in the laboratory6. Moreover, the cyst stage of E. histolytica cannot be introduced in culture or rodent models, placing a significant limitation on research into this critical process4. Other Entamoeba species have been utilized to demonstrate the fecal–oral infection route and the complete life cycle of E. histolytica. In particular, the reptile-specific E. invadens has been used to generate infection models providing critical insights into developmental changes such as encystation11.
E. muris is considered an excellent model organism when seeking to explore the life cycle of Entamoeba in hosts. It may also be used for comparative studies investigating convergence and divergence among Entamoeba species at the structural and genetic levels2. In a recent E. muris infection model, Swiss Webster (SW) mice were orally administered E. muris, and the complete life cycle of Entamoeba was then studied2. The study suggested that E. muris may be helpful in advancing research on E. histolytica. However, it did not fully describe the inflammatory response seen in E. histolytica-induced colitis of human2. To build on that work and investigate the pathogenic effects of E. muris following the fecal–oral infection route, we developed a new E. muris infection model using BALB/c mice. These were orally administered E. muris isolated from feces, which resulted in the mice developing colitis, along with changes to their intestinal microbiome. This study demonstrates that E. muris infection in BALB/c mice can mimic inflammatory conditions, suggesting a valuable tool for studying E. histolytica infection.
Results
Establishment of a mouse model for amoebic colitis using E. muris infection
In our animal facilities, we tested the mice for the presence of E. muris through PCR screening for a specific region of the E. muris 18 S rRNA gene. Positive results were obtained in some C57BL/6J mice, prompting further isolation and confirmation of the organism. Cecal contents were collected from E. muris-infected mice and analyzed using two complementary methods (Fig. 1A). Under a microscopic examination, approximately 8–30-µm-sized cystic structures containing eight nuclei were identified12, similar to the morphological characteristics of E. muris cysts (Fig. 1A). Subsequently, PCR was performed to amplify a specific region of the E. muris 18 S ribosomal RNA, generating a product of the expected size (Fig. 1A). The PCR product was then subjected to Sanger sequencing, and the resulting sequence was aligned with publicly available genomic sequences of Entamoeba species (AB445018.1). The sequence showed a complete match, confirming the identity of the organism as E. muris (Fig. 1A and Fig. S1A).
Pathology of purified E. muris-infected colons in a BALB/c mouse model following the fecal-to-oral infection route. (A) Identification of E. muris. Fecal samples were smeared onto slides and examined under the microscope. PCR was performed with specific primers targeting the 18 S ribosomal RNA sequence of E. muris. PCR products were validated through Sanger sequencing. Scale bar: 10 μm. (B) Experimental scheme. E. muris was isolated from the cecal contents of E. muris-infected mice using a Ficoll gradient. To eliminate bacterial contamination, the isolated E. muris was treated with 0.1 N HCl. BALB/c mice were orally administered with 2 × 10⁴E. muris Infection was determined via quantitative PCR (qPCR). Feces collected at 7, 14, 21, 28, 35, and 45 dpi were also used for microbiome analysis. Necropsies were performed at 9 dpi and 45 dpi to investigate the consequences of E. muris infection. (C) The standard curve was generated using Ct values obtained from qPCR with pure E. muris samples, following the equation Y = −0.8521*X + 25.37, R2 = 0.8962. (D) A graph represents the colonization rate of E. muris at weekly intervals post-infection, determined through fecal examination. (E) E. muris examination results in the cecum. Graph showing the number of E. muris detected in the cecal contents of infected mice after smear preparation and counting of cecal samples (ns, p = 0.0620). Black represents 9 dpi, and red represents 45 dpi. (F) A scoring system was used to evaluate the extent of inflammatory cell infiltration in colon tissues, ranging from 0 (none) to 3 (severe). (G) Colonic inflammation score assessed by the scoring system. Statistical significance was determined using t-tests: ***P < 0.001. (H) Eosinophil infiltration in colon tissues was quantified with manual cell counting. *P < 0.01, ***P < 0.001. (I) Representative images of H&E staining of colon tissues. Bottom panels: magnified images of boxed areas (Top panels). Scale bar: 200 μm–50 μm for enlarged. (J) Schematic diagram of colonic inflammation lesions. The diagram shows the individuals with lesions detected at specific locations in the colon of non-infected and infected groups (9 dpi, 45 dpi). Black represents no infection, red represents 9 dpi, and blue represents 45 dpi mice.
Isolated E. muris cysts were purified to remove bacterial contamination, ensuring infection with only E. muris. Each sample was first subjected to Ficoll isolation to separate and collect the E. muris cyst layer, followed by treatment with 0.1 N HCl to remove bacterial contaminants (Fig. 1B). E. muris preparation was streaked onto LB agar plates and incubated for 48 h at 37 °C under aerobic and anaerobic conditions. No bacterial growth was observed under either condition, providing strong evidence that the preparation was free of viable bacterial contamination (Fig. S1A). While LB agar may not support the growth of highly fastidious organisms, it is a standard indicator medium for common contaminants and thus provides reasonable assurance of purity.
Subsequently, BALB/c mice were infected with the purified E. muris cysts, and their infection persisted in vivo, allowing for repeated purifications and reinfections to maintain the E. muris donor. In vitro, the cysts were cultured in the Balamuth medium, and excystation led to the emergence of trophozoite forms of E. muris, further demonstrating the viability of the cysts (Fig. S1C).
BALB/c mice are frequently used in parasitic infection studies due to their robust Th2-skewed immune response, which might be critical for understanding host–pathogen interactions in parasitic diseases13. Since E. muris infection at a dose of 1 × 10⁵ in SW mice has been reported to result in asymptomatic infection without a subsequent pathology2, we infected female BALB/c mice via oral gavage with a lower dose of 2 × 10⁴ E. muris cyst, purified from the cecum of infected donor mice using the method described above (Fig. 1B). E. muris genomic DNA (gDNA) from fecal samples was collected weekly during the infection period (Fig. 1B). Using a standard curve generated from purified E. muris gDNA, quantitative PCR (qPCR) analysis was performed to quantify the amounts of E. muris gDNA in the fecal samples (Fig. 1C and D). The results showed a gradual increase in the amount of E. muris gDNA over time (Fig. 1D), which suggests that the E. muris infection model was successfully established via the fecal-to-oral route. It was further confirmed by measuring E. muris cyst number in the cecal contents (Fig. 1E).
We then examined whether E. muris in our mouse model induced an inflammatory response, according to the histopathological criteria specifically established for this study (Fig. 1F). Colon tissues were prepared using the Swiss rolling method for comprehensive histopathological examinations. The evaluation revealed that E. muris infection can cause inflammatory cell infiltration of the colonic mucosa and submucosa at 9 and 45 dpi (Fig. 1G–I). While the uninfected mucosa showed a small number of lymphocytic infiltrations in the localized lamina propria, E. muris infection developed multifocal infiltrations of mixed types and increased inflammatory cell numbers (Fig. 1G–I). We observed that polymorphonuclear cells with eosinophilic granules, identical to the eosinophil morphology, multifocally infiltrated the lamina propria in the infected mucosa, indicating an eosinophil-associated inflammatory response characteristic of parasitic infections (Fig. 1I). This inflammatory response was predominantly and multifocally observed in the middle and distal colon (Fig. 1J). Even at the lower dose of 1 × 10⁴, similar inflammation was observed (Fig. S2A and S2B). These data suggest that our mouse model for E. muris infection with BALB/c mice has the potential to serve as a representative system for studying amoebic colitis.
E. muris infection induces distinct immune responses in the colon over time
We further sought to determine which immune cell types had infiltrated the colon. Total or specific types of immune cells were manually or automatically counted with QuPath, as described in the Material & method section (Fig. S3A). Quantification was performed across the entire colon strip to ensure representative and unbiased analysis. Staining for CD45 + total immune cells supported the histopathological scoring of inflammation after E. muris infection (Fig. 2A and B). Dendritic cells are recruited as antigen-presenting cells and induce the expansion of helper T cells in parasitic infections14, and we found that CD11c + cells and CD4 + helper T cells had greatly infiltrated E. muris-infected colons at the earlier time point (Fig. 2A and B). Meanwhile, F4/80 + macrophages and CD3 + T cells were recruited to the infected mucosa more slowly (Fig. 2A and B). Furthermore, immunohistochemical staining for FOXP3 confirmed that Tregs were slowly recruited to the mucosa after infection (Fig. 2A and B).
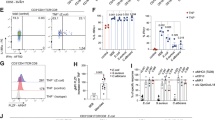
Immune cell composition in E. muris-infected colons. (A, B) Representative immunohistochemistry (IHC) images and quantification of CD45, CD11c, CD4, F4/80, CD3, and FOXP3-expressing cells in colon tissues of E. muris-infected at 9 dpi and 45 dpi and uninfected mice. Scale bar: 50 μm. *P < 0.05, **P < 0.01, and ***P < 0.001 (t-test). (C) Colonic inflammation score assessed at 14dpi by the scoring system. Statistical significance was determined using t-tests: ***P < 0.001(D) Flow cytometric analysis for quantifying the proportions of immune cell populations in the colonic lamina propria at 14 days post–E. muris infection. Shown are the proportions of CD3⁺ T cells, CD11c⁺ dendritic cells, F4/80⁺ macrophages, CD11b⁺ myeloid cells, CD19⁺ B cells, and Ly6G⁺ neutrophils among CD45⁺ cells in uninfected and infected BALB/c mice. Data represent mean ± SD; each dot represents an individual mouse. (E) qPCR analysis of colonic tissues from infected and uninfected mice at 14 dpi. Expression levels of Il4, Il5, Tnf, Il1b, Il17a, and Il10 were measured. *P < 0.05 (t-test).
At 14 days post-infection, when colonic inflammation was evident (Fig. 2C), we used flow cytometry to profile the proportions of immune cell subsets in the colonic lamina propria (Fig. 2D, Fig. S5A). Among CD45⁺ immune cells, CD3⁺ T cells showed the most significant increase in proportion, while F4/80⁺ macrophages, CD11c⁺ dendritic cells, and CD11b⁺ myeloid cells also exhibited significant increases consistent with histopathological findings (Fig. 2D, Fig. S5A). Considering our data and the fact that gastrointestinal parasitic infections are known to elicit diverse helper T-cell–mediated immune responses15, we analyzed the expression of cytokines associated with Th1, Th2, and Th17 responses, as well as Treg-related cytokines. For Th1-associated pro-inflammatory cytokines (Tnf, Il1b), no significant changes were observed, while the Treg-related immunosuppressive cytokine Il10 was significantly increased and the Th17-associated cytokine Il17a showed an increasing tendency (Fig. 2D). Among Th2 cytokines, Il5 displayed a decreasing trend with borderline statistical significance. Similar suppression of type-2 responses by Tregs has been reported in helminth infections, potentially contributing to parasite persistence16,17. These findings indicate that E. muris infection elicits a distinct host immune response characterized by Treg-associated immunomodulation.
E. muris colonization takes advantage of t-cell immunity
As shown in Fig. 2, E. muris infection induced distinct T-cell response. Therefore, we investigated the relationship between T-cell immunity and E. muris infection using a T-cell-deficient model (Fig. 3A). This model was established using BALB/c nude mice, which are genetically athymic and unable to develop mature, functional T cells18. E. muris was infected via the fecal–oral route in both BALB/c mice and BALB/c nude mice with identical genetic backgrounds (Fig. 3A). Throughout the experimental period, T-cell-deficient BALB/c nude mice infected with E. muris had significantly lower levels of E. muris gDNA in their feces than immunocompetent BALB/c mice (Fig. 3B). Furthermore, gDNA was not detected in the cecal contents from BALB/c mice depleted of CD4/CD8 + T cells using a CD4/CD8 + T-cell depletion antibody, nor was it detected in BALB/c nude mice (Fig. 3A and C). The successful depletion of CD4 and CD8⁺ T cells in this model was confirmed by IHC analysis of the spleen (Fig. S4A). Pearson correlation analysis revealed that the number of FOXP3 + cells had the strongest association with the inflammation score and also correlated with all measured innate immune cell populations. To more specifically identify Tregs, we performed CD3/FOXP3 double immunofluorescence staining and confirmed CD3⁺FOXP3⁺ Treg infiltration at 45 dpi (Fig. 3E and F). To test whether Tregs contribute to E. muris colonization, BALB/c mice were treated with an anti-CD25 antibody for Treg depletion, followed by infection with E. muris. The partial depletion of Treg cells in this model was confirmed by IHC analysis of the spleen (Fig. S4B). Anti-CD25 treatment resulted in a lower level of E. muris gDNA contents in the cecal samples compared to the control BALB/c group (Fig. 3G), although the reduction was less pronounced than that observed with CD4 and CD8 depletion, which nearly eliminated the infection. Similar phenomena have been reported in Echinococcus multilocularis and Taenia crassiceps, where Treg expansion suppresses anti-parasitic immune responses and facilitates parasite persistence19. Collectively, these findings suggest that T-cell immunity is crucial for E. muris colonization, and that Tregs may contribute to sustained colonization and inflammation in chronic infection.
The impacts of T-cell immunity on E. muris-induced colitis-like pathology. (A) Experimental scheme to investigate the relationship between E. muris infection and the host’s T-cell immunity. BALB/c (n = 10), BALB/c Nude (n = 10), and CD4- and CD8-depleted BALB/c (n = 7) mice were infected with E. muris (2 × 10⁴) and necropsied at 9 dpi and 45 dpi (B) Quantification of E. muris gDNA in fecal samples from BALB/c and BALB/c Nude mice over time, measured by q-PCR. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Quantification of E. muris gDNA in cecal contents from BALB/c, and T cell-depleted BALB/c mice at 9 dpi, measured by q-PCR. *P < 0.05. (D) Pearson correlation heatmap showing the relationships among inflammation score, various immune cell populations, and E. muris genomic DNA levels at 45 dpi in colon tissues of BALB/c mice. Inflammation scores (Fig. 1G), eosinophil counts (Fig. 1H), and immune cell numbers (Fig. 2B) were obtained from histological and immunohistochemical analyses, while E. muris gDNA levels were measured from fecal samples (Fig. 1D). The numbers in the boxes indicate Pearson r values. *P < 0.05, **P < 0.01, ***P < 0.001. (E, F) Representative immunofluorescence images and analyses of CD3⁺ and FOXP3⁺ cells in colon tissues of E. muris-infected and uninfected mice at 45 dpi. Scale bar: 200 μm–50 μm for enlarged. *P < 0.05, **P < 0.01, ***P < 0.001. (G) Quantification of E. muris gDNA in cecal contents from control and anti-CD25–treated BALB/c mice at 14 dpi, measured by q-PCR. *P < 0.05.
E. muris infection affects epithelial homeostasis
A severe form of E. histolytica infection can significantly disrupt the structural integrity of the colonic mucosa. However, most other cases of amoebic colitis induced by E. histolytica are mild20. In the current study, histopathological analyses demonstrated that E. muris infection mimics this relatively mild presentation. E. muris infection in BALB/c mice did not disrupt the structural integrity of the colonic mucosa, distinguishing it from the severe form of E. histolytica infection. We thus investigated alterations in epithelial cells, such as proliferation and mucin composition. In the uninfected group, Ki-67-positive proliferative cells were mainly located at the base of the glands (Fig. 4A and B). Meanwhile, at 9 dpi, proliferative cells were broadly distributed throughout the lower half of the mucosa, with an increased cell number (Fig. 4A and B). Then, at 45 dpi, the number of Ki-67 + cells was decreased when compared with 9 dpi, though their distribution in the mucosa was similar (Fig. 4A and B).
The alterations in colonic epithelial cells during E. muris infection. (A, B) Representative images for Ki-67 and quantification of positive cells in colon tissues of E. muris-infected and uninfected mice at 9 dpi and 45 dpi. Scale bar: 200 μm–50 μm for enlarged. The height of Ki-67 + cell distribution was measured along with the height of the colonic glands. **P < 0.01, and ***P < 0.001. (C, D) Representative AB-PAS-stained images and quantification of AB-positive goblet cell number in colon tissues of E. muris-infected (9 dpi, 45 dpi) and uninfected mice. Scale bar: 200 μm–50 μm for enlarged. Goblet cells were indicated by blue-colored droplets. (E) The graphs depict the correlations between inflammation scores and the number of Ki-67-positive cells (proliferating cells) per unit area or the number of goblet cells (mucin-producing cells) per unit area.
Our observations and histopathology revealed that mucus-containing goblet cells were decreased in the infected group (Fig. 1H). This leads us to propose that increased proliferative activity suppresses the differentiation process from stem/progenitor cells to goblet cells. We thus performed Alcian Blue-Periodic Acid-Schiff (AB-PAS) staining to evaluate the goblet cell number and the amount of mucin (Fig. 4C and D). Consistent with our proposal, a significant reduction in goblet cell numbers was observed during the early stage of infection, which persisted into the later stages, in which fewer goblet cells were observed than in the uninfected group (Fig. 4C and D). The positive correlation of the extent of inflammation with proliferative activity and its reverse correlation with the goblet cell number indicated that E. muris infection affected the epithelial composition in the colon either directly or indirectly (Fig. 4E). Collectively, these findings show that E. muris infection affects epithelial homeostasis by influencing proliferation and differentiation in the colonic epithelium.
E. muris infection disrupts the gut microbiota composition
A recent study has suggested that E. muris infection alters the cecal microbial composition7. On the basis that E. muris may directly or indirectly affect the gut microbiota by interacting with the microbiome or altering the epithelial composition, respectively, we performed metagenome amplicon sequencing with fecal samples collected from uninfected and infected groups at 45 dpi. Among the 15 most abundant bacteria, no significant changes were observed between the two groups (Fig. 5A). Similarly, microbial diversity indices, including Chao1 and Shannon indices, showed no significant differences between infected and uninfected groups (Fig. 5B). However, Principal Coordinates Analysis (PCoA) revealed that E. muris-infected mice had a clustered bacterial composition distinct from that of uninfected mice (Fig. 5C). Further analysis identified 22 bacterial species with significantly altered abundances: 18 species decreased, but 4 increased after infection (Fig. 5D). Notably, 10 of the 18 decreased species belonged to the Firmicutes phylum (Fig. 5C). Given previous reports that the abundance of Firmicutes decreases in intestinal inflammatory conditions, such as in inflammatory bowel disease (IBD)21,22, this suggests that the microbiota changes observed in E. muris infection share similarities with other inflammatory conditions.
Gut microbial changes after E. muris infection. (A) The composition of gut microbiota at the genus level in mice, comparing E. muris-infected and uninfected groups. Each color indicates a different bacterial genus, highlighting the Top 15 genera with the highest relative abundance. (B) Alpha diversity indices of gut microbiota, including Chao1 (left) and Shannon index (right), comparing E. muris-infected and uninfected groups. Statistical significance is indicated as “ns” (not significant). (C) Principal coordinate analysis (PCoA) of gut microbiota composition between E. muris-infected and uninfected groups. (D) Taxonomic representation in a cladogram (left) and Log2(FC) bar graph (right) illustrating differences in the relative abundance of bacterial species between E. muris-infected and uninfected groups. The cladogram (left) displays all bacterial species, with each letter representing a specific species. Red circles indicate species that significantly decreased in abundance after E. muris infection, while blue circles represent species that significantly increased. In the Log2(FC) bar graph (right), species with decreased abundance in the infected group are shown in red, whereas those with increased abundance are shown in blue. Taxa belonging to the phylum Firmicutes are marked with an asterisk (*). (E) Heatmap of Pearson correlation coefficients between individual microbial species, pathologic conditions, specific immune cells, and E. muris gDNA. Positive correlations are indicated in red, while negative correlations are shown in blue, with the intensity of color corresponding to the strength of the correlation. The figure in the box indicates the Pearson r. ∗P < 0.05, ∗∗P < 0.01 (Pearson correlation coefficient test).
Based on the differential abundance analysis, we conducted a Pearson correlation analysis between the 20 bacterial species significantly altered in the infected group and histopathological parameters or E. muris gDNA levels (Fig. 5E). Several species that decreased in relative abundance showed negative correlations with inflammation-associated factors. For example, Adlercreutzia caecimuris and Peptococcus simiae showed inverse correlations with the inflammation score, and Alistipes senegalensis showed negative correlations with E. muris gDNA, CD45⁺ cells, and F4/80⁺ cells. These species have been previously associated with gut health in other studies (ref). In contrast, no statistically significant correlations were observed between the increased bacterial species in the infected group and other factors.
These findings indicate that E. muris infection affects specific microbial taxa without significantly altering the overall gut microbiome diversity. Additionally, the observed correlations suggest a potential link between certain bacterial species and host immune-inflammatory responses during infection, though further studies are needed to clarify whether these changes contribute to or result from the host immune response.
Discussion
E. muris has been proposed as an experimental model for Entamoeba histolytica, effectively representing the fecal–oral transmission route and life cycle within the host2. However, the impacts of E. muris infection on mice remain largely unknown, particularly its pathological effects on the host physiology and immunity7,23. This study demonstrates that oral E. muris infection can induce inflammation in the mouse colon, characterized by variably infiltrating immune cells in the time after infection. The proliferative activity of epithelial cells significantly increased, and we found a reduced proportion of mucous goblet cells. Furthermore, E. muris infection altered the intestinal microbiota. In contrast to the immunocompetent BALB/c strain, E. muris rarely colonized mice deficient in T-cell immunity, and it did not induce inflammation in the colon. Collectively, the findings of the current study suggest that E. muris infection following the fecal-to-oral route can dynamically develop alterations in the intestinal pathophysiology, which depend on T-cell immunity.
Previous mouse models of E. muris infection using C57BL/6 or SW mice were not presented with pathological findings2,7, and the current study is the first to report that E. muris has pathogenic effects in the mouse colon. That this has not previously been found may be attributed to differences in mouse strains. For example, in Plasmodium berghei K173 infection, C57BL/6 mice are more resistant than BALB/c mice. Similarly, Leishmania major infection causes skin lesions in both strains, but with varying severities depending on the strains24,25,26. These distinct phenotypes in response to parasitic infections are likely due to differences in immune cell activity or cytokine responses during the early stages of infection27. In particular, regulatory Treg activity in BALB/c mice is much higher than in other strains. BALB/c mice are well-documented to exhibit a bias toward type 2 helper T-cell (Th2) responses, rendering them more susceptible to intracellular parasitic infections than C57BL/6 mice28. Furthermore, BALB/c mice exhibit higher expression of IL-18 compared to C57BL/6J mice, which promotes inflammation and immune cell activation, particularly by stimulating the production of pro-inflammatory cytokines like IFN-γ29. This leads to more pronounced inflammatory lesions following infection. Therefore, whereas E. muris has not caused lesions in C57BL/6J mice in previous studies2, in the BALB/c model, the higher expression of IL-18 promotes inflammation and immune cell activation29, resulting in more noticeable inflammatory lesions after infection, which may suggest that such lesions could have occurred. We here observed highly infiltrated Tregs after E. muris infection, and E. muris colonization was significantly lower in mice deficient in T cells. In addition, colonization was also reduced in mice treated with an anti-CD25 antibody to deplete Tregs, compared to immunocompetent mice. These findings suggest that T-cell immunity with Treg association may play important roles in the colonization and persistence of E. muris infection. Furthermore, human E. histolyticainfection involves complex T-cell immunity and Treg-mediated immunosuppression, with Tregs showing a significant increase in mild pediatric cases compared with moderate or severe infection30. This suggests that Tregs play a crucial role in modulating host immune responses during infection31. Given that a Treg deficiency during parasitic infection reduces the parasite burden32,33 and disease severity16,19, BALB/c mice with enhanced Treg activity offer a suitable strain for use in mechanistic studies of Treg functions associated with Entamoeba infection. In the present study, E. muris infection did not cause erosion or ulceration2. However, the infected colon exhibited an increased number of proliferative cells, which may be driven by inflammatory cytokine-mediated cell proliferation. The enhanced cell proliferation at the gland base, corresponding to the intestinal stem cell zone, could interfere with the differentiation of intestinal stem cells into goblet cells, leading to a reduction in goblet cell numbers. These findings suggest that infection-induced epithelial remodeling may disrupt epithelial function and integrity, thereby contributing to the pathophysiology of amoebic colitis.
These alterations in the gut microbiome may be driven by multiple factors. E. muris infection itself may directly influence microbial composition, similar to E. histolytica, which utilizes intestinal bacteria as an energy source34. The gut microbiome may also be shaped through its bidirectional interaction with T-cell immunity, whereby T-cell responses alter microbial communities while the microbiome in turn modulates T-cell differentiation and activity35,36,37. In addition, mucin production and glycosylation in goblet cells are closely associated with the composition of mucus-associated bacteria38, providing adhesion sites and serving as nutritional sources for microorganisms known as mucolytic bacteria39. These bacteria utilize glycosidase enzymes, specifically exoglycosidases, which sequentially cleave monosaccharides from glycans to degrade mucin40,41. Therefore, a reduction in goblet cells or mucin production in the infected colon can give rise to alterations in the gut microbiota composition. Indeed, we found distinct gut microbial environments between the infected and uninfected groups.
Several bacterial taxa were significantly reduced following E. muris infection, and most of those belonged to Firmicutes. The bacteria in this phylum are considered probiotics and are frequently reduced in inflammatory conditions in the intestine, such as IBD21,22. Further studies are required to decide whether the microbial changes are directly related to the inflammatory status in the colon after E. muris infection. However, based on the present data, it seems likely that E. muris infection negatively impacts overall gut health, including a reduction in beneficial probiotics and disruption of the microbial balance. There were notable correlations of the inflammation score, various immune cells, and the abundance of E. muris with specific bacterial species, including Adlercreutzia caecimuris (formerly known as Enterorhabdus caecimuris), Alistipes senegalensis, Bacteroides faecichinchillae, and Peptococcus simiae. In particular, Adlercreutzia caecimuris, considered a probiotic42, showed negative correlations with the inflammation score and FOXP3 + cell infiltration. Meanwhile, Alistipes senegalensis, a member of the Alistipes genus known to be a probiotic43, demonstrated negative correlations with the abundance of E. muris, F4/80, and the immune cell marker CD45, while showing a positive correlation with goblet cell abundance, suggesting that these bacterial species may reflect or influence host immune-inflammatory states.Such interactions may underlie the immune and microbial alterations observed in our E. muris infection model.
Although E. muris infection in BALB/c mice does not reproduce the severe epithelial destruction characteristic of acute fulminant amoebic colitis, it more closely resembles the asymptomatic or mild pathology that predominates in the majority of human E. histolytica infections4,44. Considering the less invasive pathological nature of our model, the increased proliferative activity of epithelial cells is likely to represent a response associated with chronic infection and inflammation. Such epithelial proliferation has also been reported to be associated with inflammatory cytokine signaling in other contexts45,46. We also noted a reduction in goblet cell numbers, a feature that has likewise been reported in mouse models of E. histolytica infection46. Taken together, these histopathological features suggest that our model reflects epithelial alterations associated with chronic infection and inflammation, rather than the acute tissue-destructive pathology of amoebic colitis.
In summary, we here put forward a new mouse model for the study of Entamoeba infection and its impacts on the colon. Although further studies revealing a precise mechanism would be required, our study demonstrates that orally infected E. muris in BALB/c mice can cause pathophysiological changes in the colon, depending on T-cell immunity. This model could be particularly useful to recapitulate the clinically relevant pathological pathological features that represent the majority of human amebiasis cases, while also providing a controlled system to dissect immune–pathogen–microbiota interactions. These findings suggest that E. muris offers a model organism for the study of amoebic colitis induced by E. histolytica. Future studies using this model may provide critical insights into the researched amoebic colitis-like pathology and its interplay with the microbial environment, advancing our understanding of host–pathogen interactions.
Materials and methods
E. muris isolation and cu lture
The cecum of BALB/cAnNTac mice was collected through dissection and suspended in DPBS (10 mM HEPES). The cecal suspension was filtered through a 40 μm cell strainer to remove large debris and washed twice with DPBS by gentle centrifugation. Ficoll centrifugation (Cytiva, 17144002) was used to isolate E. muris from the filtered suspension47. The layer containing E. muris was carefully collected using a pipette and incubated in 0.1 N HCl at 35.5 °C for 1 h to eliminate bacterial contamination48.
To culture E. muris in vitro, we followed a previously established method with modifications49. Briefly, E. muris isolated via Ficoll gradient was cultured in the Balamuth medium50 supplemented with starch, heat-inactivated FBS (incubated for 3 h), and E. coli. The culture was maintained at 35.5 °C and at an angle of 5–10°.
Quantification of E. muris by qPCR
Genomic DNA (gDNA) of E. muris was extracted using a DNA extraction kit (Bioneer, K-3032). Specific primers (F: CAGAATATCATCAAAAACAGTC, R: GAGAACCCACCAATTTCATCC) were used to detect an E. muris-specific sequence within the 18 S ribosomal RNA region by quantitative polymerase chain reaction (qPCR)51. To detect and quantify E. muris, serial dilutions of purified E. muris gDNA were prepared to generate a standard curve. The standard curve was constructed using Ct values obtained from qPCR.
gDNA was extracted from fecal or cecal content samples. The equation derived from the standard curve was used to calculate the infection rate by substituting the Ct values obtained from qPCR analysis of gDNA extracted from fecal samples.
Lamina propria tissue dissociation and flow cytometry
Distal intestinal tissue spanning from the cecum to the rectum was harvested and bisected longitudinally. One half of the tissue was subjected to enzymatic dissociation using the Lamina Propria Dissociation Kit (mouse; Miltenyi Biotec, Cat. 130−097-410) according to the manufacturer’s protocol to obtain single-cell suspensions. Immune cells were stained with fluorochrome-conjugated antibodies against CD3 (100236, Biolegend), CD19 (115506, Biolegend), F4/80 (123110, Biolegend), CD11c (117310, Biolegend), CD11b (101205, Biolegend), Ly6g (127613, Biolengend), CD45 (2093725, Invitrogen) (103106, Biolegend). Stained cells were analyzed using a NovoCyte flow cytometer (Agilent), and the proportions of immune cell subsets were determined based on surface marker expression.
Quantitative reverse transcription PCR (RT-qPCR)
To assess cytokine gene expression profiles, total RNA was extracted from dissociated colonic lamina propria tissue using the HiYield Total RNA Mini Kit (YRB100, Real Biotech Corporation, Taipei, Taiwan) according to the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized using the M-MLV cDNA Synthesis Kit (EZ006S, Enzynomics). RT-qPCR was performed on a QuantStudio PCR System (Applied Biosystems, San Francisco, CA, USA) using the ExcelTaq™ 2X Fast Q-PCR Master Mix (TQ1210, SMOBIO Technology Inc., Hsinchu, Taiwan). Expression levels of cytokine-related transcripts associated with Th1, Th2, Th17, and regulatory immune responses were analyzed. Primer sequences are listed in Supplementary Table 1.
Animal experiments of E. muris infection
The animal experiments were approved by the Animal Research Committees of Kangwon National University (approval numbers: KW-230210-1, KW-221104-1) and conducted in accordance with institutional and national guidelines for the care and use of laboratory animals. This study was carried out in compliance with the ARRIVE guidelines (https://arriveguidelines.org).
Four- to six-week-old female BALB/cAnNTac and BALB/c Nude mice used in this study were purchased from Hanabio (Hwaseong-si, South Korea) and JAbio (Suwon-si, South Korea), respectively. All animals were housed in a temperature-controlled room maintained at 25 ± 1 °C with a 12-hour light/dark cycle. The isolated E. muris was harvested by centrifugation, resuspended in 5 mL of DPBS, and enumerated under a microscope to determine the parasite concentration. Prior to administration, the E. muris suspension was treated with 0.1 N HCl for 1 h at 35.5 °C to eliminate potential bacterial contamination. The treated suspension was then directly used to orally infect each mouse with 1 × 10⁴ to 2 × 10⁴ E. muris suspended in 200 µL of DPBS. Following the experimental procedures, all mice were humanely euthanized by CO₂ inhalation in a chamber, in compliance with institutional and national animal welfare guidelines. Anesthesia was not administered in this study.
Histopathology
Lesions were evaluated histologically using H&E-stained tissue sections, focusing on tissue morphology and the degree of inflammatory cell infiltration. A semiquantitative scoring system was used to assess tissue affected and the extent of immune cell infiltration as follows: 0, none; 1, mild, localized lymphocytic infiltration; 1.5, mild to moderate, multifocal lymphocytic infiltration; 2, moderate, localized infiltration of immune cells; 2.5, moderate to severe, multifocal immune cell infiltration; 3, severe, infiltration of immune cells into the submucosa and lamina propria. Pathologic scores were determined by more than two veterinary pathologists (J.W.P. and S.H.L.).
Alcian blue staining
Paraffin-embedded sections (3 μm thick) were deparaffinized and rehydrated. Alcian Blue staining was performed with 1% Alcian Blue solution (Sigma, B8438,3% acetic acid, pH 2.5) for 30 min. The sections were incubated first with 0.5% periodic acid (Sigma, P7875) for 10 min, and then with Schiff’s reagent (Merck, 109033.0500) for 15 min. After washing with tap water, optional Hematoxylin counterstaining was applied. Sections were dehydrated through graded ethanol, cleared in xylene, and mounted. Stained sections were examined under a light microscope.
Immunohistochemistry and digital analysis
Serial paraffin sections were dewaxed, rehydrated, and subjected to antigen retrieval by heating in 0.01 mol/L citrate buffer (pH 6.0; Sigma-Aldrich, C9999) at 100 °C for 20 min. Immunostaining was performed using the ImmPRESS Peroxidase Polymer kit (Vector Laboratories, Burlingame, CA) according to the manufacturer’s instructions. Briefly, the slides were blocked with 2.5% horse serum or 2.5% goat serum before incubation with primary antibodies at room temperature for 30 min. The primary antibodies used were Rat anti-CD45 (biolegend, 103101), Rabbit anti-CD11c (Cell Signaling, 1:350), Rabbit anti-CD4 (Cell Signaling, 1:200), Rabbit anti-F4/80 (Cell Signaling, 1:500), and Rat anti-CD3 (Abcam, 1:800), Rabbit anti-Foxp3 (Cell Signaling, 1:800) and Rabbit anti-Ki-67 (Abcam, 1:400). Following washes, the slides were incubated with secondary antibodies conjugated to a peroxidase polymer for 30 min. Color development was achieved using ImmPact DAB substrate (Vector Laboratories, SK-4105), and the slides were counterstained with Meyer’s hematoxylin for 10 s. Negative controls were prepared by omitting the primary antibodies and replacing them with diluent. For digital analysis, the immunostained slides were scanned at ×40 magnification using the Pannoramic SCAN slide scanner (3D HISTECH, Budapest, Hungary). Digital quantification was performed using Qupath software (version 0.4.3, https://qupath.github.io, Edinburgh, Scotland), with parameters adjusted to align with manual cell counts conducted by a pathologist. The number of positive cells for CD45, CD11c, CD4, F4/80, CD3, Foxp3 and Ki-67 was quantified, and the total count was divided by the tissue area (mm²) to calculate the number of positive cells per mm². Positive cells detected through digital grading were classified into + 1, +2, and + 3 according to staining intensity, and all grades were summed and divided by the tissue area to calculate the number of positive cells per mm².
Immunofluorescence staining
For immunofluorescence (IF) staining, formalin-fixed paraffin-embedded (FFPE) slides underwent deparaffinization followed by stepwise rehydration using ethanol. Antigen retrieval was performed by immersing the slides in an antigen retrieval solution (DAKO, Santa Clara, United States) and applying high pressure in a cooker. After antigen retrieval, the slides were treated with Antibody Diluent with Background Reducing Components (DAKO) to block nonspecific binding and then incubated overnight at 4 °C with the primary antibodies. Rat anti-CD3 (Abcam, 1:100) and Rabbit anti-Foxp3 (Cell Signaling, 1:100) antibodies were used. Secondary detection was achieved using Alexa488- and Alexa568-conjugated secondary antibodies (Invitrogen, Waltham, United States), corresponding to the species of the primary antibodies. Nuclear counterstaining was carried out with DAPI (4′,6-Diamidino-2-phenylindole dihydrochloride) (Abbkine, Wuhan, China).
16 S Metagenomic sequencing
Fecal samples were collected from E. muris-infected and uninfected groups, and gDNA was extracted to prepare sequencing libraries following the Illumina 16S Metagenomic Sequencing Library protocols for amplifying the V3 and V4 regions of the 16S rRNA gene. The input gDNA 2ng was PCR amplified with 5x reaction buffer, 1mM of dNTP mix, 500nM each of the universal F/R PCR primer, and Herculase II fusion DNA polymerase (Agilent Technologies, Santa Clara, CA). The cycle condition for 1 st PCR was 3 min at 95 °C for heat activation, and 25 cycles of 30 sec at 95 °C, 30 sec at 55 °C and 30 sec at 72 °C, followed by a 5-min final extension at 72°C. The universal primer pair with Illumina adapter overhang sequences used for the first amplifications were as follows: V3-F: 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3’, V4-R: 5’- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3’. The 1 st PCR product was purified with AMPure beads (Agencourt Bioscience, Beverly, MA). Following purification, the 2ul of 1 st PCR product was PCR amplified for final library construction containing the index using NexteraXT Indexed Primer. The cycle condition for 2nd PCR was same as the 1 st PCR condition except for 10 cycles. The PCR product was purified with AMPure beads. The final purified product is then quantified using qPCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantificatoin kits for IlluminaSequecing platforms) and qualified using the TapeStation D1000 ScreenTape (Agilent Technologies, Waldbronn, Germany). The paired-end (2 × 300 bp) sequencing was performed by the Macrogen using the MiSeq™ platform (Illumina, San Diego, USA).
T-cell depletion
To deplete T-cells in BALB/c mice, anti-CD4 (GK 1.5, Bioxcell) and anti-CD8 (2.43, Bioxcell) antibodies were administered intraperitoneally at 100 µg each on days 1, 2, 7, 12, and 1852.
To deplete CD25⁺ cells in BALB/c mice, anti-CD25 (PC-61.5.3, Bioxcell) antibody was administered intraperitoneally at 500 µg on days on days 1, 4 and 7 days prior to infection, and on days 1, 4, 7, and 10 post-infection53.
Statistical analysis
Statistical analyses were performed with the GraphPad Prism software version 8 (San Diego, CA). Error bars indicate the standard deviations (SDs) of the mean. Comparisons between two groups were performed using the Student’s t-test. Statistical significance is indicated as follows: ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗∗P ≤ 0.001.
Data availability
The datasets generated and/or analysed during the current study are available in the NCBI Sequence Read Archive (SRA) under the BioProject accession number PRJNA1314755 (https://www.ncbi.nlm.nih.gov/bioproject/1314755).
References
Moon, T. D. & Oberhelman, R. A. Antiparasitic therapy in children. Pediatr. Clin. North Am. 52, 917–48, viii (2005).
Mendoza Cavazos, C., Heredia, M. Y., Owens, L. A. & Knoll, L. J. Using Entamoeba muris to model fecal-oral transmission of Entamoeba in mice. MBio 14, e0300822 (2023).
Ghosh, S., Padalia, J. & Moonah, S. Tissue destruction caused by entamoeba histolytica parasite: cell death, inflammation, invasion, and the gut Microbiome. Curr. Clin. Microbiol. Rep. 6, 51–57 (2019).
CDC & About Amebiasis (2024).
Ralston, K. S. & Petri, W. A. Jr. Tissue destruction and invasion by entamoeba histolytica. Trends Parasitol. 27, 254–263 (2011).
Deloer, S. et al. Mouse models of amoebiasis and culture methods of amoeba. Parasitol. Int. 65, 520–525 (2016).
Roy, M. et al. Entamoeba muris mitigates metabolic consequences of high-fat diet in mice. Gut Microbes. 16, 2409210 (2024).
Phillips, B. P. & Wolfe, P. A. The use of germfree Guinea pigs in studies on the microbial interrelationships in amoebiasis. Ann. N Y Acad. Sci. 78, 308–314 (1959).
Chadee, K. & Meerovitch, E. The pathogenesis of experimentally induced amebic liver abscess in the gerbil (Meriones unguiculatus). Am. J. Pathol. 117, 71–80 (1984).
Cleveland, L. R. & Sanders, E. P. The production of bacteria-free amoebic abscesses in the liver of cats and observations on the amoebae in various media with and without bacteria. Science 72, 149–151 (1930).
Ehrenkaufer, G. M. et al. The genome and transcriptome of the enteric parasite entamoeba invadens, a model for encystation. Genome Biol. 14, R77 (2013).
Batt, C. A. & Tortorello, M. L. (eds) Encyclopedia of Food Microbiology, 2nd edn, Vol. 1–3. (Academic Press, Amsterdam, 2014).
Shibui, A. et al. CD4(+) T cell response in early erythrocytic stage malaria: plasmodium Berghei infection in BALB/c and C57BL/6 mice. Parasitol. Res. 105, 281–286 (2009).
Corripio-Miyar, Y. et al. Functionally distinct T-helper cell phenotypes predict resistance to different types of parasites in a wild mammal. Sci. Rep. 12, 3197 (2022).
Engwerda, C. R., Ng, S. S. & Bunn, P. T. The regulation of CD4(+) T cell responses during protozoan infections. Front. Immunol. 5, 498 (2014).
Smith, K. A. et al. Low-level regulatory T-cell activity is essential for functional type-2 effector immunity to expel Gastrointestinal helminths. Mucosal Immunol. 9, 428–443 (2016).
Maizels, R. M. & McSorley, H. J. Regulation of the host immune system by helminth parasites. J. Allergy Clin. Immunol. 138, 666–675 (2016).
Flanagan, S. P. Nude’, a new hairless gene with pleiotropic effects in the mouse. Genet. Res. 8, 295–309 (1966).
Adalid-Peralta, L. et al. Regulatory T cells as an escape mechanism to the immune response in taenia crassiceps infection. Front. Cell. Infect. Microbiol. 11, 630583 (2021).
Espinosa-Cantellano, M. & Martínez-Palomo, A. Pathogenesis of intestinal amebiasis: from molecules to disease. Clin. Microbiol. Rev. 13, 318–331 (2000).
Kostic, A. D., Xavier, R. J. & Gevers, D. The Microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499 (2014).
Gyriki, D. et al. Exploring the gut microbiome’s role in inflammatory bowel disease: insights and interventions. J. Pers. Med. 14, 507 (2024).
Pritchett-Corning, K. R. & Clifford, C. B. Elsevier,. Parasitic infections of laboratory mice. in The Laboratory Mouse 503–518 (2012).
Tacchini-Cottier, F. et al. An Immunomodulatory function for neutrophils during the induction of a CD4 + Th2 response in BALB/c mice infected with leishmania major. J. Immunol. 165, 2628–2636 (2000).
Sacks, D. & Noben-Trauth, N. The immunology of susceptibility and resistance to leishmania major in mice. Nat. Rev. Immunol. 2, 845–858 (2002).
Belkaid, Y. et al. A natural model of leishmania major infection reveals a prolonged ‘silent’ phase of parasite amplification in the skin before the onset of lesion formation and immunity. J. Immunol. 165, 969–977 (2000).
Wang, H. et al. Analysis of spleen histopathology, splenocyte composition and haematological parameters in four strains of mice infected with plasmodium Berghei K173. Malar. J. 20, 249 (2021).
Chen, X., Oppenheim, J. J. & Howard, O. M. Z. BALB/c mice have more CD4 + CD25 + T regulatory cells and show greater susceptibility to suppression of their CD4 + CD25- responder T cells than C57BL/6 mice. J. Leukoc. Biol. 78, 114–121 (2005).
Takeda, K. et al. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity 8, 383–390 (1998).
Kizilbash, N., Suhail, N., Alzahrani, A. K., Basha, W. J. & Soliman, M. Natural regulatory T cells increase significantly in pediatric patients with parasitic infections: flow cytometry study. Indian J. Pathol. Microbiol. 66, 556–559 (2023).
McManus, C. M. & Maizels, R. M. Regulatory T cells in parasite infections: susceptibility, specificity and specialisation. Trends Parasitol. 39, 547–562 (2023).
Wang, J. et al. Depletion of FoxP3 + Tregs improves control of larval Echinococcus multilocularis infection by promoting co-stimulation and Th1/17 immunity. Immun. Inflamm. Dis. 5, 435–447 (2017).
Wang, J. & Gottstein, B. Foxp3⁺ T regulatory cells as a potential target for immunotherapy against primary infection with Echinococcus multilocularis eggs. Infect. Immun. 86, e00542-18 (2018).
Guillén, N. The interaction between entamoeba histolytica and Enterobacteria shed light on an ancient antibacterial response. Cell. Microbiol. 21, e13039 (2019).
Kim, K. S. Regulation of T cell repertoires by commensal microbiota. Front. Cell. Infect. Microbiol. 12, 1004339 (2022).
Zheng, D., Liwinski, T. & Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506 (2020).
Ivanov, I. I., Tuganbaev, T., Skelly, A. N. & Honda, K. T cell responses to the microbiota. Annu. Rev. Immunol. 40, 559 (2022).
Ridge, J. P., Di Rosa, F. & Matzinger, P. A conditioned dendritic cell can be a Temporal Bridge between a CD4 + T-helper and a T-killer cell. Nature 393, 474–478 (1998).
Bergstrom, K. S. B. & Xia, L. Mucin-type O-glycans and their roles in intestinal homeostasis. Glycobiology 23, 1026–1037 (2013).
Li, H. et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6, 8292 (2015).
Hansson, G. C. Mucins and the Microbiome. Annu. Rev. Biochem. 89, 769–793 (2020).
Mi, W. et al. Quercetin positively affects gene expression profiles and metabolic pathway of antibiotic-treated mouse gut microbiota. Front. Microbiol. 13, 983358 (2022).
Parker, B. J., Wearsch, P. A., Veloo, A. C. M. & Rodriguez-Palacios, A. The genus alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 11, 906 (2020).
Kantor, M. et al. Entamoeba histolytica: updates in clinical Manifestation, Pathogenesis, and vaccine development. Can. J. Gastroenterol. Hepatol. 2018, 1–6 (2018).
Newcomb, D. C. et al. IL-17A induces signal transducers and activators of transcription-6-independent airway mucous cell metaplasia. Am. J. Respir Cell. Mol. Biol. 48, 711–716 (2013).
Leon-Coria, A., Kumar, M., Moreau, F. & Chadee, K. Defining cooperative roles for colonic microbiota and Muc2 mucin in mediating innate host defense against entamoeba histolytica. PLoS Pathog. 14, e1007466 (2018).
Avron, B., Bracha, R., Deutsch, M. R. & Mirelman, D. Entamoeba invadens and E. histolytica: separation and purification of precysts and cysts by centrifugation on discontinuous density gradients of Percoll. Exp. Parasitol. 55, 265–269 (1983).
Yanagawa, Y. et al. Gene expression of axenically-isolated clinical entamoeba histolytica strains and its impact on disease severity of amebiasis. PLoS Pathog. 18, e1010880 (2022).
Kobayashi, S., Suzuki, J. & Takeuchi, T. Establishment of a continuous culture system for entamoeba muris and analysis of the small subunit rRNA gene. Parasite 16, 135–139 (2009).
Balamuth, W. Improved egg yolk infusion for cultivation of entamoeba histolytica and other intestinal protozoa. Am. J. Clin. Pathol. 16, 380–384 (1946).
Scavizzi, F. et al. A comprehensive approach for microbiota and health monitoring in mouse colonies using metagenomic shotgun sequencing. Anim. Microbiome. 3, 53 (2021).
Li, J. et al. Co-inhibitory molecule B7 superfamily member 1 expressed by tumor-infiltrating myeloid cells induces dysfunction of anti-tumor CD8 + T cells. Immunity 48, 773–786e5 (2018).
Glatigny, S., Duhen, R., Arbelaez, C., Kumari, S. & Bettelli, E. Integrin alpha L controls the homing of regulatory T cells during CNS autoimmunity in the absence of integrin alpha 4. Sci. Rep. 5, 7834 (2015).
Acknowledgements
This study was supported by the New Faculty Startup Fund from Seoul National University and by grants from the National Research Foundation (NRF) funded by the Korean government (RS-2023-00208984, 2021M3H9A1030260, RS-2024-00443043). Additional support was provided by the Research Institute for Veterinary Science and the BK21 FOUR Future Veterinary Medicine Leading Education and Research Center, Seoul National University.
Author information
Authors and Affiliations
Contributions
T.H.H. and S.H.S. performed immunohistochemistry, analyzed data, and wrote the manuscript; H.W.A., J.W.K., G.M.Y., Y.J.L., N.Y.L., and H.A.K. performed animal experiments; W.G.Y. curated data and provided resources; and S.H.L. and J.W.P. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ha, T.H., Seok, S.H., won An, H. et al. Entamoeba muris provides a mouse model for amoebic colitis pathology characterized by T cell associated colonization. Sci Rep 15, 39973 (2025). https://doi.org/10.1038/s41598-025-23596-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-23596-6